Submitted:
26 September 2023
Posted:
29 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
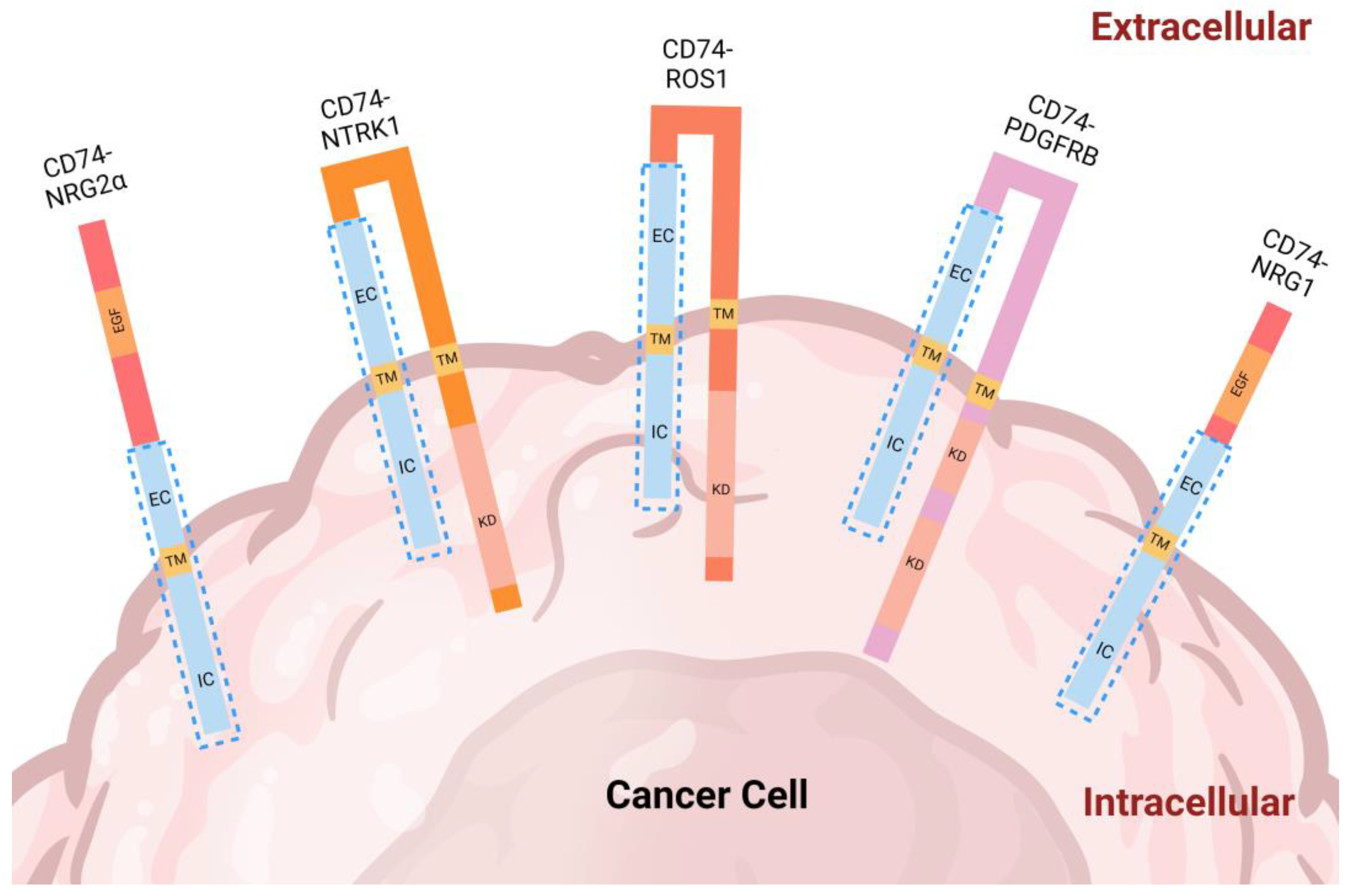
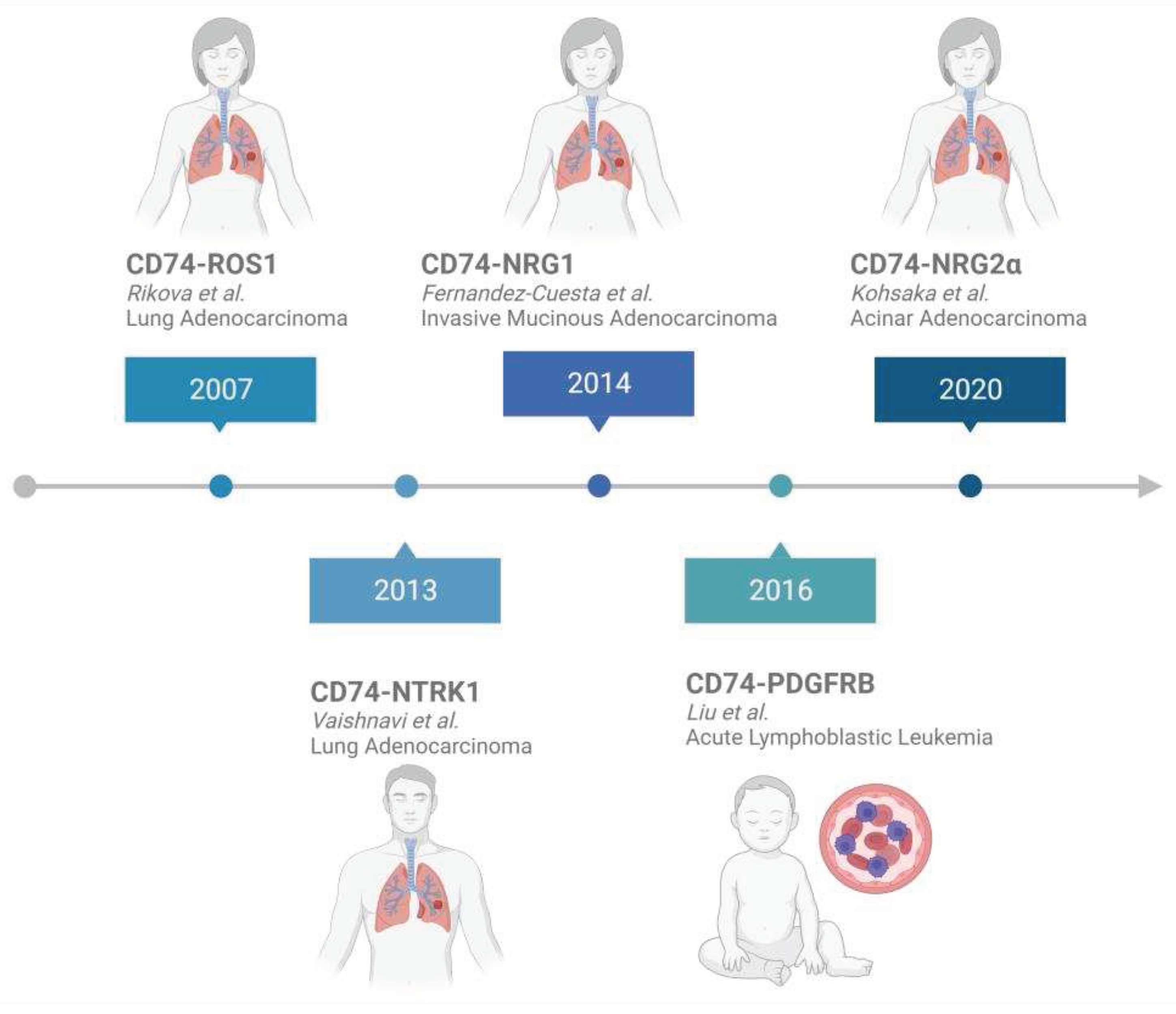
2. CD74 Fusion Proteins in Cancer
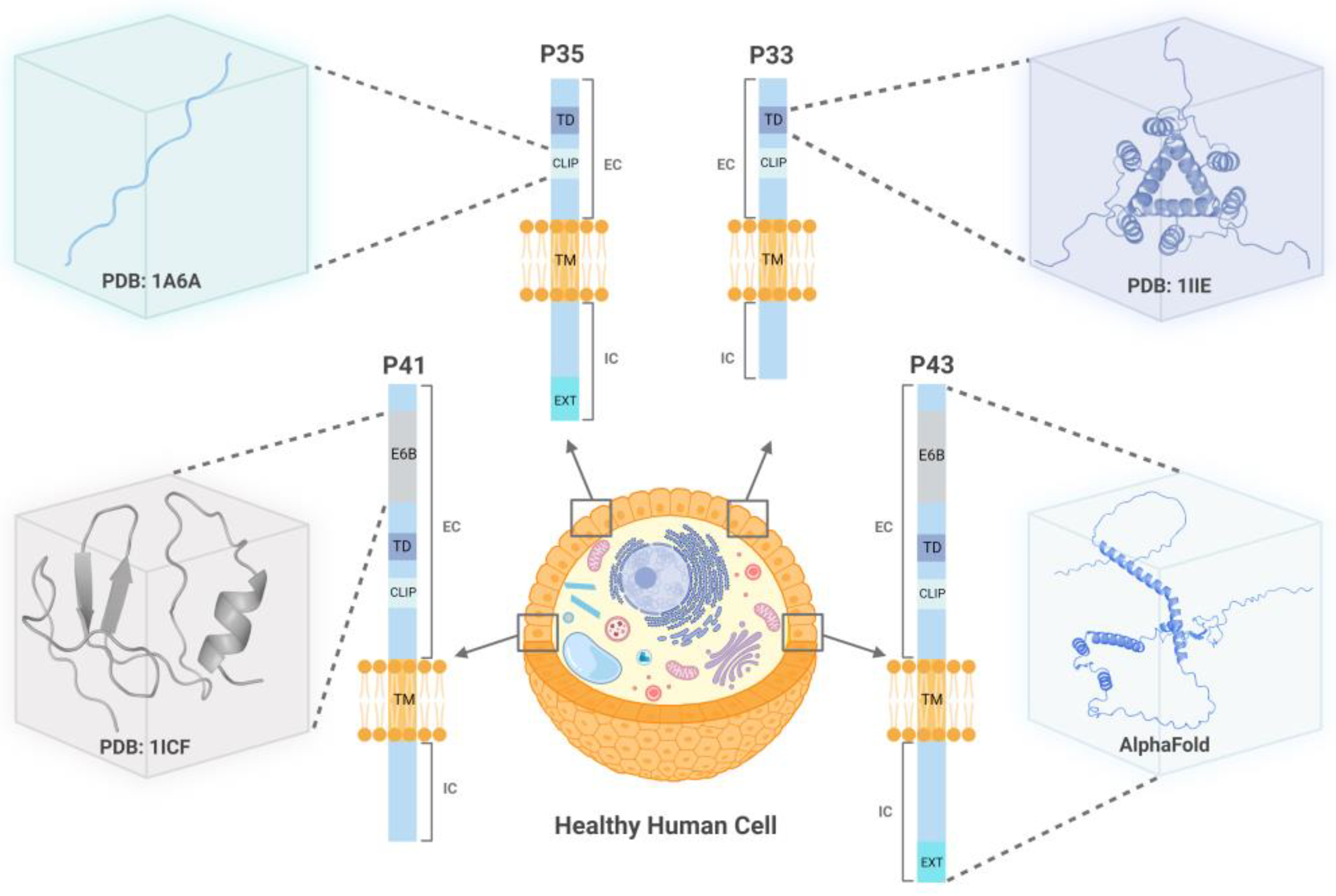
2.1. Analysis of CD74 Fusion Partners
2.2. CD74-ROS1 Expression
2.3. CD74-NTRK1 Expression
2.4. CD74-NRG1 Expression
2.5. CD74-PDGFRB Expression
2.6. CD74-NRG2a Expression
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Mendíbil, I.O.; Vizmanos, J.L.; Novo, F.J. Signatures of Selection in Fusion Transcripts Resulting from Chromosomal Translocations in Human Cancer. PLOS ONE 2009, 4, e4805. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Wang, Q.; Torres-Garcia, W.; Zheng, S.; Vegesna, R.; Kim, H.; Verhaak, R.G.W. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2014, 34, 4845–4854. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.C.; Zhang, W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin. J. Cancer 2013, 32, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis Oncol 2018. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Capitán, A.; et al. Detecting ALK, ROS1 and RET fusions and the METΔex14 splicing variant in liquid biopsies of non-small cell lung cancer patients using RNA-based techniques. Mol Oncol 2023. [Google Scholar] [CrossRef]
- Jonna, S.; Feldman, R.A.; Swensen, J.; Gatalica, Z.; Korn, W.M.; Borghaei, H.; Ma, P.C.; Nieva, J.J.; Spira, A.I.; Vanderwalde, A.M.; et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin. Cancer Res. 2019, 25, 4966–4972. [Google Scholar] [CrossRef]
- Leng, L.; Metz, C.N.; Fang, Y.; Xu, J.; Donnelly, S.; Baugh, J.; Delohery, T.; Chen, Y.; Mitchell, R.A.; Bucala, R. MIF Signal Transduction Initiated by Binding to CD74. J. Exp. Med. 2003, 197, 1467–1476. [Google Scholar] [CrossRef]
- Merk, M.; Zierow, S.; Leng, L.; Das, R.; Du, X.; Schulte, W.; Fan, J.; Lue, H.; Chen, Y.; Xiong, H.; et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF). Proc. Natl. Acad. Sci. USA 2011, 108, E577–E585. [Google Scholar] [CrossRef]
- Shi, X.; Leng, L.; Wang, T.; Wang, W.; Du, X.; Li, J.; McDonald, C.; Chen, Z.; Murphy, J.W.; Lolis, E.; et al. CD44 Is the Signaling Component of the Macrophage Migration Inhibitory Factor-CD74 Receptor Complex. Immunity 2006, 25, 595–606. [Google Scholar] [CrossRef]
- Bernhagen, J.; Krohn, R.; Lue, H.; Gregory, J.L.; Zernecke, A.; Koenen, R.R.; Dewor, M.; Georgiev, I.; Schober, A.; Leng, L.; et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 2007, 13, 587–596. [Google Scholar] [CrossRef]
- Schwartz, V.; Lue, H.; Kraemer, S.; Korbiel, J.; Krohn, R.; Ohl, K.; Bucala, R.; Weber, C.; Bernhagen, J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009, 583, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Lue, H.; et al. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal 2006, 18, 688–703. [Google Scholar] [CrossRef] [PubMed]
- Gore, Y.; Starlets, D.; Maharshak, N.; Becker-Herman, S.; Kaneyuki, U.; Leng, L.; Bucala, R.; Shachar, I. Macrophage Migration Inhibitory Factor Induces B Cell Survival by Activation of a CD74-CD44 Receptor Complex. J. Biol. Chem. 2008, 283, 2784–2792. [Google Scholar] [CrossRef] [PubMed]
- Lue, H.; et al. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene 2007, 26, 5046–5059. [Google Scholar] [CrossRef]
- Lue, H.; Dewor, M.; Leng, L.; Bucala, R.; Bernhagen, J. Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell. Signal. 2011, 23, 135–144. [Google Scholar] [CrossRef]
- Starlets, D.; Gore, Y.; Binsky, I.; Haran, M.; Harpaz, N.; Shvidel, L.; Becker-Herman, S.; Berrebi, A.; Shachar, I. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood 2006, 107, 4807–4816. [Google Scholar] [CrossRef]
- Cheng, S.-P.; Liu, C.-L.; Chen, M.-J.; Chien, M.-N.; Leung, C.-H.; Lin, C.-H.; Hsu, Y.-C.; Lee, J.-J. CD74 expression and its therapeutic potential in thyroid carcinoma. Endocrine-Related Cancer 2015, 22, 179–190. [Google Scholar] [CrossRef]
- Woolbright, B.L.; et al. Role of MIF1/MIF2/CD74 interactions in bladder cancer. J Pathol 2023, 259, 46–55. [Google Scholar] [CrossRef]
- Thavayogarajah, T.; Sinitski, D.; El Bounkari, O.; Torres-Garcia, L.; Lewinsky, H.; Harjung, A.; Chen, H.-R.; Panse, J.; Vankann, L.; Shachar, I.; et al. CXCR4 and CD74 together enhance cell survival in response to macrophage migration-inhibitory factor in chronic lymphocytic leukemia. Exp. Hematol. 2022, 115, 30–43. [Google Scholar] [CrossRef]
- Burton, J.D.; Ely, S.; Reddy, P.K.; Stein, R.; Gold, D.V.; Cardillo, T.M.; Goldenberg, D.M. CD74 Is Expressed by Multiple Myeloma and Is a Promising Target for Therapy. Clin. Cancer Res. 2004, 10, 6606–6611. [Google Scholar] [CrossRef]
- Greenwood, C.; Metodieva, G.; Al-Janabi, K.; Lausen, B.; Alldridge, L.; Leng, L.; Bucala, R.; Fernandez, N.; Metodiev, M.V. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J. Proteom. 2011, 75, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Gold, D.V.; et al. Enhanced expression of CD74 in gastrointestinal cancers and benign tissues. Int J Clin Exp Pathol 2010, 4, 1–12. [Google Scholar] [PubMed]
- McClelland, M.; Zhao, L.; Carskadon, S.; Arenberg, D. Expression of CD74, the Receptor for Macrophage Migration Inhibitory Factor, in Non-Small Cell Lung Cancer. Am. J. Pathol. 2009, 174, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Young, A.N.; Amin, M.B.; Moreno, C.S.; Lim, S.D.; Cohen, C.; Petros, J.A.; Marshall, F.F.; Neish, A.S. Expression Profiling of Renal Epithelial Neoplasms: A Method for Tumor Classification and Discovery of Diagnostic Molecular Markers. Am. J. Pathol. 2001, 158, 1639–1651. [Google Scholar] [CrossRef]
- Meyer-Siegler, K.L.; Iczkowski, K.A.; Leng, L.; Bucala, R.; Vera, P.L. Inhibition of Macrophage Migration Inhibitory Factor or Its Receptor (CD74) Attenuates Growth and Invasion of DU-145 Prostate Cancer Cells. J. Immunol. 2006, 177, 8730–8739. [Google Scholar] [CrossRef]
- Koide, N.; Yamada, T.; Shibata, R.; Mori, T.; Fukuma, M.; Yamazaki, K.; Aiura, K.; Shimazu, M.; Hirohashi, S.; Nimura, Y.; et al. Establishment of Perineural Invasion Models and Analysis of Gene Expression Revealed an Invariant Chain (CD74) as a Possible Molecule Involved in Perineural Invasion in Pancreatic Cancer. Clin. Cancer Res. 2006, 12, 2419–2426. [Google Scholar] [CrossRef]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Decker, P.A.; Morlan, B.W.; Wu, W.; Ballman, K.V.; Giannini, C.; Sarkaria, J.N. Expression of CD74 in high grade gliomas: a potential role in temozolomide resistance. J. Neuro-Oncology 2010, 100, 177–186. [Google Scholar] [CrossRef]
- Hong, W.C.; Lee, D.E.; Kang, H.W.; Kim, M.J.; Kim, M.; Kim, J.H.; Fang, S.; Kim, H.J.; Park, J.S. CD74 Promotes a Pro-Inflammatory Tumor Microenvironment by Inducing S100A8 and S100A9 Secretion in Pancreatic Cancer. Int. J. Mol. Sci. 2023, 24, 12993. [Google Scholar] [CrossRef]
- Xu, S.; Li, X.; Tang, L.; Liu, Z.; Yang, K.; Cheng, Q. CD74 Correlated With Malignancies and Immune Microenvironment in Gliomas. Front. Mol. Biosci. 2021, 8, 706949. [Google Scholar] [CrossRef]
- Rikova, K.; Guo, A.; Zeng, Q.; Possemato, A.; Yu, J.; Haack, H.; Nardone, J.; Lee, K.; Reeves, C.; Li, Y.; et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell 2007, 131, 1190–1203. [Google Scholar] [CrossRef]
- Vaishnavi, A.; Capelletti, M.; Le, A.T.; Kako, S.; Butaney, M.; Ercan, D.; Mahale, S.; Davies, K.D.; Aisner, D.L.; Pilling, A.B.; et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat. Med. 2013, 19, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cuesta, L.; et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014, 4, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-F.; Wang, B.-Y.; Zhang, W.-N.; Huang, J.-Y.; Li, B.-S.; Zhang, M.; Jiang, L.; Li, J.-F.; Wang, M.-J.; Dai, Y.-J.; et al. Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. EBioMedicine 2016, 8, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, S.; Hayashi, T.; Nagano, M.; Ueno, T.; Kojima, S.; Kawazu, M.; Shiraishi, Y.; Kishikawa, S.; Suehara, Y.; Takahashi, F.; et al. Identification of Novel CD74-NRG2α Fusion From Comprehensive Profiling of Lung Adenocarcinoma in Japanese Never or Light Smokers. J. Thorac. Oncol. 2020, 15, 948–961. [Google Scholar] [CrossRef]
- Claesson, L.; Larhammar, D.; Rask, L.; A Peterson, P. cDNA clone for the human invariant gamma chain of class II histocompatibility antigens and its implications for the protein structure. Proc. Natl. Acad. Sci. 1983, 80, 7395–7399. [Google Scholar] [CrossRef]
- Strubin, M.; Mach, B.; Long, E. The complete sequence of the mRNA for the HLA-DR-associated invariant chain reveals a polypeptide with an unusual transmembrane polarity. EMBO J. 1984, 3, 869–872. [Google Scholar] [CrossRef]
- Strubin, M.; Berte, C.; Mach, B. Alternative splicing and alternative initiation of translation explain the four forms of the Ia antigen-associated invariant chain. EMBO J. 1986, 5, 3483–3488. [Google Scholar] [CrossRef]
- O'Sullivan, D.M.; Larhammar, D.; Wilson, M.C.; A Peterson, P.; Quaranta, V. Structure of the human Ia-associated invariant (gamma)-chain gene: identification of 5' sequences shared with major histocompatibility complex class II genes. Proc Natl Acad Sci USA 1986, 83, 4484–4488. [Google Scholar] [CrossRef]
- Koch, N.; Lauer, W.; Habicht, J.; Dobberstein, B. Primary structure of the gene for the murine Ia antigen-associated invariant chains (Ii). An alternatively spliced exon encodes a cysteine-rich domain highly homologous to a repetitive sequence of thyroglobulin. EMBO J. 1987, 6, 1677–1683. [Google Scholar] [CrossRef]
- Gedde-Dahl, M.; Freisewinkel, I.; Staschewski, M.; Schenck, K.; Koch, N.; Bakke, O. Exon 6 Is Essential for Invariant Chain Trimerization and Induction of Large Endosomal Structures. J. Biol. Chem. 1997, 272, 8281–8287. [Google Scholar] [CrossRef]
- Kuwana, T.; Peterson, P.A.; Karlsson, L. Exit of major histocompatibility complex class II–invariant chain p35 complexes from the endoplasmic reticulum is modulated by phosphorylation. Proceedings of the National Academy of Sciences 1998, 95, 1056–1061. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.S.; Blum, J.S.; Cresswell, P. Invariant chain trimers are sequestered in the rough endoplasmic reticulum in the absence of association with HLA class II antigens. J. Cell Biol. 1990, 111, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Jasanoff, A.; Wagner, G.; Wiley, D.C. Structure of a trimeric domain of the MHC class II-associated chaperonin and targeting protein Ii. EMBO J. 1998, 17, 6812–6818. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; et al. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature 1995, 378, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Rudensky, A.Y.; Corper, A.L.; Teyton, L.; Wilson, I.A. Crystal Structure Of MHC Class II I-Ab in Complex with a Human CLIP Peptide: Prediction of an I-Ab Peptide-binding Motif. J. Mol. Biol. 2003, 326, 1157–1174. [Google Scholar] [CrossRef]
- Günther, S.; et al. Bidirectional binding of invariant chain peptides to an MHC class II molecule. Proc Natl Acad Sci USA 2010, 107, 22219–22224. [Google Scholar] [CrossRef]
- Nguyen, T.-B.; Jayaraman, P.; Bergseng, E.; Madhusudhan, M.S.; Kim, C.-Y.; Sollid, L.M. Unraveling the structural basis for the unusually rich association of human leukocyte antigen DQ2.5 with class-II-associated invariant chain peptides. J. Biol. Chem. 2017, 292, 9218–9228. [Google Scholar] [CrossRef]
- Gunčar, G.; Pungerčič, G.; Klemenčič, I.; Turk, V.; Turk, D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999, 18, 793–803. [Google Scholar] [CrossRef]
- Chiva, C.; et al. Synthesis and NMR structure of p41icf, a potent inhibitor of human cathepsin L. J Am Chem Soc 2003, 125, 1508–1517. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ding, N.; Zhou, H.; Wang, S.; Tang, L.; Xiao, Z. A novel CD74-ROS1 gene fusion in a patient with inflammatory breast cancer: a case report. J. Med Case Rep. 2021, 15, 277. [Google Scholar] [CrossRef] [PubMed]
- Sadras, T.; Jalud, F.B.; Kosasih, H.J.; Horne, C.R.; Brown, L.M.; El-Kamand, S.; de Bock, C.E.; McAloney, L.; Ng, A.P.; Davidson, N.M.; et al. Unusual PDGFRB fusion reveals novel mechanism of kinase activation in Ph-like B-ALL. Leukemia 2023, 37, 905–909. [Google Scholar] [CrossRef]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.; Li, H.; Liu, Y.; Xu, J.; Huang, Z.; Yan, S.; Zhang, Q.; Cheng, Y. A Novel ROS1-FBXL17 Fusion Co-Existing with CD74-ROS1 Fusion May Improve Sensitivity to Crizotinib and Prolong Progression-Free Survival of Patients with Lung Adenocarcinoma. OncoTargets Ther. 2020, 13, 11499–11504. [Google Scholar] [CrossRef] [PubMed]
- Nakaoku, T.; et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014, 20, 3087–3093. [Google Scholar] [CrossRef]
- Cai, W.; Li, W.; Ren, S.; Zheng, L.; Li, X.; Zhou, C. Coexistence of Three Variants Involving Two Different Fusion Partners of ROS1 Including a Novel Variant of ROS1 Fusions in Lung Adenocarcinoma: A Case Report. J. Thorac. Oncol. 2014, 9, e43–e46. [Google Scholar] [CrossRef]
- Hashiguchi, M.H.; Sato, T.; Watanabe, R.; Kagyo, J.; Matsuzaki, T.; Domoto, H.; Kato, T.; Nakahara, Y.; Yokose, T.; Hiroshima, Y.; et al. A case of lung adenocarcinoma with a novel CD74-ROS1 fusion variant identified by comprehensive genomic profiling that responded to crizotinib and entrectinib. Thorac. Cancer 2021, 12, 2504–2507. [Google Scholar] [CrossRef]
- Sehgal, K.; et al. Cases of ROS1 -rearranged lung cancer: when to use crizotinib, entrectinib, lorlatinib, and beyond? Precision Cancer Medicine 2020, 3. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, B.; Wang, C.; Liao, C.; Wang, S.; Cao, R.; Ma, T.; Wang, K. Case report: A novel reciprocal ROS1-CD74 fusion in a NSCLC patient partially benefited from sequential tyrosine kinase inhibitors treatment. Front. Oncol. 2022, 12, 1021342. [Google Scholar] [CrossRef]
- Xia, H.; Xue, X.; Ding, H.; Ou, Q.; Wu, X.; Nagasaka, M.; Shao, Y.W.; Hu, X.; Ou, S.-H.I. Evidence of NTRK1 Fusion as Resistance Mechanism to EGFR TKI in EGFR+ NSCLC: Results from a Large-Scale Survey of NTRK1 Fusions in Chinese Patients with Lung Cancer. Clin. Lung Cancer 2019, 21, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, S.; Liu, Y.; Ma, L.; Liu, X.; Liu, Y.; Cheng, Y. Analysis of NTRK mutation and clinicopathologic factors in lung cancer patients in northeast China. Int. J. Biol. Markers 2020, 35, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Severson, E.; Achyut, B.R.; Nesline, M.; Pabla, S.; Previs, R.A.; Kannan, G.; Chenn, A.; Zhang, S.; Klein, R.; Conroy, J.; et al. RNA Sequencing Identifies Novel NRG1 Fusions in Solid Tumors that Lack Co-Occurring Oncogenic Drivers. J. Mol. Diagn. 2023, 25, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Somwar, R.; Mangatt, B.P.; Edgren, H.; Desmeules, P.; Ruusulehto, A.; Smith, R.S.; Delasos, L.; Vojnic, M.; Plodkowski, A.J.; et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov. 2018, 8, 686–695. [Google Scholar] [CrossRef]
- Birchmeier, C.; Sharma, S.; Wigler, M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc. Natl. Acad. Sci. 1987, 84, 9270–9274. [Google Scholar] [CrossRef]
- Shih, C.-H.; Chang, Y.-J.; Huang, W.-C.; Jang, T.-H.; Kung, H.-J.; Wang, W.-C.; Yang, M.-H.; Lin, M.-C.; Huang, S.-F.; Chou, S.-W.; et al. EZH2-mediated upregulation of ROS1 oncogene promotes oral cancer metastasis. Oncogene 2017, 36, 6542–6554. [Google Scholar] [CrossRef]
- Grenier, K.; Rivière, J.-B.; Bencheikh, B.O.A.; Corredor, A.L.G.; Shieh, B.C.; Wang, H.; Fiset, P.O.; Camilleri-Broët, S. Routine Clinically Detected Increased ROS1 Transcripts Are Related With ROS1 Expression by Immunohistochemistry and Associated With EGFR Mutations in Lung Adenocarcinoma. JTO Clin. Res. Rep. 2023, 4, 100530. [Google Scholar] [CrossRef]
- Lee, H.J.; Seol, H.S.; Kim, J.Y.; Chun, S.-M.; Suh, Y.-A.; Park, Y.-S.; Kim, S.-W.; Choi, C.-M.; Park, S.-I.; Kim, D.K.; et al. ROS1 Receptor Tyrosine Kinase, a Druggable Target, is Frequently Overexpressed in Non-Small Cell Lung Carcinomas Via Genetic and Epigenetic Mechanisms. Ann. Surg. Oncol. 2012, 20, 200–208. [Google Scholar] [CrossRef]
- Birchmeier, C.; O'Neill, K.; Riggs, M.; Wigler, M. Characterization of ROS1 cDNA from a human glioblastoma cell line. Proc Natl Acad Sci U S A 1990, 87, 4799–4803. [Google Scholar] [CrossRef]
- Acquaviva, J.; Wong, R.; Charest, A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim. et Biophys. Acta (BBA) - Rev. Cancer 2009, 1795, 37–52. [Google Scholar] [CrossRef]
- Drilon, A.; et al. ROS1-dependent cancers - biology, diagnostics and therapeutics. Nat Rev Clin Oncol 2021, 18, 35–55. [Google Scholar] [CrossRef]
- Esteban-Villarrubia, J.; Soto-Castillo, J.J.; Pozas, J.; Román-Gil, M.S.; Orejana-Martín, I.; Torres-Jiménez, J.; Carrato, A.; Alonso-Gordoa, T.; Molina-Cerrillo, J. Tyrosine Kinase Receptors in Oncology. Int. J. Mol. Sci. 2020, 21, 8529. [Google Scholar] [CrossRef]
- Shaw, A.T.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014, 371, 1963–1971. [Google Scholar] [CrossRef]
- Drilon, A.; Siena, S.; Dziadziuszko, R.; Barlesi, F.; Krebs, M.G.; Shaw, A.T.; de Braud, F.; Rolfo, C.; Ahn, M.-J.; Wolf, J.; et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2019, 21, 261–270. [Google Scholar] [CrossRef]
- Barbacid, M.; Lamballe, F.; Pulido, D.; Klein, R. The trk family of tyrosine protein kinase receptors. Biochim. et Biophys. Acta (BBA) - Rev. Cancer 1991, 1072, 115–127. [Google Scholar] [CrossRef]
- Barker, P.; Lomen-Hoerth, C.; Gensch, E.; Meakin, S.; Glass, D.; Shooter, E. Tissue-specific alternative splicing generates two isoforms of the trkA receptor. J. Biol. Chem. 1993, 268, 15150–15157. [Google Scholar] [CrossRef]
- Martin-Zanca, D.; et al. Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol 1989, 9, 24–33. [Google Scholar]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Kaplan, D.R.; Hempstead, B.L.; Martin-Zanca, D.; Chao, M.V.; Parada, L.F. The trk Proto-Oncogene Product: a Signal Transducing Receptor for Nerve Growth Factor. Science 1991, 252, 554–558. [Google Scholar] [CrossRef]
- Klein, R.; Jing, S.; Nanduri, V.; O'Rourke, E.; Barbacid, M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 1991, 65, 189–197. [Google Scholar] [CrossRef]
- Klesse, L.J.; Parada, L.F. Trks: signal transduction and intracellular pathways. Microsc Res Tech 1999, 45, 210–216. [Google Scholar] [CrossRef]
- Reichardt, L.F. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 2006, 361, 1545–1564. [Google Scholar] [CrossRef] [PubMed]
- Indo, Y.; Mardy, S.; Tsuruta, M.; Karim, M.A.; Matsuda, I. Structure and organization of the humanTRKA gene encoding a high affinity receptor for nerve growth factor. J. Hum. Genet. 1997, 42, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Reuther, G.W.; Lambert, Q.T.; Caligiuri, M.A.; Der, C.J. Identification and Characterization of an Activating TrkA Deletion Mutation in Acute Myeloid Leukemia. Mol. Cell. Biol. 2000, 20, 8655–8666. [Google Scholar] [CrossRef]
- Gao, F.; Griffin, N.; Faulkner, S.; Rowe, C.W.; Williams, L.; Roselli, S.; Thorne, R.F.; Ferdoushi, A.; Jobling, P.; Walker, M.M.; et al. The neurotrophic tyrosine kinase receptor TrkA and its ligand NGF are increased in squamous cell carcinomas of the lung. Sci. Rep. 2018, 8, 8135. [Google Scholar] [CrossRef]
- Faulkner, S.; Jobling, P.; Rowe, C.W.; Oliveira, S.R.; Roselli, S.; Thorne, R.F.; Oldmeadow, C.; Attia, J.; Jiang, C.C.; Zhang, X.D.; et al. Neurotrophin Receptors TrkA, p75NTR, and Sortilin Are Increased and Targetable in Thyroid Cancer. Am. J. Pathol. 2018, 188, 229–241. [Google Scholar] [CrossRef]
- Lagadec, C.; Meignan, S.; Adriaenssens, E.; Foveau, B.; Vanhecke, E.; Romon, R.; Toillon, R.-A.; Oxombre, B.; Hondermarck, H.; Le Bourhis, X. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene 2009, 28, 1960–1970. [Google Scholar] [CrossRef]
- Faulkner, S.; Griffin, N.; Rowe, C.W.; Jobling, P.; Lombard, J.M.; Oliveira, S.M.; Walker, M.M.; Hondermarck, H. Nerve growth factor and its receptor tyrosine kinase TrkA are overexpressed in cervical squamous cell carcinoma. FASEB BioAdvances 2020, 2, 398–408. [Google Scholar] [CrossRef]
- Hong, D.; Bauer, T.; Lee, J.; Dowlati, A.; Brose, M.; Farago, A.; Taylor, M.; Shaw, A.; Montez, S.; Meric-Bernstam, F.; et al. Larotrectinib in adult patients with solid tumours: a multi-centre, open-label, phase I dose-escalation study. Ann. Oncol. 2019, 30, 325–331. [Google Scholar] [CrossRef]
- Drilon, A.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Hong, D.S.; et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 2020, 21, 531–540. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Steinthorsdottir, V.; Stefansson, H.; Ghosh, S.; Birgisdottir, B.; Bjornsdottir, S.; Fasquel, A.C.; Olafsson, O.; Stefansson, K.; Gulcher, J.R. Multiple novel transcription initiation sites for NRG1. Gene 2004, 342, 97–105. [Google Scholar] [CrossRef]
- Falls, D.L. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res 2003, 284, 14–30. [Google Scholar] [CrossRef]
- Meyer, D.; Birchmeier, C. Multiple essential functions of neuregulin in development. Nature 1995, 378, 386–390. [Google Scholar] [CrossRef]
- Kramer, R.; Bucay, N.; Kane, D.J.; E Martin, L.; E Tarpley, J.; E Theill, L. Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development. Proceedings of the National Academy of Sciences 1996, 93, 4833–4838. [Google Scholar] [CrossRef]
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef]
- Holbro, T.; et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA 2003, 100, 8933–8938. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. NEW EMBO MEMBERS' REVIEW: The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef]
- Nagasaka, M.; Ou, S.-H.I. NRG1 and NRG2 fusion positive solid tumor malignancies: a paradigm of ligand-fusion oncogenesis. Trends Cancer 2022, 8, 242–258. [Google Scholar] [CrossRef]
- Busfield, S.J.; Michnick, D.A.; Chickering, T.W.; Revett, T.L.; Ma, J.; Woolf, E.A.; Comrack, C.A.; Dussault, B.J.; Woolf, J.; Goodearl, A.D.J.; et al. Characterization of a Neuregulin-Related Gene, Don-1, That Is Highly Expressed in Restricted Regions of the Cerebellum and Hippocampus. Mol. Cell. Biol. 1997, 17, 4007–4014. [Google Scholar] [CrossRef]
- Schoeberl, B.; et al. Systems biology driving drug development: From design to the clinical testing of the anti-ErbB3 antibody seribantumab (MM-121). NPJ Systems Biology and Applications 2017, 3. [Google Scholar] [CrossRef]
- Carrizosa, D.R.; Burkard, M.E.; Elamin, Y.Y.; Desai, J.; Gadgeel, S.M.; Lin, J.J.; Waqar, S.N.; Spigel, D.R.; Chae, Y.K.; Cheema, P.K.; et al. CRESTONE: Initial efficacy and safety of seribantumab in solid tumors harboring NRG1 fusions. J. Clin. Oncol. 2022, 40, 3006–3006. [Google Scholar] [CrossRef]
- Spigel, D.; Waqar, S.; Burkard, M.; Lin, J.; Chae, Y.; Socinski, M.; Gadgeel, S.; Reckamp, K.; Leland, S.; Plessinger, D.; et al. MO01.33 CRESTONE – Clinical Study of REsponse to Seribantumab in Tumors with NEuregulin-1 (NRG1) Fusions – A Phase 2 Study of the anti-HER3 mAb for Advanced or Metastatic Solid Tumors (NCT04383210). J. Thorac. Oncol. 2021, 16, S29–S30. [Google Scholar] [CrossRef]
- Schram, A.M.; Goto, K.; Kim, D.-W.; Martin-Romano, P.; Ou, S.-H.I.; O'Kane, G.M.; O'Reilly, E.M.; Umemoto, K.; Duruisseaux, M.; Neuzillet, C.; et al. Efficacy and safety of zenocutuzumab, a HER2 x HER3 bispecific antibody, across advanced NRG1 fusion (NRG1+) cancers. J. Clin. Oncol. 2022, 40, 105–105. [Google Scholar] [CrossRef]
- Arar, M.; et al. Platelet-derived growth factor receptor beta regulates migration and DNA synthesis in metanephric mesenchymal cells. J Biol Chem 2000, 275, 9527–9533. [Google Scholar] [CrossRef]
- Hellström, M.; Kalén, M.; Lindahl, P.; Abramsson, A.; Betsholtz, C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 1999, 126, 3047–3055. [Google Scholar] [CrossRef]
- Hoch, R.V.; Soriano, P. Roles of PDGF in animal development. Development 2003, 130, 4769–4784. [Google Scholar] [CrossRef]
- Appiah-Kubi, K.; Lan, T.; Wang, Y.; Qian, H.; Wu, M.; Yao, X.; Wu, Y.; Chen, Y. Platelet-derived growth factor receptors (PDGFRs) fusion genes involvement in hematological malignancies. Crit. Rev. Oncol. 2017, 109, 20–34. [Google Scholar] [CrossRef]
- Heidaran, M.; Pierce, J.; Jensen, R.; Matsui, T.; Aaronson, S. Chimeric alpha- and beta-platelet-derived growth factor (PDGF) receptors define three immunoglobulin-like domains of the alpha-PDGF receptor that determine PDGF-AA binding specificity. J. Biol. Chem. 1990, 265, 18741–18744. [Google Scholar] [CrossRef]
- Lubinus, M.; Meier, K.; Smith, E.; Gause, K.; LeRoy, E.; Trojanowska, M. Independent effects of platelet-derived growth factor isoforms on mitogen-activated protein kinase activation and mitogenesis in human dermal fibroblasts. J. Biol. Chem. 1994, 269, 9822–9825. [Google Scholar] [CrossRef]
- Tsao, A.S.; Wei, W.; Kuhn, E.; Spencer, L.; Solis, L.M.; Suraokar, M.; Lee, J.J.; Hong, W.K.; Wistuba, I.I. Immunohistochemical Overexpression of Platelet-Derived Growth Factor Receptor–Beta (PDGFR-β) is Associated With PDGFRB Gene Copy Number Gain in Sarcomatoid Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2011, 12, 369–374. [Google Scholar] [CrossRef]
- Kim, M.S.; Choi, H.S.; Wu, M.; Myung, J.; Kim, E.J.; Kim, Y.S.; Ro, S.; Ha, S.E.; Bartlett, A.; Wei, L.; et al. Potential Role of PDGFRβ-Associated THBS4 in Colorectal Cancer Development. Cancers 2020, 12, 2533. [Google Scholar] [CrossRef]
- Nazarian, R.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Cristofanilli, M.; et al. Imatinib mesylate (Gleevec) in advanced breast cancer-expressing C-Kit or PDGFR-beta: clinical activity and biological correlations. Ann Oncol 2008, 19, 1713–1719. [Google Scholar] [CrossRef]
- David, M.; Cross, N.C.P.; Burgstaller, S.; Chase, A.; Curtis, C.; Dang, R.; Gardembas, M.; Goldman, J.M.; Grand, F.; Hughes, G.; et al. Durable responses to imatinib in patients with PDGFRB fusion gene–positive and BCR-ABL–negative chronic myeloproliferative disorders. Blood 2006, 109, 61–64. [Google Scholar] [CrossRef]
- Apperley, J.F.; Gardembas, M.; Melo, J.V.; Russell-Jones, R.; Bain, B.J.; Baxter, E.J.; Chase, A.; Chessells, J.M.; Colombat, M.; Dearden, C.E.; et al. Response to Imatinib Mesylate in Patients with Chronic Myeloproliferative Diseases with Rearrangements of the Platelet-Derived Growth Factor Receptor Beta. New Engl. J. Med. 2002, 347, 481–487. [Google Scholar] [CrossRef]
- Wang, P.; Song, L.; Ge, H.; Jin, P.; Jiang, Y.; Hu, W.; Geng, N. Crenolanib, a PDGFR inhibitor, suppresses lung cancer cell proliferation and inhibits tumor growth in vivo. OncoTargets Ther. 2014, 7, 1761–1768. [Google Scholar] [CrossRef]
- Fujino, S.; Miyoshi, N.; Ito, A.; Yasui, M.; Ohue, M.; Ogino, T.; Takahashi, H.; Uemura, M.; Matsuda, C.; Mizushima, T.; et al. Crenolanib Regulates ERK and AKT/mTOR Signaling Pathways in RAS/BRAF-Mutated Colorectal Cancer Cells and Organoids. Mol. Cancer Res. 2021, 19, 812–822. [Google Scholar] [CrossRef]
- Jun, H.J.; et al. The oncogenic lung cancer fusion kinase CD74-ROS activates a novel invasiveness pathway through E-Syt1 phosphorylation. Cancer Res 2012, 72, 3764–3774. [Google Scholar] [CrossRef]
- Neel, D.S.; Allegakoen, D.V.; Olivas, V.; Mayekar, M.K.; Hemmati, G.; Chatterjee, N.; Blakely, C.M.; McCoach, C.E.; Rotow, J.K.; Le, A.; et al. Differential Subcellular Localization Regulates Oncogenic Signaling by ROS1 Kinase Fusion Proteins. Cancer Res 2019, 79, 546–556. [Google Scholar] [CrossRef]
- Cui, M.; et al. Molecular and clinicopathological characteristics of ROS1-rearranged non-small-cell lung cancers identified by next-generation sequencing. Mol Oncol 2020, 14, 2787–2795. [Google Scholar] [CrossRef]
- Li, N.; Chen, Z.; Huang, M.; Zhang, D.; Hu, M.; Jiao, F.; Quan, M. Detection of ROS1 gene fusions using next-generation sequencing for patients with malignancy in China. Front. Cell Dev. Biol. 2022, 10, 1035033. [Google Scholar] [CrossRef]
- Muminovic, M.; et al. Importance of ROS1 gene fusions in non-small cell lung cancer. 2023.
- Bergethon, K.; Shaw, A.T.; Ou, S.-H.I.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef]
- Cai, W.; Li, X.; Su, C.; Fan, L.; Zheng, L.; Fei, K.; Zhou, C.; Manegold, C.; Schmid-Bindert, G. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann. Oncol. 2013, 24, 1822–1827. [Google Scholar] [CrossRef]
- Lu, S.; Pan, H.; Wu, L.; Yao, Y.; He, J.; Wang, Y.; Wang, X.; Fang, Y.; Zhou, Z.; Wang, X.; et al. Efficacy, safety and pharmacokinetics of Unecritinib (TQ-B3101) for patients with ROS1 positive advanced non-small cell lung cancer: a Phase I/II Trial. Signal Transduct. Target. Ther. 2023, 8, 249. [Google Scholar] [CrossRef]
- Wang, V.; Bivona, T.; Ali, S.M.; Schrock, A.B.; Miller, V.A. CD74 - ROS1 Fusion in NSCLC Detected by Hybrid Capture–Based Tissue Genomic Profiling and ctDNA Assays. J. Thorac. Oncol. 2017, 12, e19–e20. [Google Scholar] [CrossRef]
- Meng, Z.-T.; Chen, P.; Zang, F.; Liu, Y.; Xu, X.; Su, Y.; Chen, J.; Lin, L.; Zhang, L.; Zhang, T. A patient with classic biphasic pulmonary blastoma harboring CD74–ROS1 fusion responds to crizotinib. OncoTargets Ther. 2017, 11, 157–161. [Google Scholar] [CrossRef]
- Mizuno, T.; Fujiwara, Y.; Yoshida, K.; Kohno, T.; Ohe, Y. Next-Generation Sequencer Analysis of Pulmonary Pleomorphic Carcinoma With a CD74-ROS1 Fusion Successfully Treated With Crizotinib. J. Thorac. Oncol. 2019, 14, e106–e108. [Google Scholar] [CrossRef]
- Wang, G.; Gao, J.; Lv, J.; Chen, X.; Wu, J.; Wang, R.; Jiang, J. Effective Treatment with Cabozantinib in an Advanced Non-Small-Cell Lung Cancer Patient Harboring a CD74-ROS1 Fusion: A Case Report. OncoTargets Ther. 2020, 13, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Huang, S.; Ye, X.; Feng, L.; Lu, Y.; Zhou, C.; Zhao, J.; He, T.; Wang, J.; Li, B. Crizotinib resistance conferred by BRAF V600E mutation in non–small cell lung cancer harboring an oncogenic ROS1 fusion. Cancer Treat. Res. Commun. 2021, 27, 100377. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Z.; Han, X.; Li, J.; Guo, H.; Shi, J. Acquired MET D1228N Mutations Mediate Crizotinib Resistance in Lung Adenocarcinoma with ROS1 Fusion: A Case Report. Oncol. 2020, 26, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yang, J.; Wang, D.; Yan, D. ROS1 fusion lung adenosquamous carcinoma patient with short-term clinical benefit after crizotinib treatment: a case report. Ann. Transl. Med. 2022, 10, 157. [Google Scholar] [CrossRef]
- Tyler, L.C.; Le, A.T.; Chen, N.; Nijmeh, H.; Bao, L.; Wilson, T.R.; Chen, D.; Simmons, B.; Turner, K.M.; Perusse, D.; et al. MET gene amplification is a mechanism of resistance to entrectinib in ROS1 + NSCLC. Thorac. Cancer 2022, 13, 3032–3041. [Google Scholar] [CrossRef]
- Tanaka, S.; et al. A case of CD74-ROS1-positive lung adenocarcinoma diagnosed by next-generation sequencing achieved long-term survival with pemetrexed regimens. Thorac Cancer 2023. [Google Scholar] [CrossRef]
- Pizzutilo, E.G.; Agostara, A.G.; Roazzi, L.; Romanò, R.; Motta, V.; Lauricella, C.; Marrapese, G.; Cerea, G.; Signorelli, D.; Veronese, S.M.; et al. Repotrectinib Overcomes F2004V Resistance Mutation in ROS1-Rearranged Non-Small Cell Lung Cancer: A Case Report. JTO Clin. Res. Rep. 2023, 100555. [Google Scholar] [CrossRef]
- Cha, Y.J.; Lee, C.; Joo, B.; A Kim, K.; Lee, C.-K.; Shim, H.S. Clinicopathological Characteristics of NRG1 Fusion-Positive Solid Tumors in Korean Patients. Cancer Res. Treat. 2023, 55, 1087–1095. [Google Scholar] [CrossRef]
- Chen, K.; Li, W.; Xi, X.; Zhong, J. A case of multiple primary lung adenocarcinoma with a CD74-NRG1 fusion protein and HER2 mutation benefit from combined target therapy. Thorac. Cancer 2022, 13, 3063–3067. [Google Scholar] [CrossRef]
- Abe, A.; Emi, N.; Tanimoto, M.; Terasaki, H.; Marunouchi, T.; Saito, H. Fusion of the platelet-derived growth factor receptor beta to a novel gene CEV14 in acute myelogenous leukemia after clonal evolution. Blood 1997, 90, 4271–4277. [Google Scholar] [CrossRef]
- Kim, H.-G.; Jang, J.-H.; Koh, E.-H. TRIP11-PDGFRB fusion in a patient with a therapy-related myeloid neoplasm with t(5;14)(q33;q32) after treatment for acute promyelocytic leukemia. Mol. Cytogenet. 2014, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Fayiga, F.F.; Reyes-Hadsall, S.C.; Moreno, B.A.; Oh, K.S.; Brathwaite, C.; Duarte, A.M. Novel ANKRD26 and PDGFRB gene mutations in pediatric case of non-Langerhans cell histiocytosis: Case report and literature review. J. Cutan. Pathol. 2023, 50, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Nagasaka, M.; Ou, S.I. Is NRG2α Fusion a "Doppelgänger" to NRG1α/β Fusions in Oncology? J Thorac Oncol 2020, 15, 878–880. [Google Scholar] [CrossRef] [PubMed]
| CD74 Fusion Protein | Exon Breakpoint | Cancer | Reference |
|---|---|---|---|
| CD74-ROS1 | C6-R34 C6-R32 C7-R32 C7-R34 |
NSCLC NSCLC NSCLC IBC |
[30] [54] [57] [52] |
| C3-R34 | NSCLC | [58] | |
| C6-R33 | NSCLC | [59] | |
| C6-R35 | NSCLC | [60] | |
| CD74-NTRK1 | C8-N12 C7-N8 C6-N12 |
NSCLC NSCLC NSCLC |
[31] [61] [62] |
| CD74-NRG1 | C6-N6 C8-N6 C6-N4 C7-N6 |
IMA IMA NSCLC IMA |
[32] [56] [63] [64] |
| CD74-PDGFRB | C6-P11 | B-ALL | [33] |
| CD74-NRG2a | C6-N2 | NSCLC | [34] |
| Diagnosis | Variant | Detection Method |
Age | Gender | Smoker/ Pack Year (PY) |
Stage | Treatment | TKIResistance | LifespanOS | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| NSCLC | C7-R32 C6-R34 |
RT-PCR | 61 | F | 0 | IV (T4N3M1b) |
1st: pemetrexed + cisplatin*refused crizotinib for economic reason | - | - | [57] |
| IBC | C7-R34 | NGS | 64 | F | - | cT4N3M1 | 1st: paclitaxel2nd: capecitabine*refused crizotinib for economic reason | - | Died 4 mos. after dx |
[52] |
| NSCLC | C3-R34 | NGS | 38 | F | 0 | IVA (T1aN3M1A) |
1st: cisplatin, pemetrexed, + bevacizumab2nd: docetaxel + ramucirumab*3rd: crizotinib *4th: entrectinib |
Crizotinib resistance: brain metastasis | 1 mo. PFS | [58] |
| Metastatic NSCLC | - | CGP (NGS) ctDNA assay |
41 | F | 0 | - | 1st: carboplatin, pemetrexed + bevacizumab *2nd: crizotinib |
- | PFS 4 mos. after crizotinib | [129] |
| CBPB | - | NGS | 44 | F | 0 | IV (cT3N1M1b) |
*1st: crizotinib | Crizotinib resistance: ROS1 G2032R mutation |
PFS for 3 mos. PD after crizotinib resistance |
[130] |
| PPC | - | NGSRT-PCR | 56 | F | 0 | IIIA (T2aN2M0) |
1st: resection 2nd: paclitaxel, carboplatin + bevacizumab *3rd: crizotinib |
- | CR after 6 mos. | [131] |
| NSCLC | C6-R34 | NGS | 40 | F | 0 | - | 1st: gemcitabine + cisplatin 2nd: docetaxel 3rd: gefitinib *4th: cabozantinib *5th: crizotinib |
Cabozantinib resistance | SD after 2 mos. crizotinib | [132] |
| NSCLC | C6-R33 | NGSRT-qPCR | 51 | F | - | IVB (pT4N3M1c) |
1st: pemetrexed + cisplatin *2nd: crizotinib |
- | PFS 15.7 mos. after crizotinib | [55] |
| MetastaticNSCLC | C6-R33 | NGS | 60 | F | 0 | - | 1st: radiotherapy*2nd: entrectinib | - | 5 mos. SD | [59] |
| NSCLC | C6-R33 | NGS | 53 | F | - | IV | *1st: crizotinib *2nd: dabrefenib |
Crizotinib resistance: BRAF V600E mutation | 2 mos. PR after crizotinib. Dead 15 days after dabrefenib | [133] |
| Metastatic NSCLC | - | NGS | 30 | F | - | IIIC (T3N3M0) | 1st: cisplatin + pemetrexed 2nd: nedaplatin + pemetrexed *3rd: crizotinib *4th: cabozantinib |
Crizotinib resistance: MET D1228N mutation | Dead 21 months from dx | [134] |
| NSCLC- ASC | C6-R34 | NGS | 43 | F | 0 | IIIA (pT2aN2M0) | 1st: albumin-bound paclitaxel + camrelizumab *2nd: crizotinib 3rd: pemetrexed + carboplatin + bevacizumab4th: cisplatin + gemcitabine + bevacizumab + *crizotinib |
- | Dead, ~15 mos. After dx | [135] |
| NSCLC | C6-R32 C6-R35 |
DNA NGS RNA NGS FISH Sanger Seq |
44 | F | 0 | IVB | *1st: Crizotinib *2nd: Lorlatinib 3rd: chemotherapy |
Crizotinib resistance: bone metastasis Lorlatinib resistance: ROS1 G2032R mutation |
Dead, ~19 mos. After dx | [60] |
| NSCLC | - | Molecular testing- not specified | 54 | M | 0 | IV | *1st: crizotinib *2nd: entrectinib 3rd: Carboplatin + Pemetrexed 4th: pemetrexed + pembrolizumab *5th: Repotrectinib *6th: Cabozantinib |
Possible entrectinib resistance: MET amplification | Deceased ~50 months after dx | [136] |
| NSCLC | - | NGS | 54 | F | 30 | IVA (T2aN3M1a) |
1st: Cisplatin + pemetrexed 2nd: Carboplatin + paclitaxel 3rd: Pemetrexed 4th: Nivolumab 5th: Docetaxel 6th: Pemetrexed 7th: S-1 8th: Gemcitabine *9th: Entrectinib |
- | 1st: PR 2nd: SD 3rd: PR 4th: SD 5th: PR 6th: PD 7th: PD 8th: PD 9th: PR |
[137] |
| NSCLC | - | NGS | 49 | F | 0 | IV | *1st: Entrectinib *2nd: Crizotinib 3rd: Carboplatin *4th: Repotrectinib |
Possible Entrectinib resistance: ROS1 F2004V mutation | 1st: PR 2nd: SD 3rd: PD 4th: PR |
[138] |
| Diagnosis | Variant | Detection Method |
Age | Gender | Smoker/ Pack Year (PY) |
Stage | Treatment | Lifespan OS |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| NSCLC | - | Genetic Testing | 62 | F | 0 | IV | 1st: pemetrexed + cisplatin 2nd: atezolizumab + albumin-bound paclitaxel + carboplatin *3rd: afatinib + pyrotinib |
PFS 5 mos. | [140] |
| IMA | C6-N6 | NGS | 86 | M | 0 | IIA (pT2bN0M0) |
1st: carboplatin + pemetrexed 2nd: paclitaxel + bevacizumab 3rd: nivolumab 4th: GSK2849330 (anti-ERBB3 mAb) *5th: afatinib |
PR 1 year and 7 mos. On GSK2849330 | [64] |
| IMA | C8-N6 | multiplex PCR | 81 | M | Former, smoked cigars for 1 year | IIB (pT3N0M0) |
*1st: afatinib | SD at 6 wks. PD at 13 wks. |
[64] |
| IMA | C7-N6 | NGS | 51 | M | <1 | IIIB(T4N2M0) | 1st: cisplatin + pemetrexed *2nd: afatinib |
PD 8 wks. After afatinib started | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





