Submitted:
26 September 2023
Posted:
28 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study group
2.2. SNP genotyping
2.3. Statistical analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinkas, W.; Jankowski, M.; Wierzba, W. Awareness of Head and Neck Cancers: A 2021 Nationwide Cross-Sectional Survey in Poland. J. Clin. Med. 2022, 11, 538. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [CrossRef]
- Papalouka, C.; Adamaki, M.; Batsaki, P.; Zoumpourlis, P.; Tsintarakis, A.; Goulielmaki, M.; Fortis, S.P.; Baxevanis, C.N.; Zoumpourlis, V. DNA Damage Response Mechanisms in Head and Neck Cancer: Significant Implications for Therapy and Survival. Int. J. Mol. Sci. 2023, 24, 2760. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.; Tainer, J.A. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef]
- Huen, M.S.; Sy, S.M.; Chen, J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010, 11, 138–148. [Google Scholar] [CrossRef]
- El Nachef, L.; Berthel, E.; Ferlazzo, M.L.; Le Reun, E.; Al-Choboq, J.; Restier-Verlet, J.; Granzotto, A.; Sonzogni, L.; Bourguignon, M.; Foray, N. Cancer and Radiosensitivity Syndromes: Is Impaired Nuclear ATM Kinase Activity the Primum Movens? Cancers 2022, 14, 6141. [Google Scholar] [CrossRef]
- Parliament, M.B.; Murray, D. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Semin. Radiat. Oncol. 2010, 20, 232–240. [Google Scholar] [CrossRef]
- Wang, M.; Chu, H.; Zhang, Z.; Wei, Q. Molecular epidemiology of DNA repair gene polymorphisms and head and neck cancer. J. Biomed. Res. 2013, 27, 179–192. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zheng, C.C.; Huang, Y.N.; He, M.L.; Xu, W.W.; Li, B. Molecular mechanisms of chemo- and radiotherapy resistance and the potential implications for cancer treatment. MedComm. (2020) 2021, 2, 315–340. [Google Scholar] [CrossRef]
- Yang, M.H.; Chang, S.Y.; Chiou, S.H.; Liu, C.J.; Chi, C.W.; Chen, P.M.; Teng, S.C.; Wu, K.J. Overexpression of NBS1 induces epithelial-mesenchymal transition and co-expression of NBS1 and Snail predicts metastasis of head and neck cancer. Oncogene 2007, 26, 1459–1467. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Yordy, J.S.; Williams, M.D.; Giri, U.; Raju, U.; Molkentine, D.P.; Byers, L.A.; Heymach, J.V.; Story, M.D.; Lee, J.J.; Sturgis, E.M.; Weber, R.S.; Garden, A.S.; Ang, K.K.; Schwartz, D.L. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clin. Cancer Res. 2011, 17, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Chen, Y.K.; Lo, S.; Chi, T.C.; Chen, Y.H.; Hu, S.C.; Chen, Y.W.; Jiang, S.S.; Tsai, F.Y.; Liu, W.; Li, R.N.; Hsieh, Y.C.; Huang, C.J.; Yuan, S.F. MRE11 promotes oral cancer progression through RUNX2/CXCR4/AKT/FOXA2 signaling in a nuclease-independent manner. Oncogene 2021, 40, 3510–3532. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Sun, J.; Liu, Y.; Liu, D.; Du, L.; Wang, B.; Liu, W. Expression of RAD51 and Its Clinical Impact in Oral Squamous Cell Carcinoma. Anal. Cell. Pathol. (Amst). 2020, 2020, 1827676. [Google Scholar] [CrossRef] [PubMed]
- Butkiewicz, D.; Gdowicz-Kłosok, A.; Krześniak, M.; Rutkowski, T.; Krzywon, A.; Cortez, A.J.; Domińczyk, I.; Składowski, K. Association of Genetic Variants in ANGPT/TEK and VEGF/VEGFR with Progression and Survival in Head and Neck Squamous Cell Carcinoma Treated with Radiotherapy or Radiochemotherapy. Cancers 2020, 12, 1506. [Google Scholar] [CrossRef] [PubMed]
- Ensembl Database 103. Available online: http://www.ensembl.org/ (accessed on 25 February 2021).
- Teo, M.T.; Landi, D.; Taylor, C.F.; Elliott, F.; Vaslin, L.; Cox, D.G.; Hall, J.; Landi, S.; Bishop, D.T.; Kiltie, A.E. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis 2012, 33, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Naccarati, A.; Rosa, F.; Vymetalkova, V.; Barone, E.; Jiraskova, K.; Di Gaetano, C.; Novotny, J.; Levy, M.; Vodickova, L.; Gemignani, F.; Buchler, T.; Landi, S.; Vodicka, P.; Pardini, B. Double-strand break repair and colorectal cancer: gene variants within 3’ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget 2016, 7, 23156–23169. [Google Scholar] [CrossRef]
- Choudhury, A.; Elliott, F.; Iles, M.M.; Churchman, M.; Bristow, R.G.; Bishop, D.T.; Kiltie, A.E. Analysis of variants in DNA damage signalling genes in bladder cancer. BMC Med. Genet. 2008, 9, 69. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, P.; Song, C.; Wang, K.; Yan, R.; Li, J.; Dai, L. Evaluation of miRNA-binding-site SNPs of MRE11A, NBS1, RAD51 and RAD52 involved in HRR pathway genes and risk of breast cancer in China. Mol. Genet. Genomics 2015, 290, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Kim, S.Y.; Yi, S.L.; Son, S.H.; Song, D.Y.; Moon, S.Y.; Kim, J.H.; Choi, E.K.; Ahn, S.D.; Shin, S.S.; Lee, K.K.; Lee, S.W. Expression of Ku80 correlates with sensitivities to radiation in cancer cell lines of the head and neck. Oral Oncol. 2006, 42, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.F.; Tseng, H.C.; Chiu, C.F.; Liang, S.Y.; Tsai, C.W.; Tsai, M.H.; Bau, D.T. Association between DNA double strand break gene Ku80 polymorphisms and oral cancer susceptibility. Oral Oncol. 2009, 45, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mathew, D.; Bhat, S.A.; Ghoshal, S.; Pal, A. Genetic Variants of DNA Repair Genes as Predictors of Radiation-Induced Subcutaneous Fibrosis in Oropharyngeal Carcinoma. Front. Oncol. 2021, 11, 652049. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, T.; Mori, Y.; Sato, H.; Permata, T.B.M.; Uchihara, Y.; Noda, S.E.; Okada, K.; Kakoti, S.; Suzuki, K.; Ikota, H.; Yokoo, H.; Gondhowiardjo, S.; Nakano, T.; Ohno, T.; Shibata, A. Expression of non-homologous end joining factor, Ku80, is negatively correlated with PD-L1 expression in cancer cells after X-ray irradiation. Oncol. Lett. 2022, 23, 29. [Google Scholar] [CrossRef]
- Joshi, J.S.; Vora, H.H.; Ghosh, N.R.; Tankshali, R.N.; Jetly, D.H.; Trivedi, T.I. Nonhomologous end joining repair pathway molecules as predictive biomarkers for patients with oral squamous cell carcinoma. J. Cancer Res. Ther. 2021, 17, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Girard, P.M.; Kysela, B.; Härer, C.J.; Doherty, A.J.; Jeggo, P.A. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum. Mol. Genet. 2004, 13, 2369–2376. [Google Scholar] [CrossRef]
- Tseng, R.C.; Hsieh, F.J.; Shih, C.M.; Hsu, H.S.; Chen, C.Y.; Wang, Y.C. Lung cancer susceptibility and prognosis associated with polymorphisms in the nonhomologous end-joining pathway genes: a multiple genotype-phenotype study. Cancer 2009, 115, 2939–2948. [Google Scholar] [CrossRef]
- García-Lestón, J.; Roma-Torres, J.; Vilares, M.; Pinto, R.; Prista, J.; Teixeira, J.P.; Mayan, O.; Conde, J.; Pingarilho, M.; Gaspar, J.F.; Pásaro, E.; Méndez, J.; Laffon, B. Genotoxic effects of occupational exposure to lead and influence of polymorphisms in genes involved in lead toxicokinetics and in DNA repair. Environ. Int. 2013, 43, 29–36. [Google Scholar] [CrossRef]
- Mumbrekar, K.D.; Goutham, H.V.; Vadhiraja, B.M.; Bola Sadashiva, S.R. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals. DNA repair 2016, 40, 27–34. [Google Scholar] [CrossRef]
- Ribeiro, H.L., Junior; Soares Maia, A.R.; Costa, M.B.; Farias, I.R.; de Paula Borges, D.; de Oliveira, R.T.; de Sousa, J.C.; Magalhães, S.M.; Pinheiro, R.F. Influence of functional polymorphisms in DNA repair genes of myelodysplastic syndrome. Leuk. Res. 2016, 48, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, G.H.; Manjunath, V.B.; Mumbrekar, K.D.; Negi, H.; Fernandes, D.J.; Sharan, K.; Banerjee, S.; Bola Sadashiva, S.R. Polymorphisms in radio-responsive genes and its association with acute toxicity among head and neck cancer patients. PloS One 2014, 9, e89079. [Google Scholar] [CrossRef]
- Werbrouck, J.; De Ruyck, K.; Duprez, F.; Van Eijkeren, M.; Rietzschel, E.; Bekaert, S.; Vral, A.; De Neve, W.; Thierens, H. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat. Res. 2008, 656, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Xu, X.L.; Ruan, H.H.; Xu, W.Z.; Li, D.; Feng, J.G.; Han, Q.B.; Mao, W.M. The impact of functional LIG4 polymorphism on platinum-based chemotherapy response and survival in non-small cell lung cancer. Med Oncol. 2014, 31, 959. [Google Scholar] [CrossRef] [PubMed]
- Demeyer, A.; Benhelli-Mokrani, H.; Chénais, B.; Weigel, P.; Fleury, F. Inhibiting homologous recombination by targeting RAD51 protein. Biochim. Biophys Acta Rev. Cancer 2021, 1876, 188597. [Google Scholar] [CrossRef]
- Wiegmans, A.P.; Al-Ejeh, F.; Chee, N.; Yap, P.Y.; Gorski, J.J.; Da Silva, L.; Bolderson, E.; Chenevix-Trench, G.; Anderson, R.; Simpson, P.T.; Lakhani, S.R.; Khanna, K.K. Rad51 supports triple negative breast cancer metastasis. Oncotarget 2014, 5, 3261–3272. [Google Scholar] [CrossRef]
- Hasselbach, L.; Haase, S.; Fischer, D.; Kolberg, H.C.; Stürzbecher, H.W. Characterisation of the promoter region of the human DNA-repair gene Rad51. Eur. J. Gynaecol. Oncol. 2005, 26, 589–598. [Google Scholar]
- Lu, J.; Wang, L.E.; Xiong, P.; Sturgis, E.M.; Spitz, M.R.; Wei, Q. 172G>T variant in the 5’ untranslated region of DNA repair gene RAD51 reduces risk of squamous cell carcinoma of the head and neck and interacts with a P53 codon 72 variant. Carcinogenesis 2007, 28, 988–994. [Google Scholar] [CrossRef]
- Goricar, K.; Erculj, N.; Zadel, M.; Dolzan, V. Genetic polymorphisms in homologous recombination repair genes in healthy Slovenian population and their influence on DNA damage. Radiol. Oncol. 2012, 46, 46–53. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, P.; Dong, Y.; Zhu, X.; Zhang, X. Relationship between Rad51 G135C and G172T variants and the susceptibility to cancer: a meta-analysis involving 54 case-control studies. PloS One 2014, 9, e87259. [Google Scholar] [CrossRef]
- Yin, M.; Liao, Z.; Huang, Y.J.; Liu, Z.; Yuan, X.; Gomez, D.; Wang, L.E.; Wei, Q. Polymorphisms of homologous recombination genes and clinical outcomes of non-small cell lung cancer patients treated with definitive radiotherapy. PloS One 2011, 6, e20055. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, Z.Y.; Li, C.B.; Gao, S.; Ding, L.H.; Wu, X.L.; Wang, Z.Y. Genetic polymorphisms of DNA repair pathways influence the response to chemotherapy and overall survival of gastric cancer. Tumour Biol. 2015, 36, 3017–3023. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.M.; Santos, H.B.P.; de Matos, F.R.; Machado, R.A.; Coletta, R.D.; Galvão, H.C.; Freitas, R.A. Clinicopathological significance of SNPs in RAD51 and XRCC3 in oral and oropharyngeal carcinomas. Oral Dis. 2019, 25, 54–63. [Google Scholar] [CrossRef]
- HaploReg v4.2. database. Available online: https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php (accessed on 02 May 2023).
- Ward, L.D.; Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Y.; Chen, M.; Wang, Y.; Feng, Y.; Xu, Z.; Zhang, D.; Sun, Y.; Fu, Z. Association of genetic variants in ATR-CHEK1 and ATM-CHEK2 pathway genes with risk of colorectal cancer in a Chinese population. Oncotarget 2018, 9, 26616–26624. [Google Scholar] [CrossRef] [PubMed]
- Shatoff, E.; Bundschuh, R. Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures. PLoS Comput. Biol. 2020, 16, e1007852. [Google Scholar] [CrossRef] [PubMed]
- Barker, H.E.; Patel, R.; McLaughlin, M.; Schick, U.; Zaidi, S.; Nutting, C.M.; Newbold, K.L.; Bhide, S.; Harrington, K.J. CHK1 Inhibition Radiosensitizes Head and Neck Cancers to Paclitaxel-Based Chemoradiotherapy. Mol. Cancer Ther. 2016, 15, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- van Harten, A.M.; Buijze, M.; van der Mast, R.; Rooimans, M.A.; Martens-de Kemp, S.R.; Bachas, C.; Brink, A.; Stigter-van Walsum, M.; Wolthuis, R.M.F.; Brakenhoff, R.H. Targeting the cell cycle in head and neck cancer by Chk1 inhibition: a novel concept of bimodal cell death. Oncogenesis 2019, 8, 38. [Google Scholar] [CrossRef]
- Pim, D.; Banks, L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int. J. Cancer 2004, 108, 196–199. [Google Scholar] [CrossRef]
- Jeong, B.S.; Hu, W.; Belyi, V.; Rabadan, R.; Levine, A.J. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 2010, 24, 1347–1353. [Google Scholar] [CrossRef]
- Ji, X.; Neumann, A.S.; Sturgis, E.M.; Adler-Storthz, K.; Dahlstrom, K.R.; Schiller, J.T.; Wei, Q.; Li, G. p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis 2008, 29, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Niu, J.; Wei, S.; Yin, M.; Liu, Z.; Wang, L.E.; Sturgis, E.M.; Wei, Q. A novel functional DEC1 promoter polymorphism -249T>C reduces risk of squamous cell carcinoma of the head and neck. Carcinogenesis 2010, 31, 2082–2090. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Bairati, I.; Samson, E.; Cheng, D.; Mirshams, M.; Qiu, X.; Savas, S.; Waldron, J.; Wang, C.; Goldstein, D.; Xu, W.; Meyer, F.; Liu, G. Validation of genetic sequence variants as prognostic factors in early-stage head and neck squamous cell cancer survival. Clin. Cancer Res. 2012, 18, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Tommiska, J.; Eerola, H.; Heinonen, M.; Salonen, L.; Kaare, M.; Tallila, J.; Ristimäki, A.; von Smitten, K.; Aittomäki, K.; Heikkilä, P.; Blomqvist, C.; Nevanlinna, H. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin. Cancer Res. 2005, 11, 5098–5103. [Google Scholar] [CrossRef]
- Siddique, M.; Sabapathy, K. Trp53-dependent DNA-repair is affected by the codon 72 polymorphism. Oncogene 2006, 25, 3489–3500. [Google Scholar] [CrossRef]
- Vivenza, D.; Monteverde, M.; Lattanzio, L.; Tonissi, F.; Astesana, V.; Denaro, N.; Comino, A.; Russi, E.; Lo Nigro, C.; Merlano, M. Correlation of TP53 and MDM2 genotypes and clinical outcome in platinum-treated head and neck cancer patients with more than 10 years’ follow-up. Int. J. Biol. Markers 2016, 31, e183–e192. [Google Scholar] [CrossRef]
- Heikkinen, K.; Rapakko, K.; Karppinen, S.M.; Erkko, H.; Nieminen, P.; Winqvist, R. Association of common ATM polymorphism with bilateral breast cancer. Int. J. Cancer 2005, 116, 69–72. [Google Scholar] [CrossRef]
- Andreassen, C.N.; Rosenstein, B.S.; Kerns, S.L.; Ostrer, H.; De Ruysscher, D.; Cesaretti, J.A.; Barnett, G.C.; Dunning, A.M.; Dorling, L.; West, C.M.L.; Burnet, N.G.; Elliott, R.; Coles, C.; Hall, E.; Fachal, L.; Vega, A.; Gómez-Caamaño, A.; Talbot, C.J.; Symonds, R.P.; De Ruyck, K.; International Radiogenomics Consortium (RgC). Individual patient data meta-analysis shows a significant association between the ATM rs1801516 SNP and toxicity after radiotherapy in 5456 breast and prostate cancer patients. Radiother. Oncol. 2016, 121, 431–439. [Google Scholar] [CrossRef]
- Alsbeih, G.; El-Sebaie, M.; Al-Rajhi, N.; Al-Harbi, N.; Al-Hadyan, K.; Al-Qahtani, S.; Alsubael, M.; Al-Shabanah, M.; Moftah, B. Among 45 variants in 11 genes, HDM2 promoter polymorphisms emerge as new candidate biomarker associated with radiation toxicity. 3 Biotech. 2014, 4, 137–148. [Google Scholar] [CrossRef]
- Kweekel, D.M.; Antonini, N.F.; Nortier, J.W.; Punt, C.J.; Gelderblom, H.; Guchelaar, H.J. Explorative study to identify novel candidate genes related to oxaliplatin efficacy and toxicity using a DNA repair array. Br. J. Cancer 2009, 101, 357–362. [Google Scholar] [CrossRef]
- Ouimet, M.; Cassart, P.; Larivière, M.; Kritikou, E.A.; Simard, J.; Sinnett, D. Functional analysis of promoter variants in KU70 and their role in cancer susceptibility. Genes Chromosomes Cancer 2012, 51, 1007–1013. [Google Scholar] [CrossRef]
- Wang, W.; Pan, X.; Huo, X.; Yan, F.; Wang, M.; Wang, D.; Gao, Y.; Cao, Q.; Luo, D.; Qin, C.; Yin, C.; Zhang, Z. A functional polymorphism C-1310G in the promoter region of Ku70/XRCC6 is associated with risk of renal cell carcinoma. Mol. Carcinog. 2012, 51 Suppl 1, E183–E190. [Google Scholar] [CrossRef]
- Jia, J.; Ren, J.; Yan, D.; Xiao, L.; Sun, R. Association between the XRCC6 polymorphisms and cancer risks: a systematic review and meta-analysis. Medicine (Baltimore) 2015, 94, e283. [Google Scholar] [CrossRef] [PubMed]
- Guberina, M.; Sak, A.; Pöttgen, C.; Tinhofer-Keilholz, I.; Budach, V.; Balermpas, P.; Von der Grün, J.; Rödel, C.M.; Gkika, E.; Grosu, A.L.; Abdollahi, A.; Debus, J.; Belka, C.; Pigorsch, S.; Combs, S. E.; Mönnich, D.; Zips, D.; De-Colle, C.; Welz, S.; Linge, A. ERCC2 gene single-nucleotide polymorphism as a prognostic factor for locally advanced head and neck carcinomas after definitive cisplatin-based radiochemotherapy. Pharmacogenomics J. 2021, 21, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Zhong, B.Y.; Xu, H.H.; Qiao, S.Y.; Wang, G.; Huang, J.; Fan, H.Z.; Zhao, H.C. Associations between NBS1 Polymorphisms and Colorectal Cancer in Chinese Population. PloS One 2015, 10, e0132332. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, Y.; Cheng, M.; Huang, D.; Zheng, J.; Liu, B.; Ling, X.; Li, Q.; Zhang, X.; Ji, W.; Zhou, Y.; Lu, J. A functional polymorphism at microRNA-629-binding site in the 3’-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis 2012, 33, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liao, J.; Zhao, H.; Chen, F.; Zhu, X.; Li, J.; Nong, Q. NBS1 rs2735383 polymorphism is associated with an increased risk of laryngeal carcinoma. BMC Cancer 2018, 18, 175. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ma, N.; Li, M.; Tian, Q.B.; Liu, D.W. Functional variants in NBS1 and cancer risk: evidence from a meta-analysis of 60 publications with 111 individual studies. Mutagenesis 2013, 28, 683–697. [Google Scholar] [CrossRef]
- Zhu, L.; Sturgis, E.M.; Zhang, H.; Lu, Z.; Tao, Y.; Wei, Q.; Li, G. Genetic variants in microRNA-binding sites of DNA repair genes as predictors of recurrence in patients with squamous cell carcinoma of the oropharynx. Int. J. Cancer 2017, 141, 1355–1364. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, H.; Pu, F. Association between a functional variant in RAD51 gene’s 3’ untranslated region and its mRNA expression in lymphoblastoid cell lines. Springerplus 2016, 5, 1688. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Liu, Y.; Lin, Q.; Jiang, Y.; Zhou, Z.; Wen, Q.; Liang, X.; Zhou, X.; Yu, H. A functional variant in the RAD51 3’ UTR is associated with survival of hepatocellular carcinoma patients. Gene 2013, 851, 146964. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, F.; Dahlstrom, K.R.; Li, G.; Sturgis, E.M.; Zevallos, J.P.; Wei, Q.; Liu, Z. A variant at a potentially functional microRNA-binding site in BRIP1 was associated with risk of squamous cell carcinoma of the head and neck. Tumour Biol. 2016, 37, 8057–8066. [Google Scholar] [CrossRef] [PubMed]
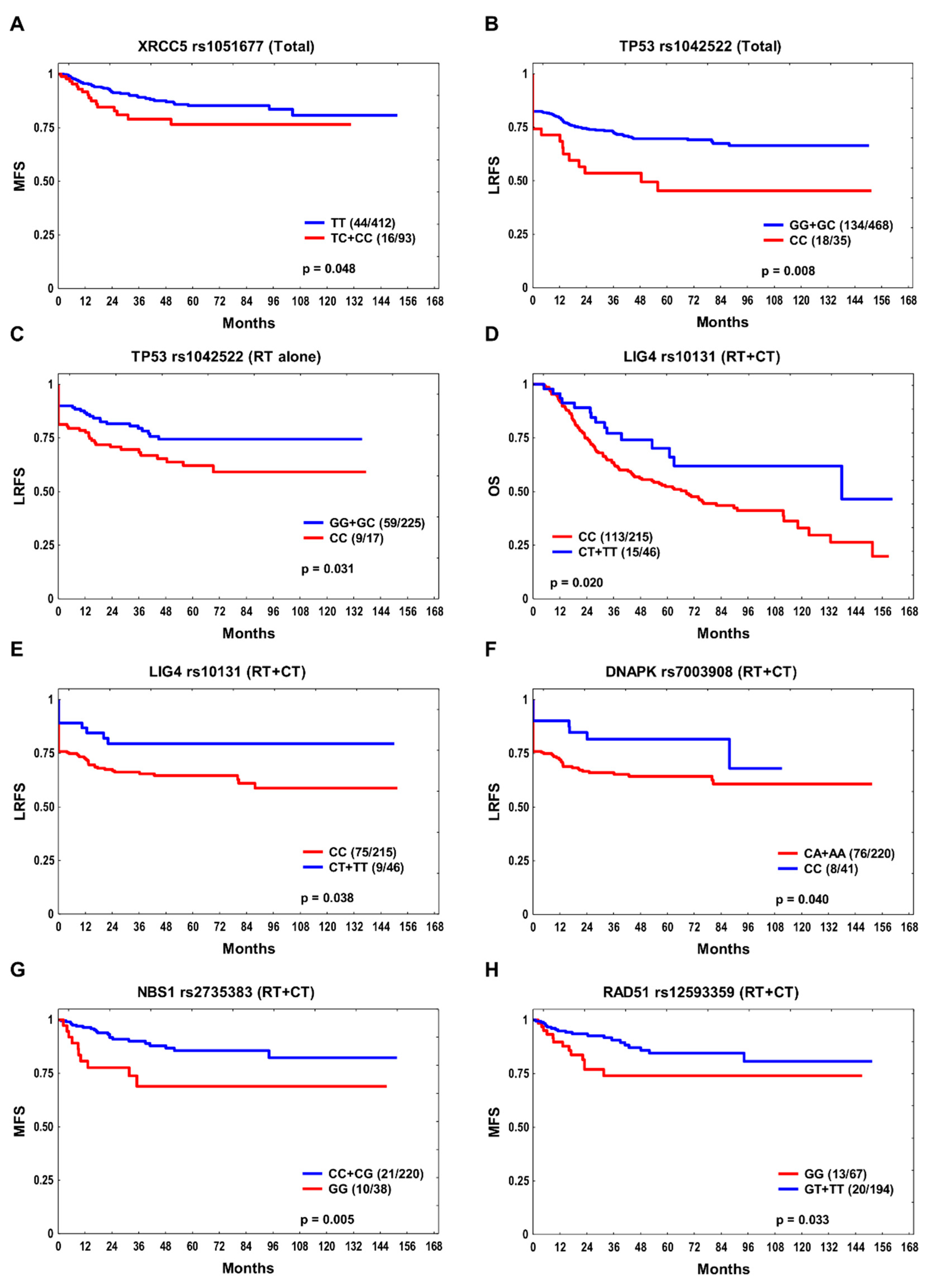
| Parameter | Total n = 505 |
RT+CT n = 261 |
RT alone n = 244 |
|---|---|---|---|
| Age at diagnosis (median) < 59 years ≥ 59 years |
235 (47%) 270 (53%) |
153 (59%) 108 (41%) |
82 (34%) 162 (66%) |
| Sex Male Female |
398 (79%) 107 (21%) |
53 (20%) 208 (80%) |
54 (22%) 190 (78%) |
| Tumor site Oropharynx Hypopharynx Larynx |
212 (42%) 63 (12%) 230 (46%) |
147 (56%) 47 (18%) 67 (26%) |
65 (27%) 16 (6%) 163 (67%) |
| T stage 1–2 3–4 |
252 (50%) 253 (50%) |
81 (31%) 180 (69%) |
171 (70%) 73 (30%) |
| N stage 0 1–3 |
207 (41%) 298 (59%) |
40 (15%) 221 (85%) |
167 (68%) 77 (32%) |
| Smoking status Never Ever |
112 (22%) 393 (78%) |
61 (23%) 200 (77%) |
51 (21%) 193 (79%) |
| Alcohol consumption a Never Ever |
124 (25%) 378 (75%) |
65 (25%) 194 (75%) |
59 (24%) 184 (76%) |
| Gene | SNP | Genotype | Total | RT+CT | RT alone | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Events/n | HR (95% CI) | p | Events/n | HR (95% CI) | p | Events/n | HR (95% CI) | p | |||
| OS | |||||||||||
| LIG4 | rs1805388 | GG | 165/322 | 1.35 (1.03–1.77) | 0.028 | - | - | - | - | - | - |
| MRE11A | rs2155209 | TT | 131/245 | 1.36 (1.05–1.75) | 0.019 | 72/134 | 1.54 (1.06–2.23) | 0.024 | - | - | - |
| XRCC5 | rs828907 | GT+TT | 182/354 | 1.41 (1.05–1.88) | 0.022 | 98/189 | 1.76 (1.13–2.72) | 0.012 | - | - | - |
| RAD51 | rs1801321 | GG | 105/203 | 1.37 (1.06–1.77) | 0.016 | 55/109 | 1.58 (1.08–2.30) | 0.018 | - | - | - |
| RAD51 | rs12593359 | GG | - | - | - | 33/67 | 1.56 (1.02–2.38) | 0.041 | - | - | - |
| CHEK1 | rs558351 | TT | - | - | - | - | - | - | 38/67 | 2.54 (1.66–3.90) | 2x10-5 |
| LRFS | |||||||||||
| TP53 | rs1042522 | CC | 18/35 | 1.89 (1.14–3.12) | 0.013 | - | - | - | 9/17 | 2.16 (1.01–4.62) | 0.047 |
| ATM | rs1801516 | GA+AA | 47/133 | 1.48 (1.04–2.12) | 0.029 | - | - | - | - | - | - |
| DNA-PKcs | rs7003908 | CA+AA | - | - | - | 76/220 | 2.14 (1.02–4.50) | 0.045 | - | - | - |
| MFS | |||||||||||
| ATM | rs189037 | GA+AA | 51/396 | 2.14 (1.00–4.57) | 0.049 | - | - | - | - | - | - |
| NBS1 | rs1805787 | CC | 38/281 | 1.81 (1.04–3.16) | 0.036 | - | - | - | - | - | - |
| XRCC6 | rs2267437 | CC | 22/128 | 1.89 (1.09–3.26) | 0.023 | 13/68 | 2.44 (1.14–5.26) | 0.022 | - | - | - |
| NBS1 | rs1805794 | CG+GG | 41/306 | 2.00 (1.12–3.58) | 0.020 | - | - | - | - | - | - |
| NBS1 | rs1805794 | GG | - | - | - | 7/32 | 3.12 (1.22–7.95) | 0.017 | - | - | - |
| NBS1 | rs2735383 | GG | - | - | - | 10/38 | 3.22 (1.42–7.32) | 0.005 | - | - | - |
| RAD51 | rs12593359 | GG | - | - | - | 13/67 | 2.88 (1.39–5.96) | 0.004 | - | - | - |
| Endpoint | Variables | HR (95% CI) | p |
|---|---|---|---|
| Total | |||
| OS | N > 0 Local recurrence Regional recurrence Metastasis/SPC MRE11A rs2155209 TT XRCC5 rs828907 GT+TT LIG4 rs1805388 GG RAD51 rs1801321 GG |
1.34 (1.02–1.76) 4.43 (3.34–5.89) 1.85 (1.32–2.58) 1.97 (1.50–2.59) 1.29 (1.00–1.65) 1.36 (1.02–1.81) 1.33 (1.01–1.74) 1.32 (1.02–1.70) |
0.038 <1x10-6 0.0003 1x10-6 0.048 0.038 0.040 0.037 |
| LRFS | T3–4 N > 0 Non-OPSCC TP53 rs1042522 CC ATM rs1801516 GA+AA |
1.78 (1.25–2.54) 1.65 (1.12–2.41) 1.71 (1.20–2.44) 1.90 (1.16–3.12) 1.47 (1.04–2.09) |
0.001 0.011 0.003 0.011 0.030 |
| MFS | HPSCC Regional recurrence XRCC6 rs2267437 CC |
3.06 (1.64–5.70) 5.14 (2.94–9.02) 1.78 (1.05–3.03) |
0.0004 <1x10-6 0.032 |
| RT+CT | |||
| OS | Alcohol: ever Local recurrence Regional recurrence Metastasis/SPC MRE11A rs2155209 TT XRCC5 rs828907 GT+TT RAD51 rs1801321 GG |
2.12 (1.31–3.43) 5.36 (3.51–8.20) 1.85 (1.22–2.81) 2.37 (1.59–3.52) 1.51 (1.05–2.18) 1.67 (1.08–2.56) 1.49 (1.03–2.16) |
0.002 <1x10-6 0.004 2x10-5 0.026 0.020 0.034 |
| MFS | Non-OPSCC Regional recurrence NBS1 rs2735383 GG RAD51 rs12593359 GG |
2.13 (1.03–4.42) 5.43 (2.51–11.75) 2.74 (1.28–5.87) 2.31 (1.10–4.86) |
0.042 2x10-5 0.010 0.027 |
| RT alone | |||
| OS | N > 0 Local recurrence Metastasis/SPC CHEK1 rs558351 TT |
2.27 (1.55–3.33) 4.27 (2.88–6.31) 2.45 (1.66–3.60) 2.47 (1.63–3.77) |
3x10-5 <1x10-6 6x10-6 2x10-5 |
| LRFS | T3–4 N > 0 Non-OPSCC TP53 rs1042522 CC |
3.12 (1.87–5.21) 1.90 (1.11–3.26) 2.01 (1.09–3.68) 2.15 (1.05–4.41) |
1x10-5 0.020 0.025 0.036 |
| MFS | Regional recurrence | 6.54 (2.81–15.24) | 1x10-5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




