Submitted:
02 October 2023
Posted:
02 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethical statement
2.2. Parasite strains
2.3. Assays design
2.4. Target selection and primers design
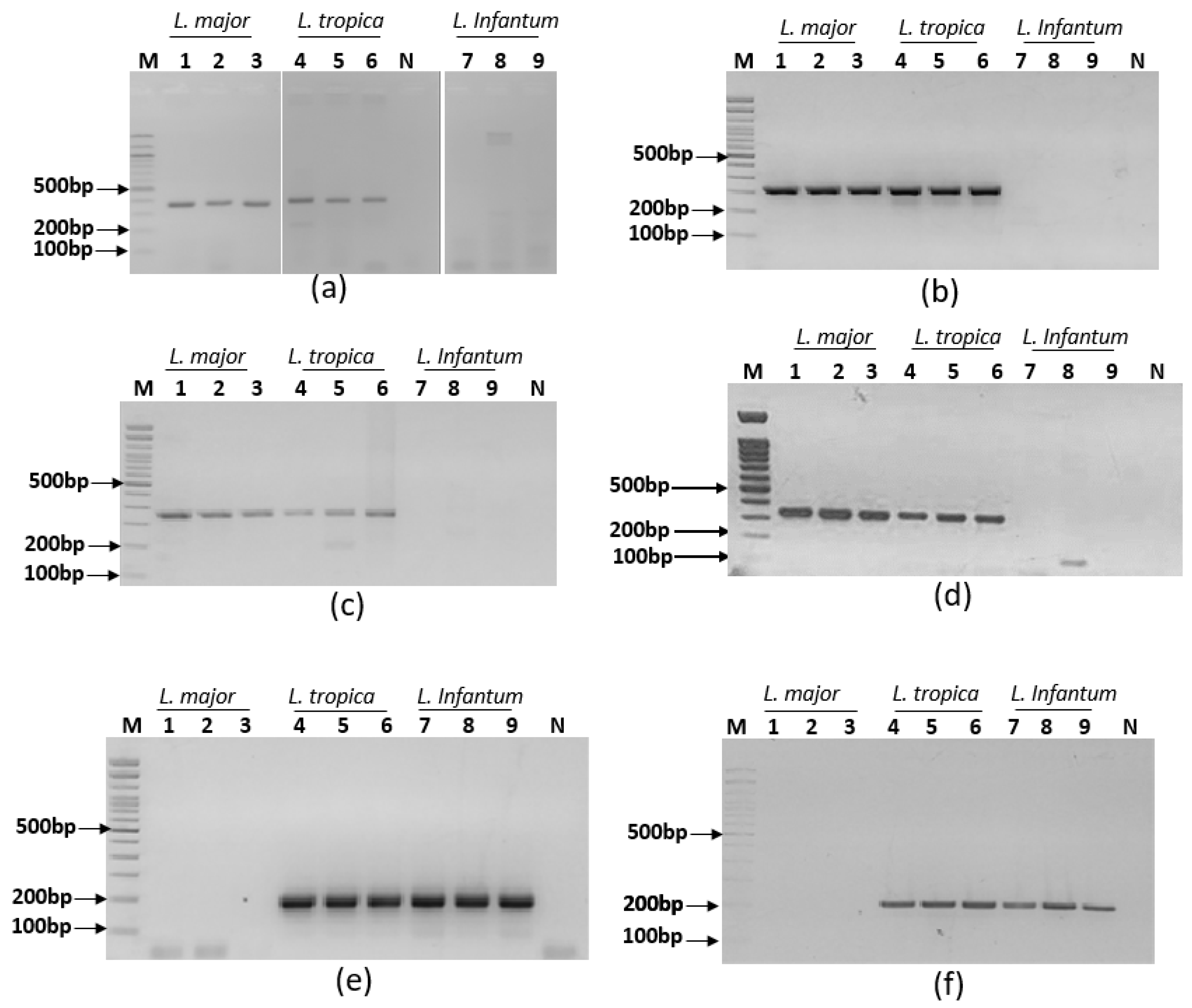
2.5. Ultra-fast simplex PCR assays for target screening
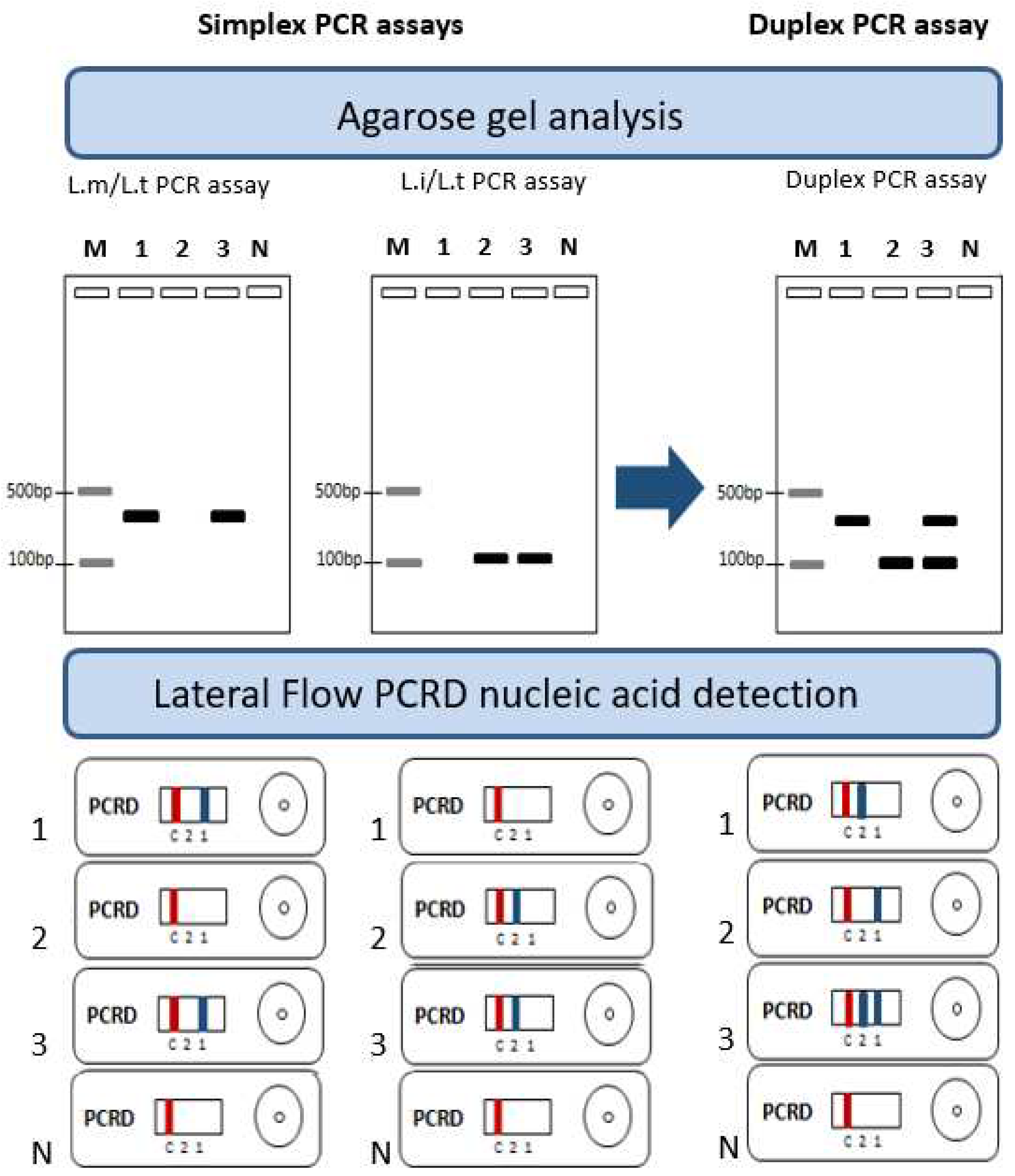
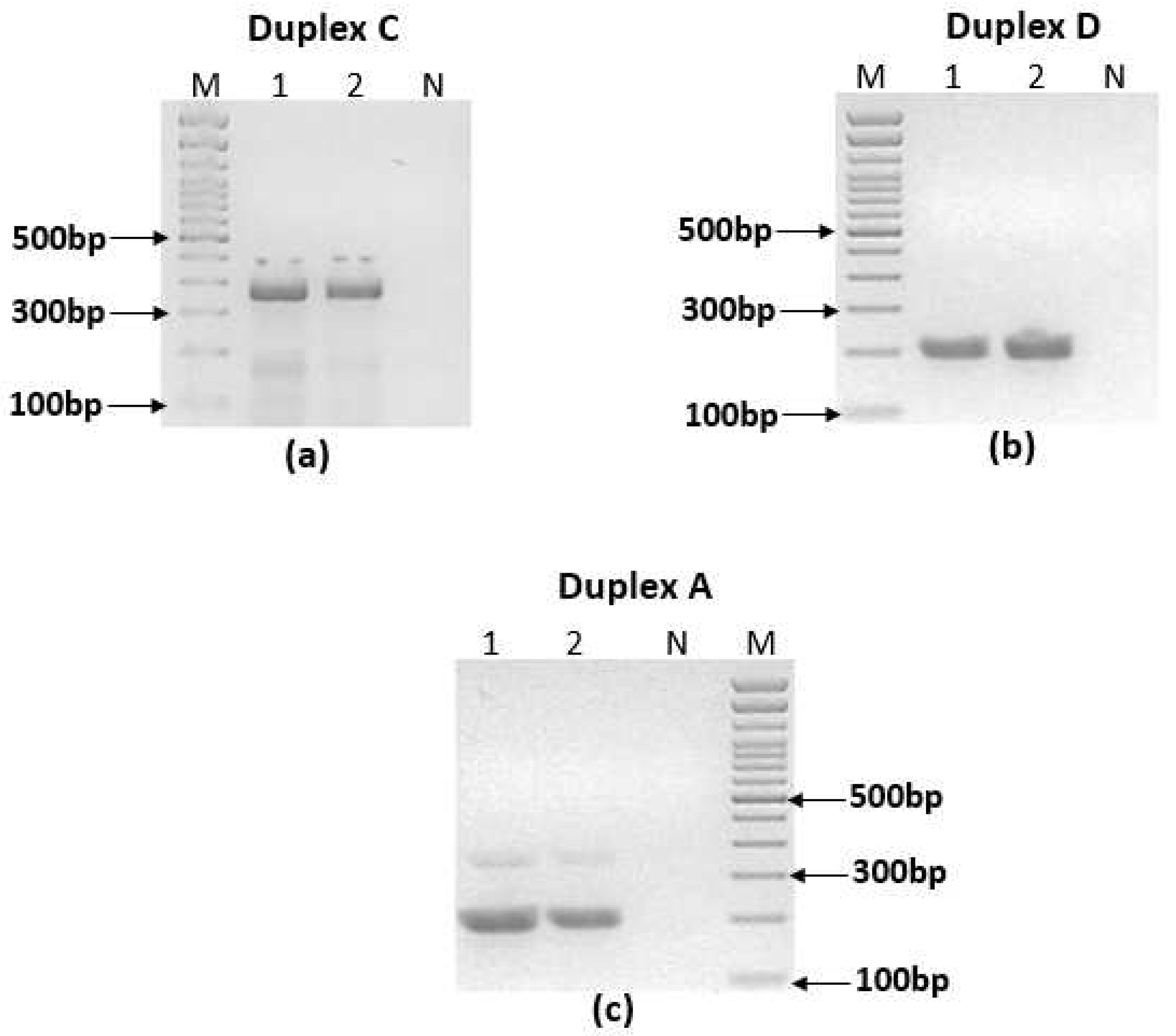
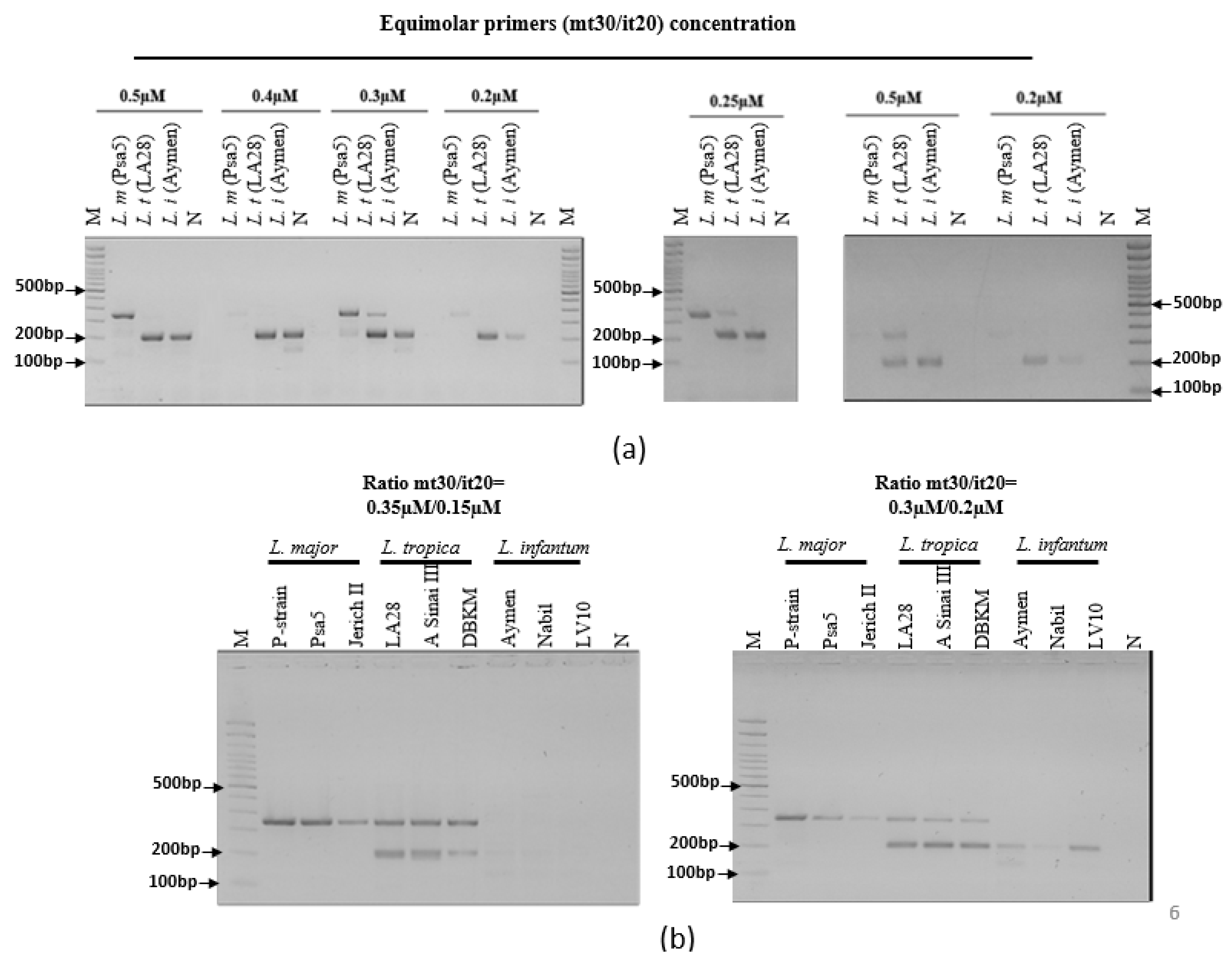
2.6. Ultra-Fast duplex PCR assays screening and optimization
2.7. Ultra- fast duplex PCR and LF reading
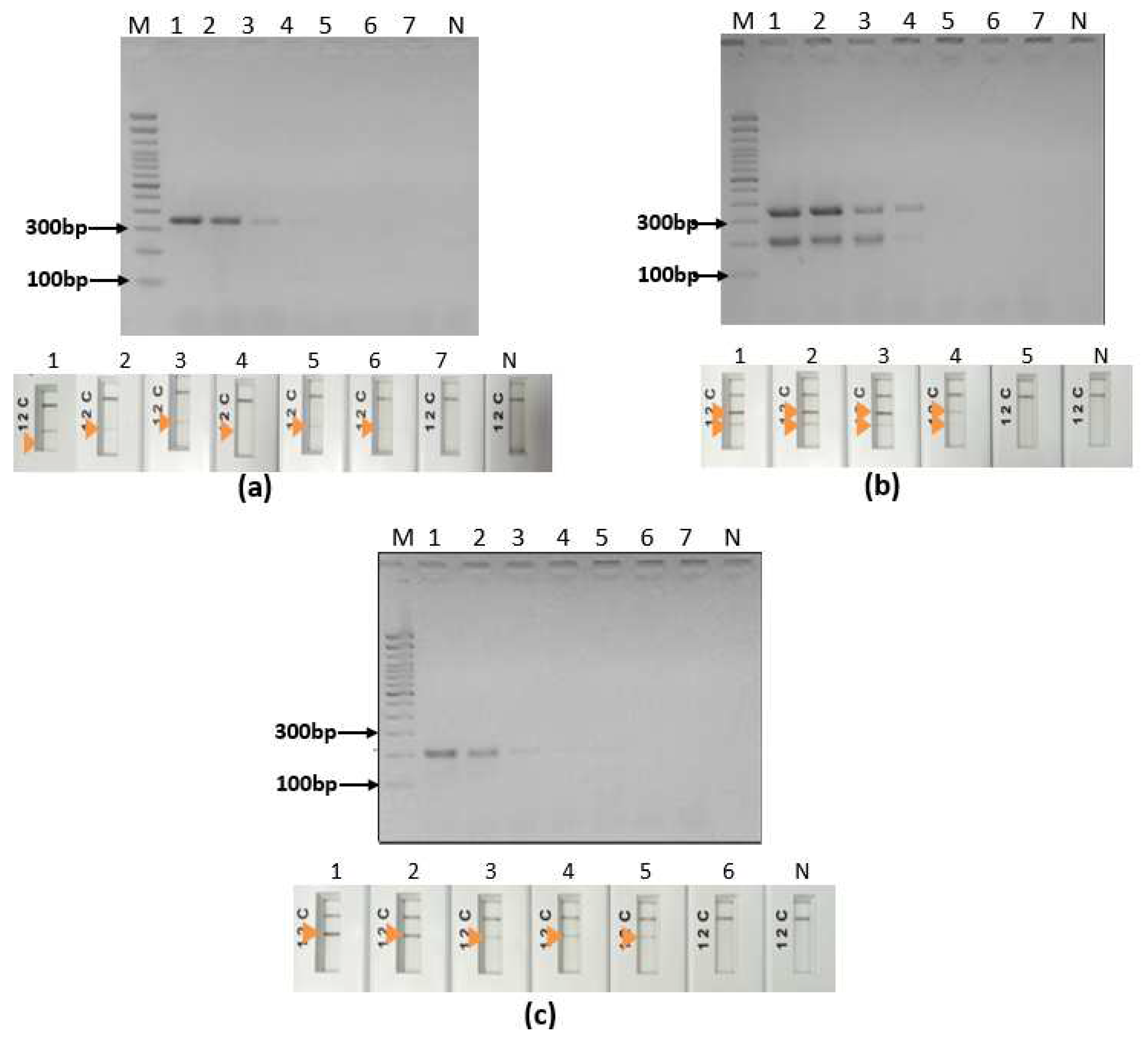
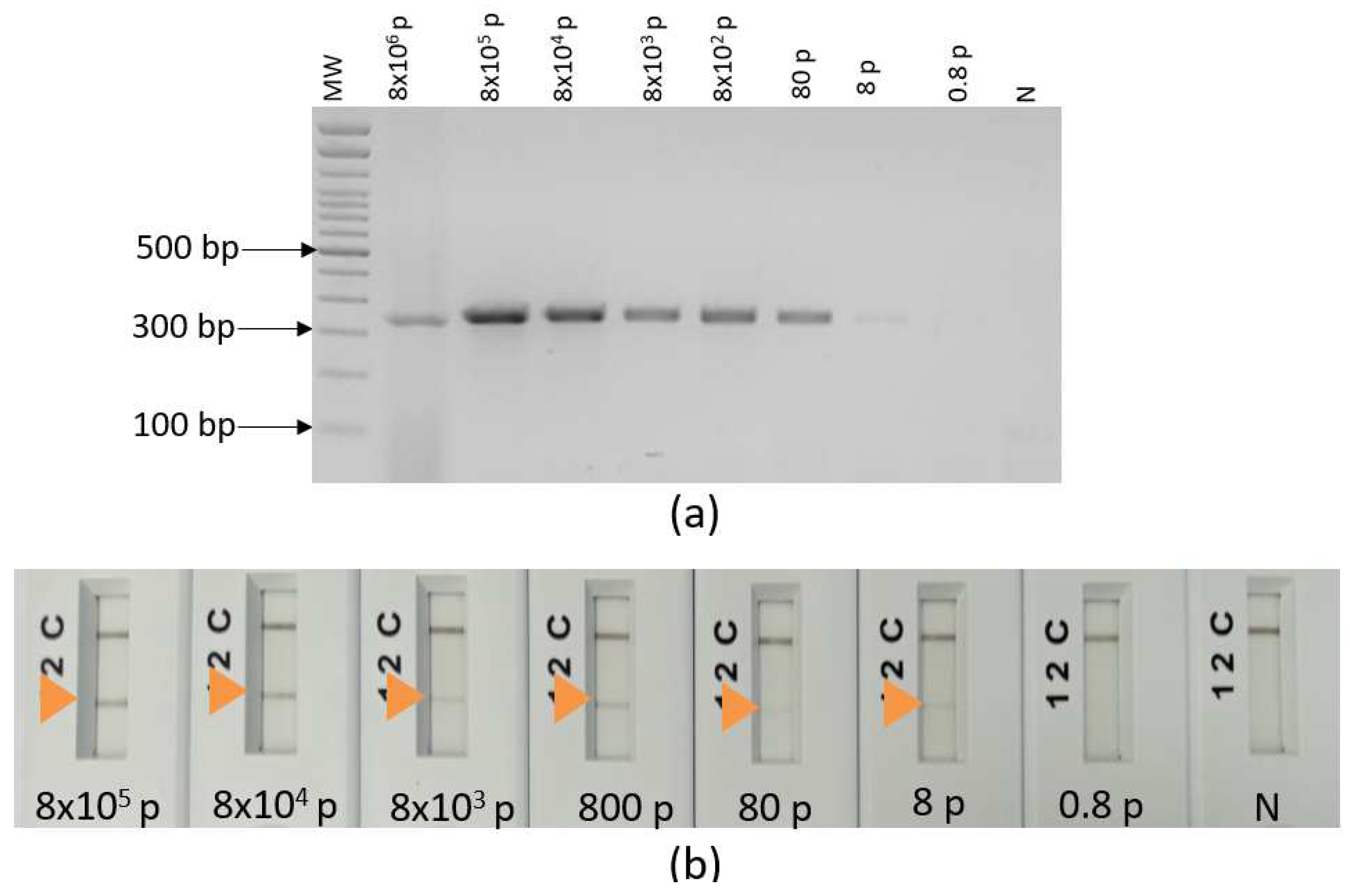
2.8. Analytical specificity and sensitivity
3. Results
3.1. Target selection and primers design
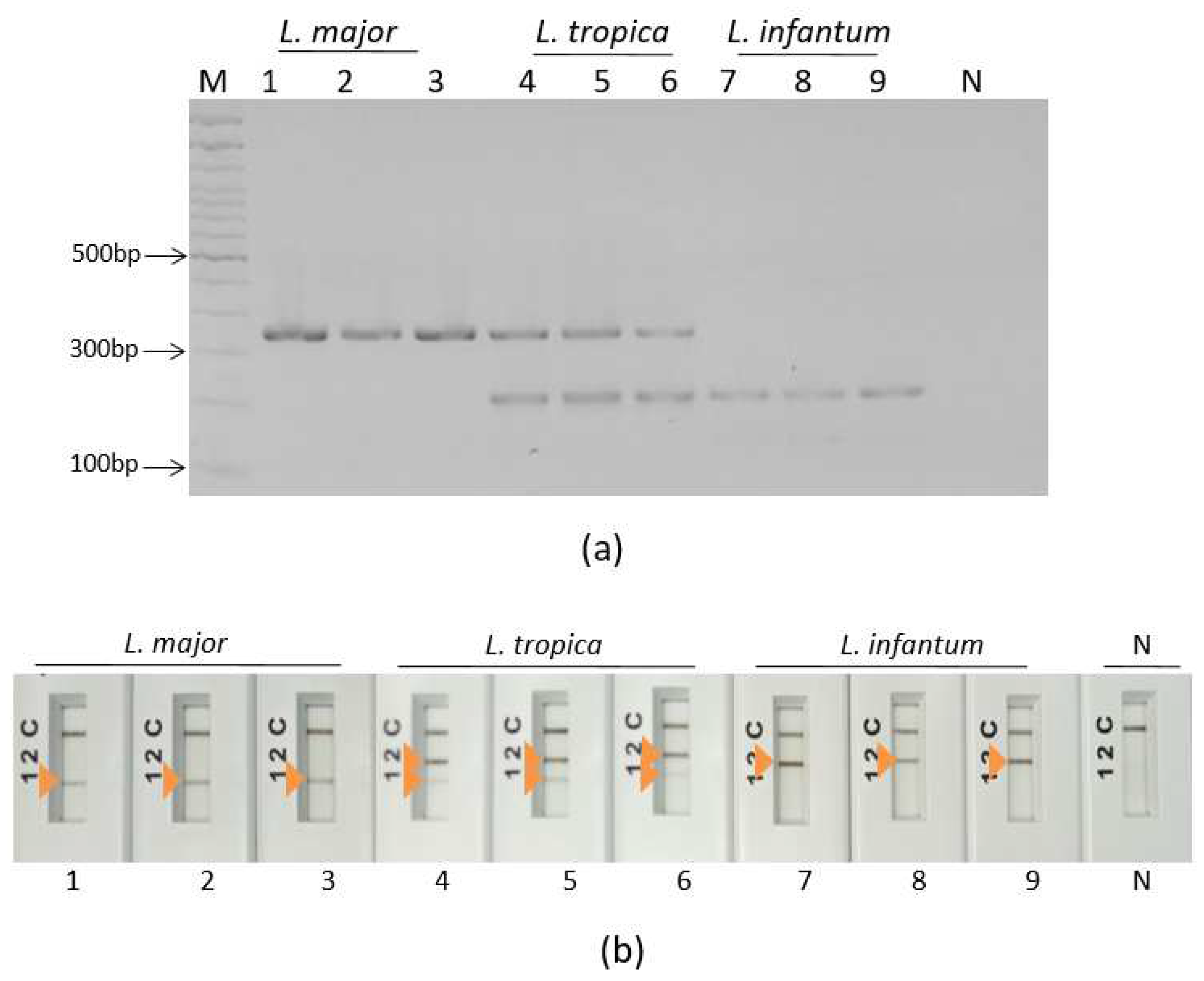
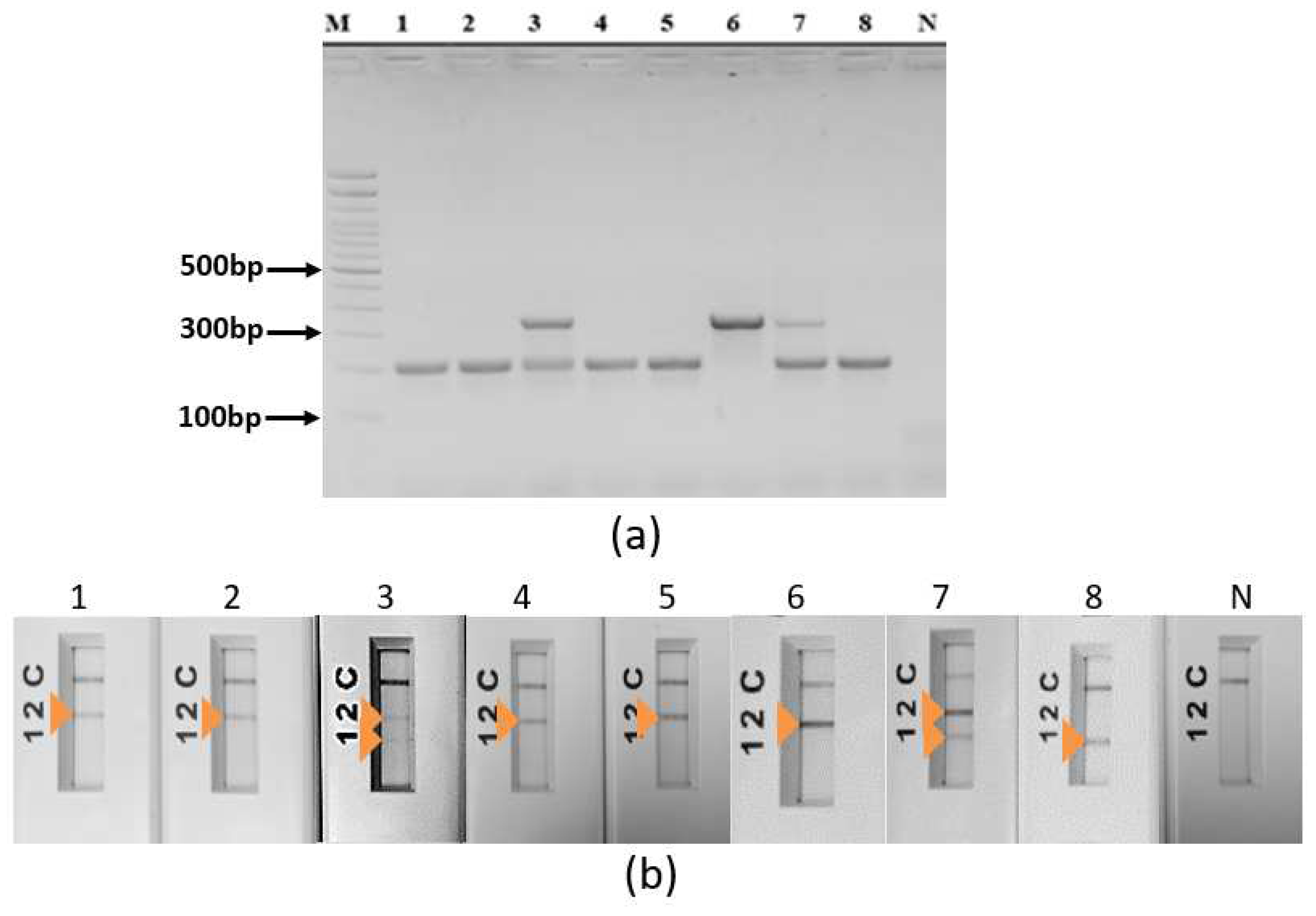
3.2. Specific simplex ultra fast PCRs screening assays
3.3. Ultra-fast duplex PCR and LF detection assays set up
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat Microbiol 2019, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Fathallah-Mili, A.; Saghrouni, F.; BenSaid, Z.; BenAoun, Y.S.-; Guizani, I.; BenSaid, M.; Mili, A.F.; Saghrouni, F.; BenSaid, Z.; BenAoun, Y.S.-; et al. Retrospective Analysis of Leishmaniasis in Central Tunisia: An Update on Emerging Epidemiological Trends; IntechOpen, 2012; ISBN 978-953-51-0274-8.
- Garni, R.; Tran, A.; Guis, H.; Baldet, T.; Benallal, K.; Boubidi, S.; Harrat, Z. Remote Sensing, Land Cover Changes, and Vector-Borne Diseases: Use of High Spatial Resolution Satellite Imagery to Map the Risk of Occurrence of Cutaneous Leishmaniasis in Ghardaïa, Algeria. Infect Genet Evol 2014, 28, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.; Saliba, H.; Altawil, A.; Villinsky, J.; Al-Nahhas, S. Cutaneous Leishmaniasis in the Central Provinces of Hama and Edlib in Syria: Vector Identification and Parasite Typing. Parasit Vectors 2015, 8, 524. [Google Scholar] [CrossRef] [PubMed]
- Rhajaoui, M.; Nasereddin, A.; Fellah, H.; Azmi, K.; Amarir, F.; Al-Jawabreh, A.; Ereqat, S.; Planer, J.; Abdeen, Z. New Clinico-Epidemiologic Profile of Cutaneous Leishmaniasis, Morocco. Emerg Infect Dis 2007, 13, 1358–1360. [Google Scholar] [CrossRef] [PubMed]
- Koff, A.; Rosen, T. Treatment of Cutaneous Leishmaniasis. Journal of the American Academy of Dermatology 1994, 31. [Google Scholar] [CrossRef]
- Arevalo, J.; Ramirez, L.; Adaui, V.; Zimic, M.; Tulliano, G.; Miranda-Verástegui, C.; Lazo, M.; Loayza-Muro, R.; De Doncker, S.; Maurer, A.; et al. Influence of Leishmania (Viannia) Species on the Response to Antimonial Treatment in Patients with American Tegumentary Leishmaniasis. J Infect Dis 2007, 195, 1846–1851. [Google Scholar] [CrossRef]
- Mosimann, V.; Neumayr, A.; Hatz, C.; Blum, J.A. Cutaneous Leishmaniasis in Switzerland: First Experience with Species-Specific Treatment. Infection 2013, 41, 1177–1182. [Google Scholar] [CrossRef]
- Schönian, G.; Nasereddin, A.; Dinse, N.; Schweynoch, C.; Schallig, H.D.F.H.; Presber, W.; Jaffe, C.L. PCR Diagnosis and Characterization of Leishmania in Local and Imported Clinical Samples. Diagn Microbiol Infect Dis 2003, 47, 349–358. [Google Scholar] [CrossRef]
- Bel Hadj Ali, I.; H, C.; Y, S.B.A.; E, H.-S.; H, S.; A, Y.; O, E.D.; Z, H.; Mm, M.; M, B.S.; et al. Dipeptidyl Peptidase III as a DNA Marker to Investigate Epidemiology and Taxonomy of Old World Leishmania Species. PLoS neglected tropical diseases 2021, 15. [Google Scholar] [CrossRef]
- van Henten, S.; Fikre, H.; Melkamu, R.; Dessie, D.; Mekonnen, T.; Kassa, M.; Bogale, T.; Mohammed, R.; Cnops, L.; Vogt, F.; et al. Evaluation of the CL Detect Rapid Test in Ethiopian Patients Suspected for Cutaneous Leishmaniasis. PLoS Negl Trop Dis 2022, 16, e0010143. [Google Scholar] [CrossRef]
- Bennis, I.; Verdonck, K.; El Khalfaoui, N.; Riyad, M.; Fellah, H.; Dujardin, J.-C.; Sahibi, H.; Bouhout, S.; Van der Auwera, G.; Boelaert, M. Accuracy of a Rapid Diagnostic Test Based on Antigen Detection for the Diagnosis of Cutaneous Leishmaniasis in Patients with Suggestive Skin Lesions in Morocco. Am J Trop Med Hyg 2018, 99, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Gonzalez, A.; Saldarriaga, O.A.; Tartaglino, L.; Gacek, R.; Temple, E.; Sparks, H.; Melby, P.C.; Travi, B.L. A Novel Molecular Test to Diagnose Canine Visceral Leishmaniasis at the Point of Care. Am J Trop Med Hyg 2015, 93, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Mondal, D.; Ghosh, P.; Khan, M.A.A.; Hossain, F.; Böhlken-Fascher, S.; Matlashewski, G.; Kroeger, A.; Olliaro, P.; Abd El Wahed, A. Mobile Suitcase Laboratory for Rapid Detection of Leishmania Donovani Using Recombinase Polymerase Amplification Assay. Parasit Vectors 2016, 9, 281. [Google Scholar] [CrossRef]
- Cossio, A.; Jojoa, J.; Castro, M.d.M.; Castillo, R.M.; Osorio, L.; Shelite, T.R.; Saravia, N.G.; Melby, P.C.; Travi, B.L. Diagnostic Performance of a Recombinant Polymerase Amplification Test—Lateral Flow (RPA-LF) for Cutaneous Leishmaniasis in an Endemic Setting of Colombia. PLOS Neglected Tropical Diseases 2021, 15, e0009291. [Google Scholar] [CrossRef]
- Khan, M.A.A.; Faisal, K.; Chowdhury, R.; Nath, R.; Ghosh, P.; Ghosh, D.; Hossain, F.; Abd El Wahed, A.; Mondal, D. Evaluation of Molecular Assays to Detect Leishmania Donovani in Phlebotomus Argentipes Fed on Post-Kala-Azar Dermal Leishmaniasis Patients. Parasit Vectors 2021, 14, 465. [Google Scholar] [CrossRef]
- Mesa, L.E.; Manrique, R.; Robledo, S.M.; Tabares, J.; Pineda, T.; Muskus, C. The Performance of the Recombinase Polymerase Amplification Test for Detecting Leishmania Deoxyribonucleic Acid from Skin Lesions of Patients with Clinical or Epidemiological Suspicion of Cutaneous Leishmaniasis. Trans R Soc Trop Med Hyg 2021, 115, 1427–1433. [Google Scholar] [CrossRef]
- Erber, A.C.; Sandler, P.J.; de Avelar, D.M.; Swoboda, I.; Cota, G.; Walochnik, J. Diagnosis of Visceral and Cutaneous Leishmaniasis Using Loop-Mediated Isothermal Amplification (LAMP) Protocols: A Systematic Review and Meta-Analysis. Parasites Vectors 2022, 15, 34. [Google Scholar] [CrossRef]
- Kim, T.-H.; Hwang, H.J.; Kim, J.H.; Department of Life and Nanopharmaceutical Sciences, Graduate School, Kyung Hee University, Seoul 02447, Republic of Korea. Optimization of Ultra-Fast Convection Polymerase Chain Reaction Conditions for Pathogen Detection with Nucleic Acid Lateral Flow Immunoassay. Intern. J. Oral Biol. 2019, 44, 8–13. [Google Scholar] [CrossRef]
- Lim, S.; Nan, H.; Lee, M.-J.; Kang, S.H. Fast On-Site Diagnosis of Influenza A Virus by Palm PCR and Portable Capillary Electrophoresis. J Chromatogr B Analyt Technol Biomed Life Sci 2014, 963, 134–139. [Google Scholar] [CrossRef]
- Mahale, P.; Warke, R.; Ramaiya, M.; Balasubramanian, D.; Shetty, S.; Mankeshwar, R.; Chowdhary, A. Assessment of Efficacy of Palm Polymerase Chain Reaction with Microscopy, Rapid Diagnostic Test and Conventional Polymerase Chain Reaction for Diagnosis of Malaria. Indian Journal of Medical Microbiology 2019, 37, 192–197. [Google Scholar] [CrossRef]
- Ben-Ismail, R.; Ben Rachid, M.S.; Gradoni, L.; Gramiccia, M.; Helal, H. Leishmaniose cutanée zoonotique en Tunisie : Étude du réservoir dans le foyer de Douara. 1987.
- Rogers, M.B.; Hilley, J.D.; Dickens, N.J.; Wilkes, J.; Bates, P.A.; Depledge, D.P.; Harris, D.; Her, Y.; Herzyk, P.; Imamura, H.; et al. Chromosome and Gene Copy Number Variation Allow Major Structural Change between Species and Strains of Leishmania. Genome Res. 2011, 21, 2129–2142. [Google Scholar] [CrossRef]
- Zelazny, A.M.; Fedorko, D.P.; Li, L.; Neva, F.A.; Steven; Fischer, H. Evaluation of 7sl Rna Gene Sequences for the Identification of Leishmania Spp. 2005.
- Tupperwar, N.; Vineeth, V.; Rath, S.; Vaidya, T. Development of a Real-Time Polymerase Chain Reaction Assay for the Quantification of Leishmania Species and the Monitoring of Systemic Distribution of the Pathogen. Diagn Microbiol Infect Dis 2008, 61, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Kariyawasam, R.; Valencia, B.M.; Lau, R.; Shao, E.; Thompson, C.A.; Stevens, M.; Kincaid, L.; Del Castillo, A.L.Q.; Cruz-Arzapalo, L.O.; Llanos-Cuentas, A.; et al. Evaluation of a Point-of-Care Molecular Detection Device for Leishmania Spp. and Intercurrent Fungal and Mycobacterial Organisms in Peruvian Patients with Cutaneous Ulcers. Infection 2021, 49, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Schönian, G.; Nasereddin, A.; Dinse, N.; Schweynoch, C.; Schallig, H.D.F.H.; Presber, W.; Jaffe, C.L. PCR Diagnosis and Characterization of Leishmania in Local and Imported Clinical Samples11Part of This Work Has Been Presented at the Second World Congress on Leishmaniosis in Crete, Greece, May 2001. Diagnostic Microbiology and Infectious Disease 2003, 47, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Budd, J.; Miller, B.S.; Weckman, N.E.; Cherkaoui, D.; Huang, D.; Decruz, A.T.; Fongwen, N.; Han, G.-R.; Broto, M.; Estcourt, C.S.; et al. Lateral Flow Test Engineering and Lessons Learned from COVID-19. Nat Rev Bioeng 2023, 1, 13–31. [Google Scholar] [CrossRef]
- Ahmed, M.; Pollak, N.M.; Hugo, L.E.; van den Hurk, A.F.; Hobson-Peters, J.; Macdonald, J. Rapid Molecular Assays for the Detection of the Four Dengue Viruses in Infected Mosquitoes. Gates Open Res 2022, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, N.J.; Menting, S.; Wentink-Bonnema, E.M.S.; Broekhuizen-van Haaften, P.E.; Withycombe, E.; Schallig, H.D.F.H.; Mens, P.F. Laboratory Evaluation of the Miniature Direct-on-Blood PCR Nucleic Acid Lateral Flow Immunoassay (Mini-DbPCR-NALFIA), a Simplified Molecular Diagnostic Test for Plasmodium. Malar J 2023, 22, 98. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, Q.; Yuan, W.; Shi, Y.; Dong, S.; Luo, X. Visual and Rapid Identification of Chlamydia Trachomatis and Neisseria Gonorrhoeae Using Multiplex Loop-Mediated Isothermal Amplification and a Gold Nanoparticle-Based Lateral Flow Biosensor. Front Cell Infect Microbiol 2023, 13, 1067554. [Google Scholar] [CrossRef]
- Sint, D.; Raso, L.; Traugott, M. Advances in Multiplex PCR: Balancing Primer Efficiencies and Improving Detection Success. Methods Ecol Evol 2012, 3, 898–905. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and Application in Diagnostic Virology. Clin Microbiol Rev 2000, 13, 559–570. [Google Scholar] [CrossRef]
- Fernández-Arévalo, A.; El Baidouri, F.; Ravel, C.; Ballart, C.; Abras, A.; Lachaud, L.; Tebar, S.; Lami, P.; Pratlong, F.; Gállego, M.; et al. The Leishmania Donovani Species Complex: A New Insight into Taxonomy☆. International Journal for Parasitology 2020, 50, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Krayter, L.; Schnur, L.F.; Schönian, G. The Genetic Relationship between Leishmania Aethiopica and Leishmania Tropica Revealed by Comparing Microsatellite Profiles. PLoS ONE 2015, 10, e0131227. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.M.; Welburn, S.C. Leishmaniasis Beyond East Africa. Frontiers in Veterinary Science 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Li, Z.; Kim, T.-H.; Wang, X.; Bu, Z.; Li, S.; Hwang, H.J.; Kim, J.H. Ultra-Fast Detection and Differentiation of Brucella Genus Bacteria, B. Abortus, B. Melitensis, and B. Suis, Using Multiplex Convection Polymerase Chain Reaction 2019.
- Wu, Y.; Jiang, M.; Li, S.; Waterfield, N.R.; Yang, G. Establish an Allele-Specific Real-Time PCR for Leishmania Species Identification. Infectious Diseases of Poverty 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Cássia-Pires, R. de; Melo, M. de F.A.D. de; Barbosa, R. da H.; Roque, A.L.R. Multiplex PCR as a Tool for the Diagnosis of Leishmania Spp. KDNA and the Gapdh Housekeeping Gene of Mammal Hosts. PLoS ONE 2017, 12, e0173922. [Google Scholar] [CrossRef]
- Saldarriaga, O.A.; Castellanos-Gonzalez, A.; Porrozzi, R.; Baldeviano, G.C.; Lescano, A.G.; Santos, M.B. de L.; Fernandez, O.L.; Saravia, N.G.; Costa, E.; Melby, P.C.; et al. An Innovative Field-Applicable Molecular Test to Diagnose Cutaneous Leishmania Viannia Spp. Infections. PLOS Neglected Tropical Diseases 2016, 10, e0004638. [Google Scholar] [CrossRef]
- Phuektes, P.; Mansell, P.D.; Browning, G.F. Multiplex Polymerase Chain Reaction Assay for Simultaneous Detection of Staphylococcus Aureus and Streptococcal Causes of Bovine Mastitis. Journal of Dairy Science 2001, 84, 1140–1148. [Google Scholar] [CrossRef]
| WHO Code | Lab Code | Species | Zymodem | Clinical manifestation |
| MMER/TN/87/Ron114 | R114 | L. major | MON-25 | NA |
| MPSA/TN/87/Ron99 | R99 | L. major | MON-25 | NA |
| MPSA/TN/87/Ron44 | R44 | L. major | MON-25 | NA |
| MPSA/TN/87/Ron155 | R155 | L. major | MON-25 | NA |
| MPSA/TN/87/Ron 102 | R102 | L. major | MON-25 | NA |
| MRHO/SU/59/P-Strain | P-strain | L. major | MON-4 | NA |
| MHOM/IL/83/IL24 | IL24 | L. major | MON-66 | CL |
| MHOM/IL/83/IL53 | IL53 | L. major | MON-67 | CL |
| MHOM/IL/67/Jericho II | Jerichll | L. major | MON-26 | CL |
| MPSA/TN/89/Psa1 | Psa1 | L. major | NT | NA |
| MPSA/TN/89/Psa5 | Psa5 | L. major | NT | NA |
| MHOM/TN/11/EMPA12 | EMPA12 | L. major | NT | CL |
| MHOM/TN/80/IPT1 | IPT1 | L. infantum | MON-1 | VL |
| MHOM/TN/88/Aymen | Aymen | L. infantum | MON-1 | VL |
| MHOM/TN/88/Nabil | Nabil | L. infantum | MON-1 | VL |
| MHOM/TN/92/LV08 | LV08 | L. infantum | NT | VL |
| MHOM/TN/92/LV10 | LV10 | L. infantum | MON-80 | VL |
| MHOM/TN/94/LV49 | LV49 | L. infantum | MON-24 | VL |
| MHOM/TN/94/LV50 | LV50 | L. infantum | MON-1 | VL |
| MHOM/TN/97/Drep 13 | D13 | L. infantum | MON-24 | CL |
| MHOM/TN/98/Drep16 | D16 | L. infantum | MON-24 | CL |
| MHOM/TN87/KA412 | KA412 | L. infantum | MON-1 | VL |
| MHOM/BR/74/PP75 | PP75 | L. infantum | MON-1 | VL |
| MHOM/IQ/76/BAG17 | Bag17 | L. tropica | LON-24 | CL |
| MRAT/IQ/73/Adhanis I | Adhanis | L. tropica | MON-5 | NA |
| MCAN/IN/71/DBKM | DBKM | L. tropica | MON-62 | NA |
| MHOM/IL/00/Gabaï159 | Gabai 159 | L. tropica | LON-9 | CL |
| MHOM/GR/00/LA28 | LA28 | L. tropica | LON-16 | CL |
| MHOM/IQ/73/A Sinaï III | A Sinai III | L. tropica | LON-11 | CL |
| MHOM/IQ/76/BAG9 | Bag 9 | L. tropica | MON-53 | CL |
| MHOM/SU/74/SAF K27 | K27 | L. tropica | MON-60 | CL |
| MHOM/IQ//73/Bumm30 | Bumm30 | L. tropica | LON-17 | VL |
| MHOM/IL/78/Rachnan | Rachnan | L. tropica | MON-60 | CL |
| MHOM/ET/72/GEBRE1 | L1005 | L. donovani | MON-82 | VL |
| MPSA/SA/84/Jisha 238 | J238 | L. arabica | LON-64 | NA |
| MHOM/ET/72/L100 | L100 | L. aethiopica | MON-14 | CL |
| MRHO/SU/74/95-A | 95A | L. turanica | MON-64 | NA |
| Target | Gene | Protein | ||
| L. major | L. infantum | L. tropica | ||
| mt30 | Intergenic region between LmjF30.0190 & LmjF30.0200 | Intergenic region between LINF_300006850 & LINF_300006900 |
Intergenic region between LTRL590_300007200 & LTRL590_300007300 |
None |
| mt22 | Non coding sequence | LinJ.22.0300 | LTRL590_220009300 | Hypothetical protein in L. infantum and L. tropica Non coding sequence in L. major |
| it20 | Absent | LinJ.20.0040 | LTRL590_200005300 | Phosphate-Repressible Phosphate Permease-like protein Absent in L. major |
| mt7SL | LmjF.05.SRP.RNA | LINJ_05_snRNA1 | 7SL gene Partial sequence | 7SL RNA |
| Primer pairs | Sequences (5'-3') | Size (bp) | Expected specificity | ||
|---|---|---|---|---|---|
| L. major | L. tropica | L. infantum | |||
| mt22F1 | ACCGAACCCAACGCTGAAG | 366 | + | + | - |
| mt22R1 | AGTGCATGAGGCGTGTATGG | ||||
| mt22F2 | CACTCATGCGTGTCCATTCT | 319 | + | + | - |
| mt22R2 | GTATGGGAAGGTGGGGGT | ||||
| mt22F1 | 352 | + | + | - | |
| mt22R2 | |||||
| mt22F2 | 333 | + | + | - | |
| mt22R1 | |||||
| mt30F | GGTGCAATGTGCGCATG | 350 | + | + | - |
| mt30R | GCTTGGCGCTCTCGAAAAG | ||||
| mt7sLF | TTGGTGGTGGTGGGATGGAC | 191 | + | + | - |
| mt7sLR | CACCACGTCAACGCAGCAAA | ||||
| it20F1 | TCTGGATTGCAGTCGTCGG | 209 | - | + | + |
| it20R1 | CTTGGCGATACCTCCTGAT | ||||
| it20F2 | AGCCTTGGTGGTGTCTTTTG | 195 | - | + | + |
| it20R2 | CAAAGAAGACGGCAGACACA | ||||
| Duplex PCRs | Primer pairs | Size (bp) | Expected specificity | ||
|---|---|---|---|---|---|
| L. major | L. tropica | L. infantum | |||
| A | mt22F1 | 366 | + | + | - |
| mt22R1 | |||||
| it20F1 | 209 | - | + | + | |
| it20R1 | |||||
| B | mt22F2 | 319 | + | + | - |
| mt22R2 | |||||
| it20F2 | 195 | - | + | + | |
| it20R2 | |||||
| C | mt22F1 | 366 | + | + | - |
| mt22R1 | |||||
| it20F2 | 195 | - | + | + | |
| it20R2 | |||||
| D | mt22F2 | 319 | + | + | - |
| mt22R2 | |||||
| it20F1 | 209 | - | + | + | |
| it20R1 | |||||
| E | mt22F2 | 333 | + | + | - |
| mt22R1 | |||||
| it20F1 | 209 | - | + | + | |
| it20R1 | |||||
| F | mt22F2 | 333 | + | + | - |
| mt22R1 | |||||
| it20F2 | 195 | - | + | + | |
| it20R2 | |||||
| G | mt30F | 350 | + | + | - |
| mt30R | |||||
| it20F1 | 209 | - | + | + | |
| it20R1 | |||||
| H | mt30F | 350 | + | + | - |
| mt30R | |||||
| it20F2 | 195 | - | + | + | |
| it20R2 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





