1. Introduction
Pork ranks second as the most consumed meat in the world with an estimated 106.3 million tons consumed annually per capita as of 2020 [
1]. According to the National Pork Producers Council, the U.S. exports more than 2.2 million metric tons of pork and pork-related products annually [
2] resulting in U.S. pork and pork product exports having a value of approximately
$7.7 billion [
3]. Based on numbers from the USDA, in 2019, 129.9 million farmed pigs were slaughtered for food in the U.S. In the U.S., pork is consumed in the form of fresh pork cuts such as chops, ribs, roasts, or hams and the remaining is consumed in the form of processed pork such as sausage, hot dogs, and bacon [
4].
Salmonella is the number one cause of foodborne illness in the U.S. [
5]. Every year in the U.S., non-typhoidal
Salmonella is responsible for approximately 1,027,561 cases, 19,336 hospitalizations, and 378 deaths [
6], resulting in a
$3.7 billion economic burden annually [
7]. According to the foodborne illness attribution estimates for 2019, published by the Interagency Food Safety Analytics Collaboration (IFSAC) in October 2021 in conjunction with the Centers for Disease Control and Prevention (CDC), the U.S. Food and Drug Administration (FDA), and the USDA’s FSIS, 75.9% of
Salmonella cases were attributed to seven major food categories: chicken, fruits, pork, seeded vegetables, other produce, turkey, and eggs. Additionally, it was estimated that 12.8% of cases are attributed to pork [
8]. These data indicate that pork is the second highest contributor to salmonellosis cases from FSIS-regulated products, and the third highest contributor from all food products.
Previous studies have biomapped microorganisms in pork processing facilities to determine the prevalence of
Salmonella, Generic
E. coli, and other indicator organisms at various stages of the pork processing chain. These studies reported that pathogen and indicator organism prevalence were reduced throughout the processing line, but increased again in trim and further processing stages, creating a U-shaped curve of the biomapped organism [
4]. The facility in this study was operating as a HIMP facility prior to NSIS for over 20 years.
Salmonella prevalence, as determined by the previously developed baseline biomapping study, is 19% within the entire facility. Out of 650 samples collected during the baseline biomapping study, 125 samples were positive at 24 h for prevalence testing. However, only 47 out of the 125 were positive at the 6-hour SalQuant time point, meaning that only these select samples were quantifiable [
4]. Two samples were selected in this facility for sample collection: Boneless Picnic Trim and Final Product Brick Sausage. The previous study concluded that the selected testing locations for further processed products had a 24% prevalence for
Salmonella positives. This increase in prevalence when compared to the facility average indicates a need for intervention strategies at the later pork processing stages. This rise in bacterial prevalence at the further processing stages of pork production indicates a need for intervention strategies to control pathogens later in production. To mitigate
Salmonella in trimmings and ground pork products, novel intervention strategies must be studied for efficacy and cost effectivity for industry applications because to date, there are few interventions available.
Lymph nodes are embedded within various muscle tissue groups, these glands are used to filter lymphatic fluid during the lifespan of the animal. According to recent research, lymph nodes on both pork and beef have been shown to carry a high load of
Salmonella after harvest [
9]. Currently, pork trim and ground products are processed with the following lymph nodes and glands included in the muscle tissue.
-
Topical
- o
This group includes superficial popliteal lymph nodes located in the back legs, as well as superficial inguinal lymph nodes located in the fat on the medio-ventral surface of the hind leg. This group may also include various accessory glands.
-
Jowl
- o
The pork jowl contains 2-3 salivary glands and several lymph nodes down the jowl, cheeks, and neck. This group also notably includes sub iliac nodes
-
Internal
- o
These deep tissue lymph nodes are located in the fat in the crease between the semitendinosus (eye of round) and gastrocnemius (knuckle). This group also includes the gluteal and ischiadic lymph nodes located on the sarcotuberal ligament. This group also contains various glands from the loin region.
The removal of glands and lymph nodes from boneless picnic hams prior to grinding for sausage production is a possible means to reduce Salmonella prevalence in further processed pork products, but little data exist on node/gland removal as a mitigation strategy.
2. Materials and Methods
2.1. Sample collection and treatment
Pork trim and ground sausage samples were collected on-line from a large-scale USDA inspected hog processing facility. The facility where samples were collected is a large hog processing facility that is currently processing approximately 10,400 head per day and 1250 head per hour on average, located on the east coast of the United States. This facility is USDA-inspected and is currently operating under the New Swine Inspection System (NSIS), as proposed in 2019. To operate under NSIS, the processing plant was required to implement specific worker safety measures, including an agreement with a workers’ union to represent their employees [
10]. These safety measures allow the processing plant to work at higher line speeds under supervision and regular testing requirements to ensure safety and quality standards while increasing productivity. According to a constituent update published by USDA FSIS in 2021, facilities that are not operating under the time-limited trial of NSIS can only process up to 1,106 head per hour. Picnic trim is lean muscle and fat trimming that come from the picnic shoulder of the carcass. The shoulder is a muscle of the hog that typically is tougher due to the amount of work put on the muscle. This trim, along with formulated fat and spices, is most often used at the meat component for ground sausage products within this facility. Ground sausage samples are taken after final packing in vacuum brick packages with easy open seals. This sausage is formulated according to the company’s recipe independent of the treatment group. A total of 15 samples were taken from each sampling location for each treatment group (n = 90) per repetition. The entire study was replicated 5 times over a period of 4 months to account for natural variation and seasonality (n = 450). The individual sampling dates (March 23rd, 2022, March 29th, 2022, April 4th, 2022, May 20th, 2022, June 1st, 2022, and June 16th, 2022) were used to account for seasonality and natural variation within the product. For trim and ground samples, sample collection was conducted following the protocols for Whole Pork Cuts (Intact and Non-Intact) and Comminuted Pork Aseptic Grab Sample Not in Final Packaging, respectively, per FSIS Directive Number 65–20 from the Raw Pork Products Sampling Program. This consisted of using fresh, not frozen, raw pork, using a single Whirl-Pak bag (Millipore Sigma, Burlington, MA, USA) to aseptically collect approximately 375 g of the pork trim to fill the bag leaving 5 to 8 centimeters (cm) of space at the top. Ground pork sausage was collected as 1 pound (453.6 g) bricks in final packaging. To conduct the raw material sample collection, 15 samples of each of the three treatment groups were taken after gland removal on each day of sampling for five replications.
Samples were immediately chilled and shipped overnight to the International Center for Food Industry Excellence (ICFIE) Food Microbiology laboratory at Texas Tech University for microbiological analysis. Samples were processed and evaluated for EB, AC, and Salmonella concentrations. Additionally, retainer samples were kept at plant for further evaluation. Chilled pork carcasses were fabricated through standard process. As the shoulder passed through the production line, the foot, jowl, and neck bone were removed. The butt was then separated from the picnic to generate a bone in, skin on picnic. The picnic was then transferred to the boning department through conveyors and equipment. Samples treatment groups 2 and 3 were run on belts, conveyors, and other equipment that only received gland-free product during processing to avoid cross-contamination. Between treatments, the belts and equipment were thoroughly sanitized by trained personnel in order to reduce any potential cross contamination. All picnic trim samples were collected from a singular vat per treatment group. The treatment groups are defined as follows:
-
Treatment 1 – Standard Trim on Boneless Picnic Hams – Control
- o
Standard Trim without removing additional glands /defects. Remove Skin, Bone, Meat, Trim and Inedible. Collect the skin, meat, bone, trim and inedible weight for the vat for yield information. Identify the Vat of Picnics and send to sausage production.
-
Treatment 2 – Retail Trim on Boneless Picnic Hams
- o
Additional Trimming Required - remove blood clots, and all surface / exposed glands regardless of color. Remove Skin, Bone, Meat, Trim, and Inedible tissue. Collect the skin, meat, bone, trim and inedible weight for the vat for yield information. Identify the vat of picnics and send to sausage production.
-
Treatment 3 – Export Trim on Boneless Picnic Hams
- o
Bone out Picnics to Standard Trimming with the addition of removing exposed glands and surface blood clots regardless of size and color. Remove glands associated with the Jowl and glands inside the boneless picnic remove Skin, bone, meat, trim and inedible tissues. Collect the skin, meat, bone, trim and inedible weight for the vat for yield information Identify the vat of picnics and send to sausage production.
These treatment groups were consistent for both the trim and group samples.
2.2. Processing Methodology
Upon arrival at Texas Tech University, the samples were evaluated for any leaking, damage, or potential temperature abuse. For pork trim, 50 g of the specific cut were weighed into a 55 oz. filtered Whirl-Pak bag and 200 mL prewarmed (45°C) BAX MP media (Hygiena™, Camarillo, CA, USA) was added. Trim samples were homogenized using a stomacher (Model 400 Circulator, Seward, West Sussex, UK) at 230 rpm for 30 s. For ground pork sausage samples, 50 g were weighed using a sterile scoopula into a 55 oz. filtered Whirl-Pak bag and 200 mL BAX MP Media was added. Ground pork samples were homogenized using a stomacher at 230 rpm for 1 min. From the primary bag, 30 mL of homogenate was aseptically transferred into a 24 oz. filtered Whirl-Pak bag using a 50 mL disposable serological pipette (Fisher Scientific, Foods 2022, 11, 2580 5 of 20 Waltham, MA, USA). Additionally, 30 mL BAX MP Media, containing 1 mL Quant solution (Hygiena™, Camarillo, CA, USA) was added to the 30 mL pure sample. A 10-mL aliquot of each sample type was transferred using a 10-mL disposable serological pipette into sterile tubes for microbial indicators enumeration, prior to sample incubation for Salmonella enumeration and prevalence. This study was replicated 5 times over a period of four months (n= 450).
2.3. Microbial Analyses
The TEMPO® system (BioMérieux, Paris, France) was utilized for the enumeration of indicator organisms. For AC, the Association of Official Agricultural Chemists (AOAC) 121204 method was used, where TEMPO cards were incubated for 22–28 h at 35 ± 1 °C. For EB enumeration, AOAC 050801 method was followed, and cards were incubated for 22–27 h at 35 ± 1 °C. For the TEMPO enumeration of indicator organisms, the original samples was diluted to a 1/20 dilution on all sample and indicator types. To prepare this dilution, 3 mL of water and 1 mL of the sample rinsate was added to the dehydrated media vial. This dilution was then filled into the correlating TEMPO card for each indicator, EB or APC, and incubated according to the directions for each organism. Once incubated appropriately, the cards were read by the TEMPO Reader. Results were converted to Log10 values for interpretation and evaluation.
Pork trim samples were immediately incubated at 42 °C for 6 h for quantification purposes. Ground pork sausage samples were incubated at 42 °C for a 7-h period for quantification. After incubation, the AOAC 081201 protocol for enumeration of Salmonella using the BAX® System SalQuant™ (Hygiena, Camarillo, CA, USA) was followed. Additionally, the SalQuant™ protocols for pork trim and ground pork are part of the AOAC validation Level 2 modification to the BAX® System Real-Time PCR Assay for Salmonella and BAX® System SalQuant™ (Certification No. 081201). After obtaining a sample for enumeration protocol, samples were placed back to continue incubation at 42 °C for a period of 18–24 h (prevalence testing). If samples were not positive for BAX® System SalQuant™, the BAX® System RT-Salmonella Assay protocol for detection was followed.
The BAX® System Real-Time PCR Assay for Salmonella can be subdivided into 3 stages involving preparation, lysis, and PCR. The first stage consisted of prepping the lysis reagent and pre-heating thermal blocks to 37 °C and 95 °C. The lysis step involved the 5-µL sample transfer to cluster tubes, a heating step to 37°C for 20 min, a subsequent heating step to 95 °C for 10 min, and cooling for 5 min. The PCR stage entailed hydrating PCR tables with 30 µL lysate and running the BAX® Q7 thermocycler.
2.4. Data Analysis
All data were analyzed using R (Version 4.1.2) statistical software to evaluate the reduction of microbial loads as a result of each treatment when compared to the control. Indicator counts were converted to LogCFU/g and Salmonella counts were reported as LogCFU/sample. A one-way ANOVA was performed, comparing counts from each of the treatment groups, followed by pairwise multiple comparison T-tests, and adjusted by the Bonferroni method. A p-value of 0.05 or less was selected prior to the analysis to determine significant differences.
Data were arranged into boxplots, where the horizontal line crossing the box represents the median, the top and bottom lines of the box represent the lower (0.25) and upper (0.75) quartiles, the vertical top lines represent 1.5 times the interquartile range, and the vertical bottom line represents 1.5 times the lower interquartile range. The dots represent the actual data points. [a,b,c] For each matrix, boxes with different letters are significantly different between treatments according to t-test analysis at p-value < 0.05.
3. Results
The LogCFU/g counts APC indicate significant differences between the control samples, the retail trim, and the export trim. Enterobacteriaceae results show a statistically significant difference among the control trim and both treatment groups, but there was not a significant difference between EB counts obtained from retail trim and export trim. Salmonella counts were recorded and presented in Log10CFU/sample using a 50-g sample basis. Trim samples had overall higher counts of both indicator organisms and Salmonella for all treatment groups when compared to ground samples from the same treatment groups. Indicator organisms, especially EB, show a wide range of variance for each set of samples, this indicates an overall need for better process control methods within this facility in order to reduce the variation.
3.1. Detection and Quantification of Salmonella
A total of of 72/450 samples tested positive for Salmonella (16%). Prevalence was evaluated using BAX® System Real-Time
Salmonella assay.
Table 1 shows
Salmonella prevalence at from each treatment group. From those
Salmonella-positive samples, 38 samples (52.7%) were suitable for enumeration using the BAX® System SalQuant™, the majority detected from treatment group 1, the control group. The breakdown of collected positives from each treatment group and matrix can be found in table 1.
Overall, 72 of the 450 samples tested positive for Salmonella on the prevalence assay after 24 hours of enrichment (n = 72). Of these, 43 samples were part of the control group, 22 of the positive samples were from the retail trim (topical gland removal only) group, and 7 positives were a part of the export trim category which had the topical, jowl, and internal glands removed. Overall, more Salmonella positives were detected in ground samples as opposed to trim samples, this was displayed across each treatment group.
Salmonella counts were very low when analyzed on per-g basis in most samples, thus when transformed to Log10CFU/g, counts resulted in negative values, making analysis and visualization more difficult for interpretation. Thereby, all data were transformed to LogCFU/sample which is equivalent to LogCFU/50 g to facilitate data interpretation. The Limit of Quantification (LOQ) for SalQuant™ on pork trim and ground pork is 1 CFU/g. When using SalQuant™, counts can be extrapolated below the LOQ since counts are obtained from a regression equation specific to each matrix, provided by the methodology. To better accommodate for this extrapolation process, a new LOQ was established as 10% of the real LOQ, 0.1 CFU/mL and 0.1 CFU/g or 0.70 LogCFU/sample. Samples with a value of <0.70 LogCFU/sample were reported as 50% of the new LOQ (0.35 LogCFU/sample), which will be referred to as the new Limit of Detection (LOD). This value was also used for samples that were not quantifiable during the 6 and 7-h time points but found positive for Salmonella to be recorded as prevalence only. Samples that were not quantifiable or detectable were reported as 0 LogCFU/sample but are not reflected on figure 1 as including these values would alter the mean values of positive samples.
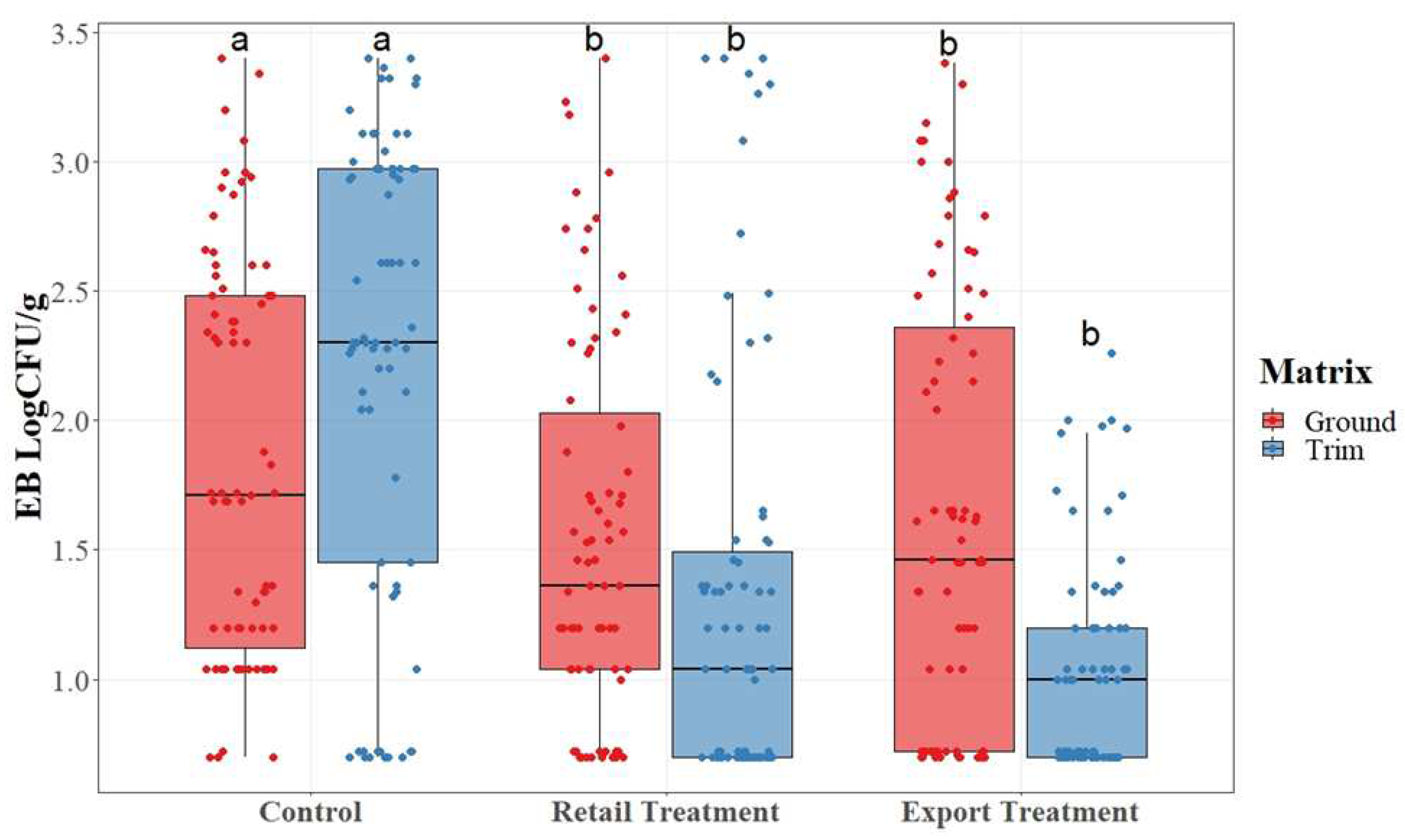
Figure 1.
Salmonella Quantification as determined by SalQuant on pork trimmings and ground product subjected to gland and node removal. In each boxplot, the horizontal line crossing the box represents the median, the top and bottom lines of the box represent the lower (0.25) and upper (0.75) quartiles, the vertical top lines represent 1.5 times the interquartile range, and the vertical bottom line represents 1.5 times the lower interquartile range. The dots represent the actual data points. a,b.c For each matrix, boxes with different letters are significantly different between treatments according to t-test analysis at p-value < 0.05.
Figure 1.
Salmonella Quantification as determined by SalQuant on pork trimmings and ground product subjected to gland and node removal. In each boxplot, the horizontal line crossing the box represents the median, the top and bottom lines of the box represent the lower (0.25) and upper (0.75) quartiles, the vertical top lines represent 1.5 times the interquartile range, and the vertical bottom line represents 1.5 times the lower interquartile range. The dots represent the actual data points. a,b.c For each matrix, boxes with different letters are significantly different between treatments according to t-test analysis at p-value < 0.05.
Of the 450 collected samples across 5 replications, 38 samples were positive at the SalQuant time point for quantification (n = 38). The recorded quantitative values are displayed in
Figure 3. The control group averaged 2.5 Log CFU/Sample and 3.8 Log CFU/Sample of
Salmonella in ground and trim samples, respectively. The export trim group held the lowest average of
Salmonella counts for both matrices at less than 1 Log CFU/sample. There were statistical differences among each of the three treatment groups for both detection and quantification methodologies. Of the 31 quantifiable samples, 3 were from the export trim, 12 were from retail trim, and 16 were from the control trim. The mean of each sample point was used to determine significant difference between the sample groups. There was a significant difference (P<0.05) between the control samples and each of the treatment groups, however there was not a significant difference between the retail trim and export trim treatment groups.
3.2. Enumeration of Enterobacteriaceae and Aerobic Count Bacteria
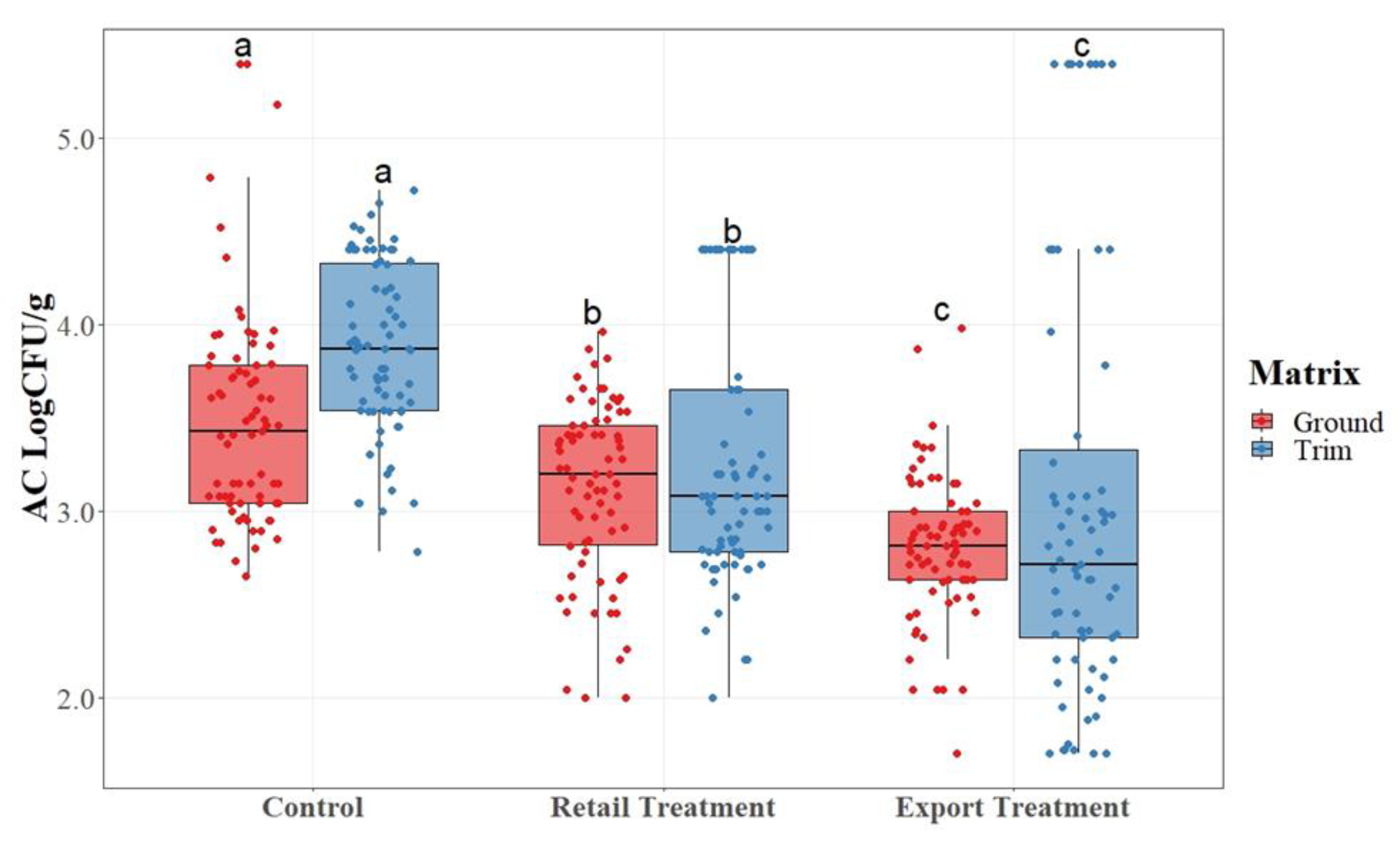
APC counts ware described in
Figure 2. Total APCs were statistically compared across mean values for each treatment group and matrix.
As shown in
Figure 1, there was a statistical difference (P<0.05) among treatment groups, for both matrices with the control having the highest, retail having the lower than the control but higher than the export and export having the lowest. Export trim which was composed of boneless picnic trim with topical, jowl, and internal glands removed, had the lowest average APC counts for aerobic plate counts for both the ground and trim matrices. The lowering of APC counts indicates the reduction of overall microbial activity within the samples collected from each treatment group as the glands were removed.
EB counts, as detailed in
Figure 3, were compared by mean value for each treatment group and matrix.
Figure 3.
Enterobacteriaceae (EB) results for pork trim and ground pork collected from a commercial pork facility with and without lymph node removal.
Figure 3.
Enterobacteriaceae (EB) results for pork trim and ground pork collected from a commercial pork facility with and without lymph node removal.
Enterobacteriaceae counts were determined using the TEMPO system and converted into Log10CFU/g values. While the range for each treatment group remains wide across treatments, the median value for each group decreases as the lymph nodes and glands are removed. The means of each sample point were used to determine significant difference among treatments. There was a statistical difference (P< 0.05) among EB counts collected from the control group, and counts collected from each of the treatment groups. Unlike the APC counts, there was not a statistical difference between treatment retail trim and export trim. The reduction of EB microorganism is beneficial to the product as it indicates a lower amount of potential pathogenic presence.
The pattern of the detected and quantified Salmonella correlates closely with the pattern of the EB and APC indicator organisms measured within this study. While these values correlate in pattern, there is not an exact ratio between the relationships. Since the organisms follow similar patterns, indicator organisms can be observed to suggest the presence of Salmonella in pork products, however, the observation of indicator organisms cannot be utilized in place of Salmonella testing in this particular operation. The similar pattern followed by Salmonella and the indicator organisms additionally suggest that the removal of lymph nodes effected the organisms directionally.
4. Discussion
The quantification of Salmonella in pork samples from industry settings may be limited due to difficulties with pathogen recovery, pathogen stress caused by the processing environment, and the application of antimicrobial interventions. The quantification techniques utilized within this project have been studied and proven to recover pathogens from positive samples as a result of a recovery stage through the implementation of short enrichment steps that increase the likelihood of collecting quantification estimation data. Additionally, Salmonella quantification may offer an opportunity to make risk-based and data-driven decisions based on prevalence and overall concentration in specific stages in the process, rather than the presence or absence of the pathogen. Quantification can benefit the pork industry as displayed by the results of this study which provides evidence for emerging technologies to be applying in the industry for pathogen quantification and indicator microorganism levels in pork samples. The utilization of a rapid PCR-based enumerative method for Salmonella, in conjunction with the enumeration of indicator microorganism, provides the pork industry with a tool to make data-driven decisions to reduce pathogenic prevalence in trim and further processed pork products and to mitigate the risk to public health of foodborne illness.
Furthermore, the results of this study indicate that the removal of topical lymph nodes and glands from boneless picnic trim was an effective method for the reduction of Salmonella in boneless picnic pork trim and ground sausage products in this operation. Furthermore, the results of this study indicate that the removal of topical, jowl, and internal glands and lymph nodes further reduced the prevalence of Salmonella and other indicator organisms in boneless picnic trim and ground sausage when compared to the removal of topical glands and lymph nodes alone. It is important to note that in this study, strict sanitary measures were used in node and gland removal and the amount of time taken to remove the nodes and glands was significant and thus it may be difficult to implement in commercial operations. There could be a risk of cross contamination if the nodes are not carefully removed and proper sanitation protocols aren’t implemented. If this method is chosen to mitigate Salmonella, effective and efficient methods for gland and lymph node removal should be determined and developed before implementation of these strategies within the industry. It is also critical to understand which nodes contributed to the most reduction. Nodes were not isolated and it could be a single node of combination that resulted in the reductions. Finally, the serotype and the pathogenicity of the Salmonella was not determined. In order to make an impact on public heath, the serotypes of highest concern that are related to human illnesses should be considered. Additional mitigation strategies should also be observed, such as pre-harvest strategies to prevent node contamination, or chemical/physical applications of interventions for the reduction of pathogenic prevalence within further processed pork products. This study clearly establishes that lymph nodes contribute to Salmonella presence in ground product.
Pork facilities also face proposed performance standards from the FSIS that must be met. While not yet implemented, the current proposed performance standards determine the “pass or fail” status of a processing plant are based on the total number of
Salmonella positives taken from samples over a 52-week rolling window. However, the positives are based on detection methodology that determines if any amount of
Salmonellais present within the tested sample. Pork processing plants are currently being tested on the presence or absence of
Salmonella alone and not on quantification or determination or serotype or pathogenicity. It is currently estimated that the infectious dose of
Salmonella is relatively high when compared to other pathogens, estimated between 10
5 and 10
6 cells [
11]. The quantifiable
Salmonella positives detected within this study showed levels far below 10
6 CFU. Thus suggesting that pathogenic loads of the product might be below or far below the number of cells that would cause human infection in a healthy adult upon consumption of a fully cooked product. Therefore, this data could be used to inform future decisions regarding performance standards and their dependency on quantification based methodologies for
Salmonella testing rather than presence-absence alone in order to make more informed decisions about the safety of a product. Implementing quantification methodologies within the industry for pathogenic testing may provide further insight and data to make risk-based public health decisions.
Author Contributions
Conceptualization, M.X.S.-P. and M.M.B; methodology, R.L.J., R.B.L., D.A.V., and M.X.S.-P.; validation, R.L.J and R.B.L..; formal analysis, R.L.J. and D.A.V.; investigation, R.L.J., R.B.L., and D.A.V.; resources, M.X.S.-P.; data curation, R.L.J. and D.A.V.; writing—original draft preparation, R.L.J..; writing—review and editing, R.J.J., R.B.L., D.A.V., M.M.B. and M.X.S.-P; supervision, M.X.S.-P.; project administration, R.L.J. and M.X.S.-P.; funding acquisition, M.M.B and M.X.S.-P. All authors have read and agreed to the published version of the manuscript.
Funding
The current study was funded by the International Center for Food Industry Excellence (ICFIE) at Texas Tech University and supported by in-kind distributions from Hygiena and BioMérieux.
Data Availability Statement
Data available upon request from the corresponding author. The data are not publicly available due to privacy from the pork processing partner which allowed the project to be conducted within their facility.
Acknowledgments
We would like to acknowledge the help of all ICFIE Food Microbiology personnel that helped with sample processing and helped in any other way to make this project a reality.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shahbendeh, M. Meat Consumption Worldwide from 1990 to 2021, by Meat Type; 2021;
- National Pork Producers Council New report highlights U.S. pork industry contributions to American jobs and the economy. Available online https://nppc.org/press-releases/2021_economic_impact_report/ (accessed on 3 May 2023).
- United States Department of Agriculture - Food Safety and Inspection Service Sampling Requirements to Demonstrate Process Control in Slaughter Operations; 2020;
- Bueno López, R., Vargas, D. A., Jimenez, R. L., Casas, D. E., Miller, M. F., Brashears, M. M., & Sanchez-Plata, M. X. (2022). Quantitative Bio-Mapping of Salmonella and Indicator Organisms at Different Stages in a Commercial Pork Processing Facility. Foods, 11(17), 2580. [CrossRef]
- Center for Food Safety and Applied Nutrition. 2019 Annual Report on the Sources of Foodborne iIllness. Available online https://www.fda.gov/food/cfsan-constituent-updates/release-2019-annual-report-sources-foodborne-illness-interagency-food-safety-analytics-collaboration. (accessed on 26 August 2022).
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R. v.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerging Infectious Diseases 2011, 17, 7–15. [CrossRef]
- Hoffmann, S.; Maculloch, B.; Batz, M. Economic Burden of Major Foodborne Illnesses Acquired in the United States; 2015;
- Interagency Food Safety Analytics Collaboration Foodborne Illness Source Attribution Estimates for 2019 for Salmonella, Escherichia Coli O157, Listeria Monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States; Atlanta, Georgia and Washington, District of Columbia, 2021;
- Webb, H. E., Brichta-Harhay, D. M., Brashears, M. M., Nightingale, K. K., Arthur, T. M., Bosilevac, J. M., Kalchayanand, N., Schmidt, J. W., Wang, R., Granier, S. A., Brown, T. R., Edrington, T. S., Shackelford, S. D., Wheeler, T. L., & Loneragan, G. H. (2017). Salmonella in Peripheral Lymph Nodes of Healthy Cattle at Slaughter. Frontiers in M icrobiology, 8, 2214. https://doi.org/10.3389/fmicb.2017.02214. [CrossRef]
- United States Department of Agriculture - Food Safety and Inspection Service Modernization of Swine Slaughter Inspection 2019.
- Kurtz, J. R., Goggins, J. A., & McLachlan, J. B. (2017). Salmonella Infection: Interplay Between the Bacteria and Host Immune System. Immunology Letters, 190, 42–50. https://doi.org/10.1016/j.imlet.2017.07.006. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).