1. Introduction
The current worldwide increase in resistant bacteria has led to a search for novel antimicrobial agents. Thus, bacteriocin and bacteriocin-like inhibitory substances are promising tools against pathogenic bacteria with potential applications in both the food industry and the medical field.
Listeria monocytogenes causes listeriosis, is predominantly foodborne, in particular ready-to-eat food, and most often affecting susceptible groups such as pregnant women, newborn infants, children, elderly and immunocompromised individuals. However, not always human listeriosis are directly attributed to eating contaminated food, having several potential ways for transmission, including neonatal cross-infection [
1]. Antibiotics are the most common treatment choice of listeriosis, although antibiotic resistance in
L. monocytogenes has been reported over the past few decades [
2]. Listeriosis is an emerging infection of public health concern worldwide that has a high incidence in pregnant women [
3,
4,
5,
6,
7,
8]. Infection during pregnancy can cause fetal loss, premature birth and illness or death in newborn [
9].
Enterococcus sp. are indigenous species in the gastrointestinal tracts of humans and animals.
Enterococcus faecalis and
Enterococcus faecium are the most common species isolated from the human gut [
10]. In addition to their role in well-being and health as part of the commensal gut microbiota, significant attention has been focused on the development of these strains as probiotics because of their beneficial health effects in the host [
11].
Breastfeeding can be a significant source of enterococci to the infant gut [
12] which may affect the overall composition of the neonate gut microbiota and exert biological functions. In a previous work, a total of 41 enterococci (26
Enterococcus faecalis and 15
Enterococcus faecium) were isolated from healthy breast-fed infants younger than 6 months. The presence of virulence factors, resistance against antibiotics and biogenic amine production was evaluated [
13]. Here, the evaluation of enterocin production by those enterococci was carried out, which can be of interest as alternatives to antibiotics due to the increased the number of resistances in clinical and commensal bacteria [
14,
15,
16]. The use of bacteriocins as therapeutic agents as well as part of a multiple hurdle approach with antibiotics have been recently explored as viable alternative [
17]. Moreover, the fact that
Enterococcus strains have been isolated from infant faeces could be an advantage over other strains isolated from foods [
18,
19] for their potential use as probiotics [
20,
21] since they are able to compete with pathogenic bacteria for nutrients and colonize GIT effectively [
22].
The aim of this in vitro study was to acquire information on the distribution of structural genes encoding the production of bacteriocins, and on the occurrence of bacteriocin activity among enterococci isolated from breast-fed infants. In addition, the expression of these genes and the antimicrobial spectrum against virulent L. monocytogenes and potential beneficial bacteria was evaluated.
2. Materials and Methods
2.1. Bacterial cultures and media
Twenty-six
E. faecalis and 15
E. faecium isolates from faecal samples from 23 healthy breast-fed infants younger than 6 months were used in this study [
13]. Isolates were cultured in MRS broth (Scharlau Chemie SA, Barcelona, Spain) at 37°C in aerobic conditions.
Listeria monocytogenes strains were grown in TSB (Scharlau Chemie SA, Barcelona, Spain) at 37°C in aerobic conditions. Lactobacilli strains were grown in MRS at 37°C in aerobic conditions. Whereas, Bifodobacterium strains were grown in RCM broth (BD, Le Pont de Claix, France) at 37°C in anaerobic conditions in sealed jars using AnaeroGen sachets (Oxoid, Ltd. Basingstoke, UK).
2.2. L. monocytogenes antibiotic susceptibility testing
Antimicrobial susceptibility testing of
L. monocytogenes was performed on Müller-Hinton agar (Condalab) supplemented with 5% defibrinated horse blood (Thermo-Fisher Scientific) and 20 mg/mL β-NAD (Sigma-Aldrich) according to the disk diffusion method recommended by the European Committee on Antimicrobial susceptibility Testing (EUCAST) [
23]. The following antibiotics, chosen for using in the veterinary and human medicine for treatment of listeriosis, were tested: penicillin G (10 IU/disc), ampicillin (10 µg/disc), amoxicillin/clavulanic acid (30 µg/disc), oxacillin (1 µg/disc), gentamicin (10 µg/disc), chloramphenicol (30µg/disc), vancomycin (30 µg/disc), tetracyclin (30 µg/disc), ciprofloxacin (5 µg/disc), clindamycin (2µg/disc), erythromycin (15 µg/disc), cefoxitin (30 µg/disc), trimethoprim-sulphamethoxazole (25 µg/disc), meropenem (10 µg/disc) and rifampicin (5 µg/disc) (Oxoid). Control strain used in this study was
Staphylococcus aureus ATCC 29213.
2.3. Detection of enterocin genes
Enterocin-encoding genes in enterococci above mentioned was detected by PCR using the specific oligonucleotide primers listed in
Table 1. The PCR were sequenced and compared with known sequences in the BLASTN database (National Center for Biotechnology Information).
2.3. Bacteriocin assay and spectrum of activity
The antimicrobial activity of enterococcal strains harbouring enterocin gene was tested using Listeria monocytogenes Scott A, Listeria monocytogenes OHIO and Listeria monocytogenes ATCC 19115 as indicator strains. Enterocin extracts were obtained by centrifuging at 10,000 g for 15 min at 4 ªC a 18 h culture of the enterocin producer, followed by filtering the supernatant through 0.22-mm pore size low protein binding filter (Millex GV, Millipore, Molsheim, France), and raising the pH to 6.5 with 2 M NaOH. Enterocin extracts were aliquoted and stored at -70 ºC until use. In the agar diffusion assay, a volume of 25 mL of each enterocin extract was placed in duplicate into wells (5 mm diameter) made in plates of TSA inoculated with 0.1% (v/v) of a 24 h culture of each Listeria strain. After aerobic incubation at 37 ªC for 24 h, the bacterial lawns were checked for inhibition zones. The titre (expressed in enterocin units [EU] per millilitre) was defined as the reciprocal of the highest dilution causing a clear zone of inhibition in the indicator lawn. Two independent trials were performed and each strain was assayed in duplicate.
Later, enterocin assays were also carried out against Lacticaseibacillus rhamnosus GG, Limosilactobacillus reuteri Biogaia, Limosilactobacillus reuteri INIA P572, Lacticaseibacillus paracasei INIA P272 Lacticaseibacillus rhamnosus INIA P344, Bifidobacterium longum BB536, Bifidobacterium animalis BB12 Bifidobacterium pseudolongum INIA P2 and Bifidobacterium breve INIA P18. Plates of MRS, RCM or TSA were used for the different strains.
2.4. Total RNA isolation and reverse transcription (RT)-PCR analysis
The
Enterococcus that harbour enterocin genes were grown in MRS broth (10 mL) overnight under aerobic conditions at 37 ºC. The total RNA was isolated using the High Pure RNA isolation kit (Roche, Mannheim, Germany) as specified by the manufacturer. The RNA was quantified by measuring its optical density at 260 nm. The total RNA quality was assessed spectrophotometrically and with gel electrophoresis. Using GeneAmpR EZ r
Tth RNA PCR kit (Applied Biosystems, Branchburg, New Jersey, USA) according to the manufacturer’s instructions the total RNA was then used as a template to generate first strand cDNA and PCR amplification. RNA and cDNA samples obtained above were used for the PCR amplification of the enterocin A, enterocin B, enterocin P genes with oligonucleotides listed in
Table 1. The constitutively expressed 16S rRNA (63F/1387R) served as an internal control gene for these RT-PCR experiments.
The following PCR conditions were used: denaturation at 95°C for 5 min; 35 cycles of denaturation at 94°C for 15 s, annealing at 57°C for 30 s, and extension at 72°C for 60 s; and a final extension cycle at 72°C for 5 min. The amplification products were resolved by electrophoresis in 2% agarose gels. To confirm the absence of contaminating DNA, similar experiments were conducted without reverse transcriptase.
2.5. Overlay agar spot assay
Enterococcus strains with enterocin genes were grown overnight on MRS broth (Condalab) at 37 ºC under aerobic conditions. Two-microliter of inoculums were spot inoculated onto the MRS agar plates and grown for 18 h at 37°C under aerobic conditions. The MRS agar plates containing the growth of Enterococcus strains in spot form were then overlaid with 0.75% brain-heart infusion agar (Condalab) inoculated with 106 log cfu/mL of the different L. monocytogenes strains, and incubated at 37 ºC for 24 h. Two independent trials were performed and each pathogenic strain was assayed in duplicate. The diameter of inhibition were measured and expressed in mm as the mean of n=4.
3. Results
3.1. Detection of enterocin genes
A total of 41 enterococci (26 E. faecalis and 15 E. faecium) were examined for the presence of enterocin genes by means of PCR. The detected enterocins are listed in
Table 2. Only E. faecalis INIA P290 showed the presence the enterocin gene AS-48. Whereas 10 out of 15 E. faecium isolates harbouring one (Ent A or Ent P) or two (Ent A + Ent B or Ent P + Ent P) enterocin-encoding genes (
Table 2).
3.2. Antibiotic susceptibility of L. monocytogenes
Zones of inhibition were measured in opened plates and with reflected light and interpreted according the EUCAST criteria. The breakpoints of Staphylococcus spp. resistance were considered if no resistance criteria exist in the EUCAST or Clinical and Laboratory Standards Institute (CLSI) guidelines for Listeria susceptibility testing. Based on the results, the strains were classified as sensitive, intermediate resistant or resistant.
Fifteen antibiotics belonging to penicillins, aminoglycosides, phenicols, glycopeptides, tetracyclines, fluoroquinolones, lincosamides, macrolides, cephalosporins, sulfonamides, and carbapenems were tested. The three strains of L. monocytogenes resulted sensitive to most antibiotics tested, specifically to penicillins (penicillin G, ampicillin and amoxicillin/clavulanic acid), aminoglycosides (gentamycin), phenicols (chloramphenicol), glycopeptides (vancomycin), tetracyclines (tetracycline), fluoroquinolones (ciprofloxacin), macrolides (erythromycin), sulfonamides (trimethoprim-sulphamethoxazole) and carbapenems (meropenem). L. monocytogenes is intrinsically resistant to cephalosporin antibiotics (cefoxitin). However, the three strains studied also exhibited resistance to oxacillin and clindamycin (intermediate resistant to clindamycin in the case of strain Ohio).
3.2. Antimicrobial activity
Plate diffusion bioassays, using strains of L. monocytogenes as indicator organism, showed that only E. faecalis INIA P290 (Ent AS-48), E. faecium P442 (Ent A and Ent B), E. faecium P445 (Ent A and Ent B) and E. faecium P545 (Ent B and Ent P) were able produced an inhibitory zone active against L. monocytogenes strains. The highest inhibitory zone was produced by E. faecium P545 which produced enterocin B and enterocin P.
On the other hand, L. rhamnosus LGG, L. reuteri Biogaia, L. reuteri INIA P572, Lb. paracasei INIA P272, L. rhamnosus INIA P344, Bifidobacterium animalis BB12, Bifidobacterium pseudolongum INIA P2 and Bifidobacterium breve INIA P18 did not show inhibition for any of the enterocins produced individually or in combination. Only Bifidobacterium longum BB536 showed inhibition by enterocin AS-48 and the joint production of enterocins A and B or B and P.
3.3. Transcriptional analysis of enterocins
In some cases, enterocin gene were detected by PCR, however we did not observed inhibition of indicator microorganisms, thus RT-PCR were carrying out to checking the expression of these genes (
Table 2).
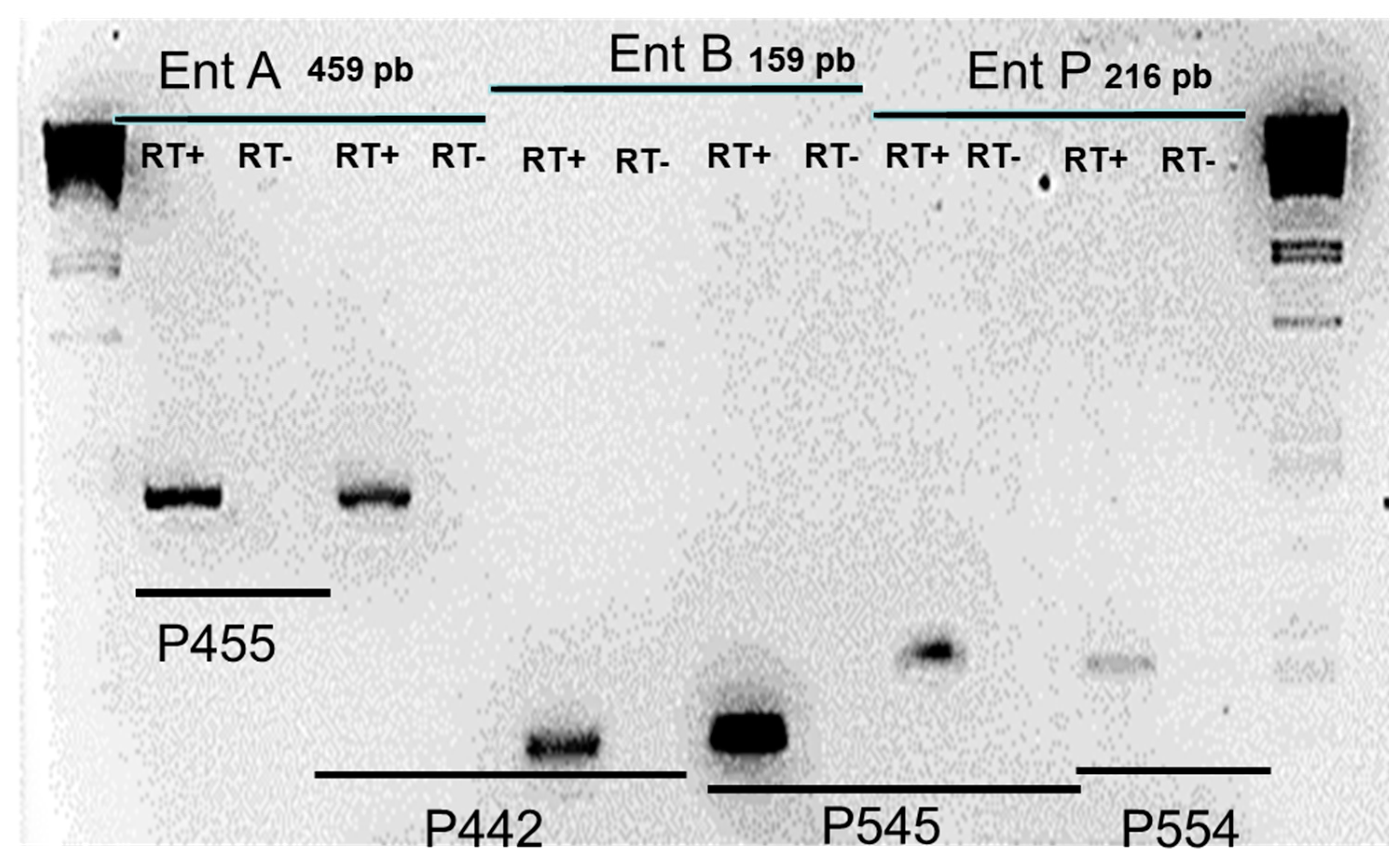
RT-PCR analysis showed that enterocin A and enterocin B cDNA were amplified from
E. faecium INIA P442 and
E. faecium INA P445 (
Figure 1), and enterocin B and enterocin P cDNA were amplified from strain
E. faecium INIA P545, being the expression of B higher in both cases. Enterocin AS-48 cDNA was amplified from strain
E. faecalis INIA P290. In all these cases, the expression of the genes was correlated with the inhibition of
L. monocytogenes strains.
On the other hand, E. faecium INIA P553, P554 and P555 showed expression of enterocin P, however, they did not show inhibition of L. monocytogenes strains and E. faecium INA P125 and E. faecium INA P552 did not show expression of enterocin A nor inhibition of L. monocytogenes strains. Finally, E. faecium INA P454 showed expression of enterocin A, but did not show expression of enterocin B or inhibition of any L. monocytogenes strains.
3.4. Overlay agar spot assay
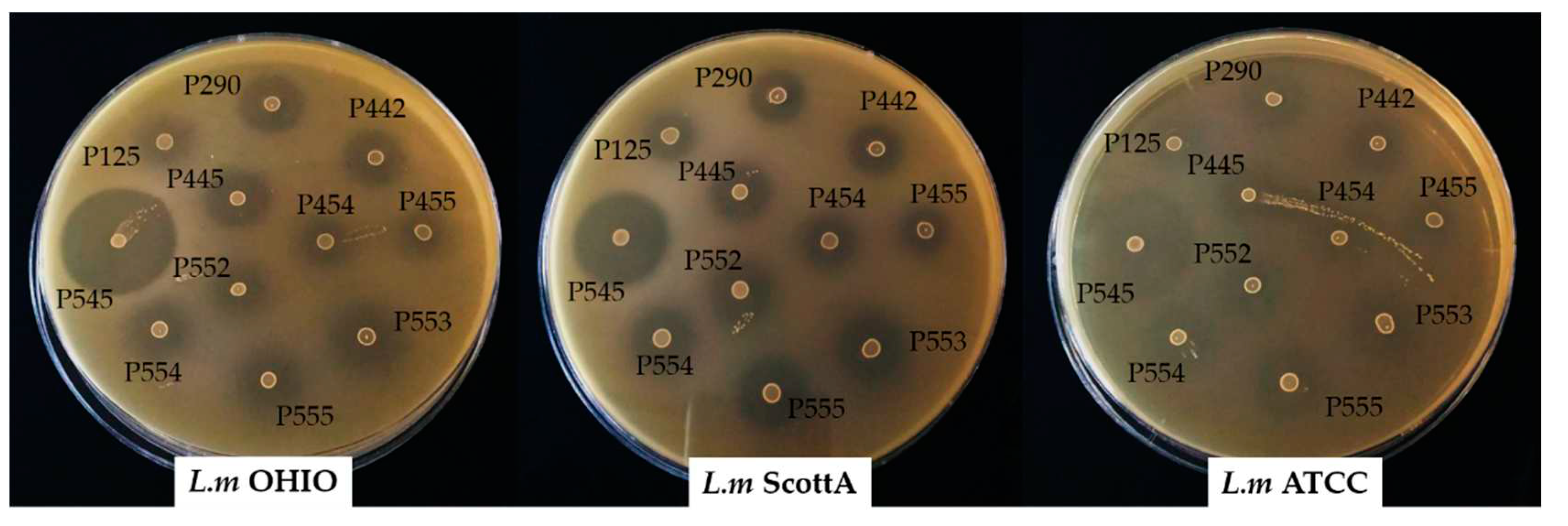
All enterococci assayed showed a clear inhibitory antimicrobial activity around the spot against the three strains of
L. monocytogenes (
Figure 2). The enterocin B and enterocin P-producing
E. faecium INIA P545, showed the highest diameters of inhibition (between 32 and 37 mm). Enterocin AS-48-producing
E. faecalis INIA P290 and enterocin A and B-producing strains
E. faecium P442 and
E. faecium P445 resulted in values between 17 and 22 mm. Enterocin P-producing strains
E. faecium P553,
E. faecium P554 and
E. faecium P555 showed diameters of inhibition between 16 and 21. The lowest measurements were obtained for Enterocin A-producing or non enterocin-producing strains
E. faecium INIA P125,
E. faecium INIA P454,
E. faecium INIA P455 and
E. faecium INIA P552 (between 10 and 18 mm).
4. Discussion
E. faecalis and
E. faecium are the predominant especies of
Enterococcus strains found in breast-fed infants [
13], being the frequency of enterocin genes higher in
E. faecium than in
E. faecalis isolates [
30,
31]. In this study, a combination of enterocin A and B genes was frequently found among, which is in accordance with previous studies [
30,
32,
33]. According to these authors, the association between enterocins A and B may be due to the absence of transport or accessory protein genes in some
Enterococcus strains and both enterocins may act synergistically when expressed in the same culture [
34,
35].
The inhibitory activity against
L. monocytogenes was dependent on single enterocin genes (AS-48) or enterocin genes occurring in combinations (A and B or B and P). However, the presence of multiple enterocin genes does not assured that all of the genes were expressed at the same time, as this was demonstrated in the case of
E. faecium INIA P454 (
Table 2). According with our data, if two enterocins were present in the same supernatant, their antimicrobial activity is higher, as it has been previously reported [
34]. As far as we know this is the first study in which the inhibition of
L. monocytogenes strains was correlated with the expression of enterocin genes under the conditions tested and not with the presence of these genes. Overlay agar spot assay data are in concordance with the previous results with neutralized supernatants. The highest diameter of inhibition was produced by
E. faecium P545 which produced simultaneously enterocin B and enterocin P. Acid could be responsible of the inhibition observed in the non enterocin-producing strains and of the previous absence of inhibition observed in strains able to produce enterocin P or enterocin A individually, since their activity are usually higher at low pH [
36].
Enterocins have exhibited activity against different foodborne pathogens in different studies [
37,
38,
39,
40,
41], however there are not too much information about their effect against potential beneficial bacteria. In this work, no lactobacilli was inhibited and
B. longum BB536 was the only bifidobacteria sensible to the enterocins produced by the
Enterococcus isolated from breast-fed infants.
The bacteriocinogenic enterococci strains are potential candidates for food, human and animal health applications for their interesting properties such as multi-bacteriocin production and viability in different environments, including food and gastrointestinal tract. They are commonly used as starter cultures as well as probiotics for therapeutic treatments without reported adverse effects [
42]. Enterocins showed in this paper can be considered target specific, safe, heat stable and able to exert a synergistic effect with antibiotics, which are interesting features for the development of alternative treatments against antibiotic-resistant bacterial infections. In this sense, it is believed that multiple bacteriocin productions can help the producer strain to abolish resistance problem of some target strains [
43]. The use of broad-spectrum antibiotics alter the host microbiota and therefore its functional capacity, which can have negative effects such as altered metabolic activity and the selection of antibiotic-resistant organisms [
44]. Exposure to broad-spectrum antibiotics is particularly unfavourable during infancy and early childhood since microbiota lacks diversity and stability, making it more sensitive to environmental incursions. This can be particularly important in the development and education of the host immune system [
45]. The use of bacteriocins could avoid these problems because of their target cell specificity.
On the other hand, it has been shown that some
Enterococcus strains could reduce the infection of
L. monocytogenes [
46,
47,
48,
49]. The use of probiotic bacteria isolated from breast-fed infants represent a valuable strategy to reduce the risk of disease due to their prophylactic and therapeutic potential [
50]. No virulence determinants or hemolysin activity was detected in any of the
E. faecium studied. The gelatinase gene, when present, was silent in
E. faecium, and none of the isolates was resistant to vancomycin. These traits together with their antilisterial activity and the presence of genes linked to colonization [
13] could contribute for their use as probiotics able to exert a protective effect against bacterial infections.
Author Contributions
J.M.L., R.M., E.R-M., and J.L.A. have made a substantial, direct, and intellectual contribution to the work, and approved it for publication. All authors have read and agreed to the published version of the manuscript
Funding
This work has received financial support from project RTA2017-00002-00-00 (Spanish Ministry of Science and Innovation).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare no conflict of interest.
References
- McLauchlin, J.; Amar, C.F.L.; Grant, K.A. Neonatal cross-infection due to Listeria monocytogenes. Epidemiol Infect 2022, e77. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr Rev Food Sci Food Saf 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Wilking, H.; Lachmann, R.; Holzer, A.; Halbedel, S.; Flieger, A.; Stark, K. Ongoing high incidence and case-fatality rates for invasive listeriosis, Germany, 2010–2019. Emerg Infect Dis 2021, 27, 2485–2488. [Google Scholar] [CrossRef]
- Pohl, A.M.; Pouillot, R.; Bazaco, M.C.; Wolpert, B.J.; Healy, J.M.; Bruce, B.B.; Laughlin, M.E.; Hunter, J.C.; Dunn, J.R.; Hurd, S.; Rowlands, J.V.; Saupe, A.; Vugia, D.J.; Van Doren, J.M. Differences among incidence rates of invasive listeriosis in the U.S. FoodNet population by age, sex, race/ethnicity, and pregnancy status, 2008–2016. Foodborne Pathog Dis 2019, 16, 290–297. [Google Scholar] [CrossRef]
- Herrador, Z.; Gherasim, A.; Lopez-Velez, R.; Benito, A. Listeriosis in Spain based on hospitalisation records, 1997 to 2015: need for greater awareness. Euro Surveill 2019, 24, 1800271. [Google Scholar] [CrossRef]
- Li, W.; Bai, L.; Ma, X.; Zhang, X.; Li, X.; Yang, X.; Huang, J.Y.; Fanning, S.; Guo, Y. Sentinel listeriosis surveillance in selected hospitals, China, 2013–2017. Emerg Infect Dis 2019, 25, 2274–2277. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, Y.; Liu, Y.; Lu, Z.; Wang, D.; Cui, X.; Chen, Q.; Ma, X. Isolation and characterization of clinical Listeria monocytogenes in Beijing, China, 2014–2016. Front Microbiol 2019, 10, 981. [Google Scholar] [CrossRef]
- Ke, Y.; Ye, L.; Zhu, P.; Sun, Y.; Zhu, Z. Listeriosis during pregnancy: a retrospective cohort study. BMC Pregnancy Childbirth 2022, 22, 261. [Google Scholar] [CrossRef]
- Wang, Z.; Tao, X.; Liu, S.; Zhao, Y.; Yang, X. An update review on Listeria infection in pregnancy. Infect Drug Resist 2021, 14, 1967–1978. [Google Scholar] [CrossRef]
- Layton, B.A.; Walters, S.P.; Lam, L.H.; Boehm, A.B. Enterococcus species distribution among human and animal hosts using multiplex PCR. J Appl Microbiol 2020, 109, 539–547. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: between probiotic potential and safety concerns—An update. Front Microbiol 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langa, S.; Reviriego, C.; Jiménez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr 2003, 143, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Peirotén, A.; Medina, M.; Arqués, J.L.; Rodriguez, E. Virulence and antibiotic resistance of enterococci isolated from healthy breastfed infants. Microb Drug Resist 2018, 24, 63–69. [Google Scholar] [CrossRef]
- Francino, M.P. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 2016, 6, 1543. [Google Scholar] [CrossRef]
- Lebreton, F.; van Schaik, W.; McGuire, M.A.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; Zeng, Q.; Wortman, J.; Birren, B.; Willems, R.J.L.; Earl, A.M.; Gilmore, M.S. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 2013, 4, e00534-13. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V. Editorial for Special Issue “Alternatives to Antibiotics: Bacteriocins and Antimicrobial Peptides”. Antibiotics 2022, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Cavera, V.L.; Arthur, T.D.; Kashtanov, D.; Chikindas, M.L. Bacteriocins and their position in the next wave of conventional antibiotics. Int J Antimicrob Agents 2015, 46, 494–501. [Google Scholar] [CrossRef]
- Cavicchioli, V.Q.; Camargo, A.C.; Todorov, S.D.; Nero, L.A. Novel bacteriocinogenic Enterococcus hirae and Pediococcus pentosaceus strains with antilisterial activity isolated from Brazilian artisanal cheese. J Dairy Sci 2017, 100, 2526–2535. [Google Scholar] [CrossRef]
- Moraes, P.M.; Perin, L.M.; Todorov, S.D.; Franco, S.D.; Silva, A.; Franco, B.D.G.M.; Nero, L.A. Bacteriocinogenic and virulence potential of Enterococcus isolates obtained from raw milk and cheese. J Appl Microbiol 2012, 113, 318–28. [Google Scholar] [CrossRef] [PubMed]
- Im, E.J.; Lee, H.H.; Kim, M.; Kim, M.K. Evaluation of enterococcal probiotic usage and review of potential health benefits, safety, and risk of antibiotic-resistant strain emergence. Antibiotics (Basel) 2023, 12, 1327. [Google Scholar] [CrossRef]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Enterococcus spp. as a producer and target of bacteriocins: A double-edged sword in the antimicrobial resistance crisis context. Antibiotics 2021, 10, 1215. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Langa, S.; Martin, V.; Maldonado, A.; Jimenez, E.; Martin, R.; Rodríguez, J.M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- EUCAST Disk Diffusion Method for Antimicrobial Susceptibility Testing. Version 11.0 (January 2023). www.eucast.org.
- Martínez-Bueno, M.; Valdivia, E.; Gálvez, A.; Coyett, e J.; Maqueda, M. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Molecular Microbiol 1998, 27, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hidalgo, M.; Maqueda, M.; Gálvez, A.; Abriouel, H.; Valdivia, E.; Martínez-Bueno, M. The genes coding for enterocin EJ97 production by Enterococcus faecalis EJ97 are located on a conjugative plasmid. Appl Environ Microbiol 2003, 69, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Aymerich, T.; Holo, H.; Havarstein, L.S.; Hugas, M.; Garriga, M.; Nes, I.F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol 1996, 62, 1676–1682. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, M.; Franz, C.; Dicks, L.M.T; Holzapfel, W.H. Preliminary characterization of bacteriocins produced by Enterococ,cus faecium and Enterococcus faecalis isolated from pig faeces. J Applied Microbiol 2000, 88, 482–494. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, M.; Martínez-Bueno, M.; Martín-Vivaldi, M.; Valdivia, E.; Soler, J.J. Bacteriocins with a broader antimicrobial spectrum prevail in enterococcal symbionts isolated from the hoopoe’s uropygial gland. FEMS Microbiol Ecol 2013, 85, 495–502. [Google Scholar] [CrossRef]
- Yousif, N.M.K.; Dawyndt, P.; Abriouel, H.; Wijaya, A.; Schillinger, U.; Vancanneyt, M.; Swing, s J.; Dirar, H.A. Molecular characterization, technological properties and safety aspects of enterococci from ‘Hussuwa’, an African fermented sorghum product. J Appl Microbiol 2005, 98, 216–228. [Google Scholar] [CrossRef]
- De Vuyst, L.; Moreno, M.F.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int J Food Microbiol 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Pangallo, D.; Harichová, J.; Karelová, E.; Drahovská, H.; Chovanová, K.; Ferianc, P. Timko, J. Molecular investigation of enterococci isolated from different environmental sources. Biologia 2004, 59, 829–837. [Google Scholar]
- Özdemir, G.B.; Oryasm, E.; Biyik, H.H.; Özteber, M.; Bozdoğan, B. Phenotypic and genotypic characterization of bacteriocins in enterococcal isolates of different sources. Ind J Microbiol 2011, 51, 182–187. [Google Scholar] [CrossRef]
- Poeta, P.; Costa, D.; Rojo-Bezares, B.; Zarazaga, M.; Klibi, N.; Rodrigues, J.; Torres, C. Detection of antimicrobial activities and bacteriocin structural genes in faecal enterococci of wild animals. Microbiol Res 2007, 162, 257–263. [Google Scholar] [CrossRef]
- Casaus, F.; Nilsen, T.; Cintas, L.M.; Nes, L.F.; Hernández, P.E.; Holo, H. Enterocin B, a new bacteriocin from Enterococcus faecium TI36 which can act synergistically with enterocin A. Microbiology 1997, 143, 2287–2294. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Worobo, R.W.W.; Quadri, L.E.N.; Schillinger, U.; Holzapfel, W.H.; Vederas, J.C.; Stiles, M.E. Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE 900. Appl Environ Microbiol 1999, 65, 2170–2178. [Google Scholar] [CrossRef] [PubMed]
- Foulquié Moreno, M.R.; Callewaert, R.; Devreese, B.; Van Beeumen, J.; De Vuyst, L. Isolation and biochemical characterisation of enterocins produced by enterococci from different sources. J Appl Microbiol 2003, 94, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Arqués, J.L.; Rodríguez, E.; Langa, S.; Landete, J.M.; Medina, M. Antimicrobial activity of lactic acid bacteria in dairy products and gut: effect on pathogens. Biomed Res Int 2015, 584183. [Google Scholar] [CrossRef] [PubMed]
- Ogaki, M.B.; Rocha, K.R.; Terra, M.R.; Furlaneto, M.C.; Furlaneto-Maia, L. Screening of the enterocin-encoding genes and antimicrobial activity in Enterococcus species. J Microbiol Biotechnol 2016, 26, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Darbandi, A.; Asadi, A.; Ari, M.M.; Ohadi, E.; Talebi, M.; Zadeh, M.H.; Emamie, A.D.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J Clin Lab Anal 2022, 36, 24093. [Google Scholar] [CrossRef]
- Wu, Y.; Pang, X.; Wu, Y.; Liu, X.; Zhang, X. Enterocins: Classification, Synthesis, Antibacterial Mechanisms and Food Applications. Molecules 2022, 27, 2258. [Google Scholar] [CrossRef]
- Zdolec, N; Kiš, Marta. Antimicrobial properties of food enterococci. In Lactic Acid Bacteria in Food Biotechnology. Innovations and Functional Aspects.; Ray, R.C., Paramithiotis, P., Azevedo, V.A.C., Montet, D., Eds.; 2022; Volume 11, pp. 195–203. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 2011, 151, 125–40. [Google Scholar] [CrossRef]
- Perez, R.H.; Himeno, K.; Ishibashi, N.; Masuda, Y.; Zendo, T.; Fujita, K.; Wilaipun, P.; Leelawatcharamas, V.; Nakayama, J.; Sonomoto, K. Monitoring of the multiple bacteriocin production by Enterococcus faecium NKR-5-3 through a developed liquid chromatography and mass spectrometry-based quantification system. J Biosci Bioeng 2012, 114, 490–6. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol 2020, 10, 572912. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- He, Y.; Yang, Q.; Tian, L.; Zhang, Z.; Qiu, L; Tao, X.; Wei, H. Protection of surface layer protein from Enterococcus faecium WEFA23 against Listeria monocytogenes CMCC54007 infection by modulating intestinal permeability and immunity. Appl Microbiol Biotechnol 2021, 105, 4269–4284. [Google Scholar] [CrossRef] [PubMed]
- Popović, N.; Djokić, J.; Brdarić, E.; Dinić, M.; Terzić-Vidojević, A.; Golić, N.; Veljović, K. The influence of heat-killed Enterococcus faecium BGPAS1-3 on the tight junction protein expression and immune function in differentiated Caco-2 cells infected with Listeria monocytogenes ATCC 19111. Front Microbiol 2019, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Rajput, K.; Dubey, R.C.; Kumar, A. Probiotic potential and immunomodulatory properties in Enterococcus faecium GMB24 and Enterococcus hirae SMB16 isolated from goat and sheep milk. Arch Microbiol 2022, 204, 619. [Google Scholar] [CrossRef]
- Salvucci, E.; Saavedra, L.; Hebert, E.M.; Haro, C.; Sesma, F. Enterocin CRL35 inhibits Listeria monocytogenes in a murine model. Foodborne Pathog Dis 2012, 9, 68–74. [Google Scholar] [CrossRef]
- Song, MW; Kim, K-T; Paik, H-D. Probiotics as a functional health supplement in infant formulas for the improvement of intestinal microflora and immunity. Food Rev Intl. 2021, 39, 858–74. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).