1. Introduction
Colletotrichum, a member of the Ascomycete phylum, is a kind of widespread phytopathogen that can infect various crops worldwide [
1]. In 2012, it was recognized as one of the top 10 fungal pathogens due to its scientific and economic significance [
1]. Anthracnose caused by
Colletotrichum higginsianum poses a significant threat to Chinese flowering cabbage (
Brassica parachinensis) cultivation in South China, leading to reduced crop quality and yield [
2]. The annual yield losses due to anthracnose outbreaks can reach up to 40%, severely impacting the development of the Chinese flowering cabbage industry in this region [
2].
C. higginsianum has a hemibiotrophic lifestyle and has been found to infect various cruciferous plants including model plant
Arabidopsis [
3,
4,
5]. Moreover, genomic and transcriptomic data for
C. higginsianum have been published and are readily accessible for genetic manipulation studies [
6,
7], making significant contributions to our understanding of the mechanisms underlying interactions between plant pathogenic fungi and host plants.
Melanin is a macromolecular pigment produced by various fungi through three pathways: polyketide synthase pathway, tyrosine degradation synthetic pathway and tyrosine degradation pathway [
8,
9,
10]. Previous studies have demonstrated that
C. higginsianum produces DHN-melanin through the polyketide synthase pathway [
5]. The biosynthetic pathway involves the polymerization of acetyl-CoA and malonyl-CoA by polyketide synthase (Pks) to form 1,3,6,8-Tetrahydroxynaphthalene (T4HN). T4HN then undergoes enzymatic conversions catalyzed by tetrahydroxynaphthalene reductase (Thnr), scytalone dehydratase (Scd), and trihydroxynaphthalene reductase (Thr1) to produce vermelone. Finally, vermelone is dehydrated and polymerized to form DHN-melanin [
10,
11]. It has been observed in related fungal species that deficiencies or loss of enzymes involved in this pathway can result in blocked biosynthesis of DHN-melanin [
8,
12,
13]. However, the specific role played by these enzymes in DHN-melanin biosynthesis in
C. higginsianum remains poorly understood and requires further research.
Melanin is an integral component of the cell wall rather than an independent entity [
14], capable of covalently cross-linking with polysaccharide components within it [
15,
16]. In fungi, there exists a direct interaction between melanin and chitin - a crucial constituent of the fungal cell wall [
17]. Reports suggest that chitin acts as a scaffold for melanin deposition within the fungal cell wall contributing to its fixation [
18,
19]. Additionally, melanin deposited on the fungal cell wall effectively enhances its integrity against various environmental stresses such as heavy metals, ultraviolet (UV) irradiation, and enzymatic lysis [
8,
9,
20]. For example, mutants lacking melanin in
Botrytis cinerea exhibit increased sensitivity to agents that interfere with cell wall integrity [
21]. In terms of pathogenicity, proper appressorium formation and maintenance of high turgor pressure are crucial for successful infection by fungi like
C. higginsianum and
Magnaporthe oryzae [
22,
23,
24]. Melanization primarily depends on melanin deposition in the cell wall [
25]. Impaired melanin synthesis in
M. oryzae inhibits proper appressorium melanization, resulting in reduced intracellular turgor pressure and decreased pathogenicity due to failure in penetrating host plant epidermal cells [
24]. Similarly, deficiency of the gene
THR1 in
C. lagenarium impairs DHN-melanin production and nonmelanized appressoria formation leading to a notable reduction in pathogenicity [
26]. Conversely, in
C. gloeosporioides, deletion of
CgCmr1 and
CgPks1 lead to unmelanization and decreased turgor pressure in the appressoria, while had no effect in pathogenicity [
27]. Given the uncertainty regarding the connection between melanin and pathogenicity, it is therefore particularly important to study the relationship between melanin, cell wall integrity, and pathogenicity in
C. higginsianum.
In this study, we aimed to investigate the role of two enzymes (ChPks and ChThr1) involved in DHN-melanin biosynthesis in C. higginsianum. The expression patterns of genes ChPKS and ChTHR1 were analyzed using RT-qPCR technique. Gene knockout and complementation strains were constructed for both genes to assess their impact on DHN-melanin biosynthesis as well as growth, development, and pathogenicity in C. higginsianum. The main goal of this research is to gain a better understanding of the molecular mechanisms underlying DHN-melanin biosynthesis and its relationship with cell wall integrity and pathogenicity in C. higginsianum. This knowledge will contribute to improve our understanding of the pathogenesis in anthracnose disease of cruciferous plants. Ultimately, it can aid in developing more effective control strategies against this disease.
3. Discussion
Melanin is a critical component of fungal cell wall that helps to maintain cell wall integrity and is involved in defending environmental stresses, accumulation of turgor pressure and pathogenicity [
9,
16]. Previous studies have reported that the genus
Colletotrichum synthesizes DHN-melanin through the polyketide synthase pathway [
5,
27,
28]. Deficiencies in these enzymes can inhibit or block DHN-melanin biosynthesis in many fungi [
25,
26,
27]. Pks and Thr1 are key enzymes that play a crucial role in the biosynthetic processes of DHN-melanin in several fungi [
29,
30]. For example, in
C. graminicola, CgPKS1 gene encoding polyketide synthase is expressed during appressorial melanization [
25]. Similarly, the expression level of
BRN2, a homolog of THR1 in
Monilinia fructicola and
M. fructigena, is noticeably up-regulated during melanin synthesis [
31]. In our study, we observed a significant up-regulation of both genes
ChPKS and
ChTHR1 during hyphal and appressorial melanization processes of
C. higginsianum. This finding indicates that ChPks and ChThr1 are essential players in DHN-melanin production. Interestingly, we also found that the expression level of gene
ChTHR1 was significantly up-regulated at 40 hours post-infection (hpi), suggesting its potential involvement beyond appressorial melanization processes during the biotrophic infection phase of
C. higginsianum.
Polyketide synthase (Pks) has been defined as the initial step in the DHN-malanin biosynthetic process [
32,
33,
34]. Deletion of the
PKS1 gene in
C. graminicola and
C. gloeosporioides resulted in colonies that were yellow to light orange in color, indicating a lack of melanization. In addition, appressoria lacking
CgPKS1 in
C. graminicola showed sensitivity to externally applied cell-wall-degrading enzymes [
25,
27]. Similarly, deletion of the
ChPKS gene led to an orange-yellow colony and unmelanized appressoria. Furthermore, TEM observation revealed that the
ChpksΔ mutant did not exhibit any deposits of DHN-melanin dots on its cell wall. This disruption affected the integrity of the cell wall and made it sensitive to agents interfering with cell wall function. Melanin deposition is known to be essential for maintaining turgor pressure within appressoria. For instance,
CgPks1Δ mutants exhibited significantly lower turgor pressure compared to WT strains of
C. gloeosporioides; however,
CgPks1 was found not to play an essential role in penetration and pathogenicity [
27]. In contrast, knockout mutants lacking
CgPKS1 did not affect turgor pressure but noticeably reduced penetrance and pathogenicity in
C. graminicola [
25]. These findings indicate that Pks plays different roles among fungi species. In our study with
ChpksΔ mutants from
C. higginsianum, we observed a significant decrease in turgor pressure within appressoria similar to what was seen with deletion of
CgPks1 in
C. gloeosporioides due to disrupted cell wall integrity resulting from unmelanized appressoria formation by
ChpksΔ mutants. This loss of turgor pressure accumulation led to a loss of penetrance and pathogenicity.
Trihydroxynaphthalene reductase (Thr1) is also a key enzyme in DHN-melanin biosynthesis [
35,
36,
37]. In our study, we observed that the knockout of the gene
ChTHR1 resulted in colony and appressoria with limited melanization, as well as a significant reduction in turgor pressure in the
Chthr1Δ mutant. This phenotype is similar to that observed in
M. oryzae mutants lacking the
Buf1 gene encoding trihydroxynaphthalene reductase; however, it should be noted that these Δ
buf1 mutants exhibit reduced penetrance [
24]. Similarly, disruption of gene
THR1 also leads to loss of penetrance in
C. lagenarium [
26]. Interestingly, our results revealed that the
Chthr1Δ mutant was still capable of penetrating Chinese flowering cabbage and
Arabidopsis leaves. TEM observation revealed the cell wall of
Chthr1Δ mutant fails to form a dense melanin layer, resulting in a notably reduction in turgor pressure and increased sensitivity to cell wall interfering agents compared to the WT strains. However, some melanin dots were still deposited on the cell wall of the
Chthr1Δ mutant, leading to turgor pressure compared to
ChpksΔ mutant. Notably though, this decreased turgor pressure did not affect penetrance. Furthermore, we observed a significant reduction in pathogenicity of
Chthr1Δ mutants on
Arabidopsis plants when compared to the WT strains. This finding suggests that while ChThr1 may play a role during biotrophic infection phase and contribute to pathogenesis in
C. higginsianum, its exact mechanisms require further investigation.
During DHN-melanin biosynthesis of
C. higginsianum, the absence of both ChPks and ChThr1 inhiinhibinhibit melanin production. However, compare to the
ChpksΔ mutant where DHN-melanin biosynthetic processes is completely blocked, the
Chthr1Δ mutant is still able to produce melanin. These results demonstrate that enzymes upstream in the DHN-melanin biosynthesis pathway may have a greater role in DHN-melanin synthesis. Additionally, it has been reported that both reductases Thnr and Thr1 belong to the short-chain de dehydrogenase/reductase (SDR) family and share common conserved core structural motifs during DHN-melanin biosynthesis [
29]. Studies conducted on
C. lagenarium have revealed that Thnr and Thr1 can co-mediate the reduction of T4HN, suggesting functional complementarity between them [
38]. However, our results indicate that expression levels of gene
ChTHNR was down-regulated in the
Chthr1Δ mutant (
Figure S3), suggesting that additional mechanisms may be present in
C. higginsianum to partially replace the role of ChThr1.
In summary, the deficiency of ChPks completely blocks DHN-melanin biosynthesis, thereby compromising the integrity and rigidity of the cell wall. Consequently, this results in a significant decrease in sensitivity to cell wall interfering agents as well as turgor pressure while also reducing pathogenicity in C. higginsianum. It is important to note that despite these effects on melanin production being observed in the Chthr1Δ mutant with limited DHN-melanin biosynthesis occurring within its hyphae and appressoria region; complete loss of pathogenicity has not been observed thus far.
4. Materials and Methods
4.1. Strains, Plant and Culture Conditions
The C. higginsianum IMI 349063 strain used in this study for whole genome sequencing was generously provided by Prof. Junbin Huang of Huazhong Agricultural University (Hubei, People’s Republic of China). All fungal strains were cultured at a temperature of 27°C.
To assess colony morphology after 5 days cultivated in the dark, the wild type (WT) and all fungal strains were cultivated on PDA medium containing potato extract (200 g/L), dextrose (20 g/L), and agar (20 g/L).
For pathogenicity tests on Arabidopsis thaliana Col-0, the plants were grown for 4 weeks under controlled conditions with a light/dark cycle of 12 hours/22°C during the light period and 12 hours/20°C during the dark period.
Similarly, Chinese flowering cabbage were used for pathogenicity tests after incubating for 4 weeks under controlled conditions with a light/dark cycle of 12 hours/26°C throughout both periods.
4.2. Bioinformatics Analysis of ChPks and ChThr1
ChPks protein sequence (XP_018155915) and ChThr1 protein sequence (XP_018155910) were retrieved from the genome database of
C. higginsianum IMI 349063 using BLASTp search with
C. graminicola PKS1 protein sequence (XP_008093079) and
Sclerotinia sclerotiorum Thr1 protein sequence (XP_001586798) as queries. The conserved domains within ChPks and ChThr1 proteins were predicted using Pfam analysis available at
http://pfam.xfam.org/. A neighbor-joining phylogenetic tree was constructed based on amino acid alignments using MEGA X software.
4.3. Expression Pattern Analysis of Genes ChPKS and ChTHR1
The WT strain of C. higginsianum was inoculated in PDB (potato 200 g/L, dextrose 20 g/L) medium, and the hyphae was collected at 7 d, 8 d, 9 d and 10 d for RNA extraction. The expression levels of genes ChPKS and ChTHR1 were determined using real time quantitative PCR (RT-qPCR), with ACTIN serving as the endogenous reference gene. Additionally, conidia from the WT strain of C. higginsianum were collected and adjusted to a concentration of 1×106 mL-1. Four-week-old A. thaliana Col-0 seedlings were used for inoculation.
Samples were taken at various time points including: 0 hours post-inoculation (hpi), 8 hpi, 22 hpi, 40 hpi and 60 hpi for RNA extraction. RT-qPCR was used to analysis the expression pattern of genes
ChPKS and
ChTHR1 during infection. Relative expression levels were calculated using the 2
-ΔΔCt method. The primers used are shown in
Table 1.
4.4. Deletion and Complementation of ChPKS and ChTHR1
To knockout genes
ChPKS and
ChTHR1, we constructed knockout plasmids p821-ChPKS-KO and p821-ChTHR1-KO according to the methods described in
Figure S1A and S1B. The knockout plasmid was then transformed into the WT strain using the ATMT technique to replace genes
ChPKS and
ChTHR1 with
HPH1 (hygromycin phosphotransferase gene), respectively. Hygromycin-resistant transformants were isolated as candidate mutants for
ChpksΔ and
Chthr1Δ. PCR analysis was used to confirmation the
ChpksΔ and
Chthr1Δ mutant (
Figure S2). The
ChpksΔ and
Chthr1Δ mutant were further confirmed by Southern blotting.
Sma I and
EcoR I were digested to gDNA of WT and
ChpksΔ mutant, and gDNA of WT and
Chthr1Δ mutant were digested with
Acc65 I. Then agarose gel electrophoresis to separate the DNA fragment, and transferred to a nylon membrane and assayed using DNA probes specific for
ChPKS and
ChTHR1, respectively.
Subsequently, for complementation of genes
ChPKS and
ChTHR1 at their original position, we constructed complementation plasmids pSFZY-ChPKS-COM and pSFZY-ChTHR1-COM according to the methods described in
Figure S1C and S1D. Similarly, we used the ATMT technique to transform pSFZY-ChPKS-COM and pSFZY-ChTHR1-COM into the
ChpksΔ and
Chthr1Δ mutant respectively. Neomycin-resistant transformants were isolated as candidate complementation strains for
ChpksΔ-C and
Chthr1Δ-C. The presence of these complemented strains was confirmed through PCR analysis (
Figure S2A-F). The primers used are shown in
Table 1.
4.5. Phenotypic Analysis
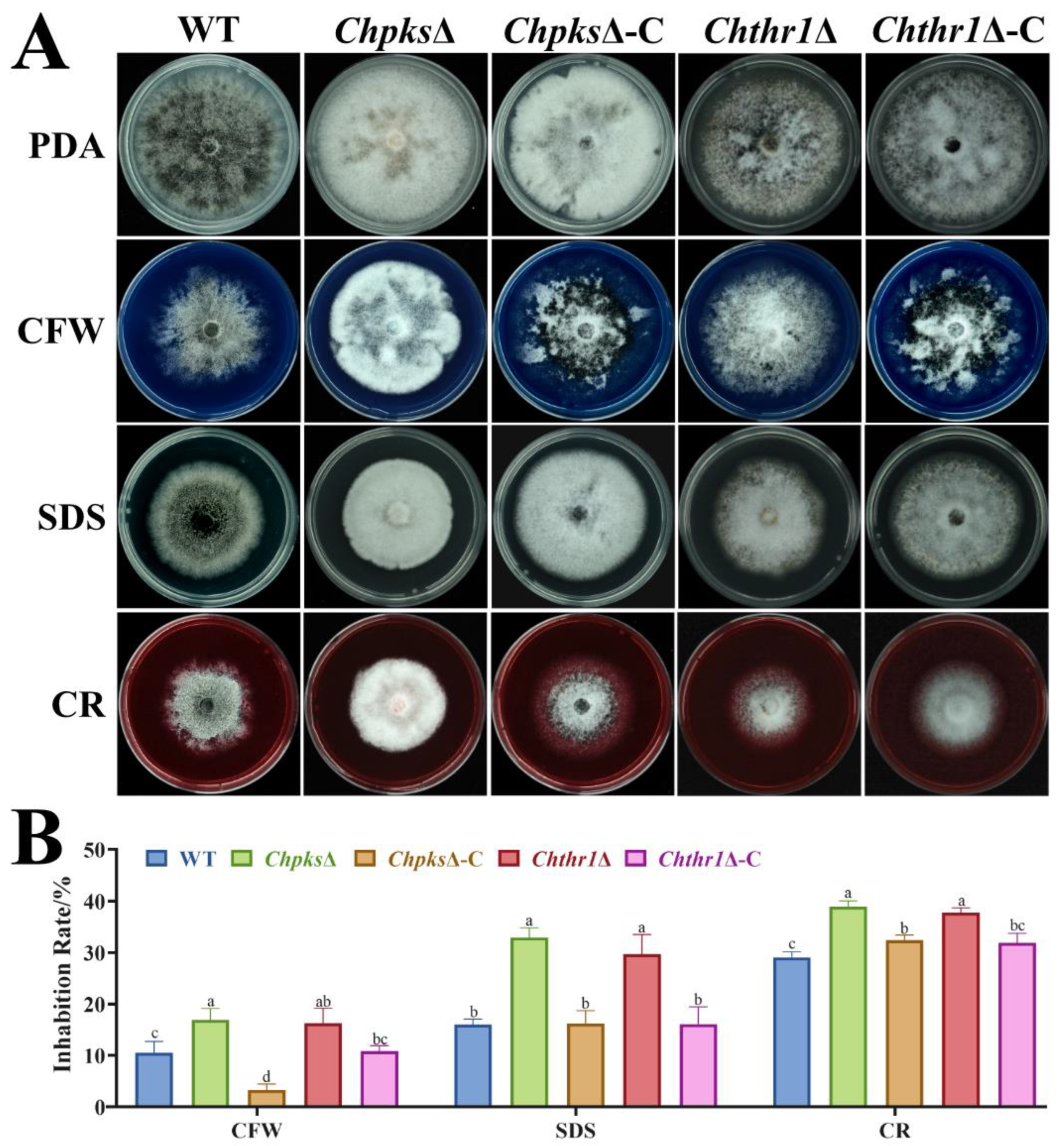
All fungal strains were cultured on 6 cm PDA plates at 27°C under dark conditions, and the colony diameter was measured after 5 d of growth. Additionally, all fungal strains were grown on PDA plates supplemented with specific compounds: 200 μg·mL-1 calcofluor white (CFW), 0.01% sodium dodecyl sulfate (SDS) and 200 μg·mL-1 congo red (CR). The inhibition rate was calculated by measuring the colony diameter after 5 d of growth.
To determine conidiation, the concentration of conidia suspensions for all strains was adjust to a density of 1 × 106·mL-1, and spread them onto 9 cm Mathur plates containing glucose (2.8 g·L-1), MgSO4·7H2O (1.22 g·L-1), KH2PO4 (2.72 g·L-1), oxoid mycological peptone (2.18 g·L-1), and agar (30 g·L-1). After incubation in darkness at 27°C for 5 days, conidia were washed down with sterile distilled water to determine the conidiation.
For appressorial formation analysis, the concentration of conidia suspensions for all strains was adjust to 1 × 105·mL-1. Then, a volume of 20 μL conidia suspensions from each strains was placed on hydrophobic cover glass. After incubating in darkness at 27°C for 22 h, the formation rate of at least 100 appressoria per strain was determined using microscopy.
Appressoria of all strains were obtained as described above. Subsequently, replace the sterile distilled water containing appressoria with a glycerol solution of concentration raging from 1 to 4 M. The collapse rate of at least 100 appressoria was calculated after a 10-minute incubation period.
4.6. Transmission Electron Microscopy
To observe melanin dots on the cell wall. The WT, ChpksΔ and Chthr1Δ strains were cultured in PDB liquid medium for a duration of 10 days prior to conducting transmission electron (TEM) analysis. Hypahe of WT, ChpksΔ and Chthr1Δ strains were fixed in 2.5 (vol/vol) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.3) at 4°C for 24 h, and were encapsulated in 3% (w/vol) low melting point agarose and then processed in Spurr resin on a Lynx tissue processor on a 24 h schedule. Provide additional permeation under vacuum at 60°C before embedding samples and polymerising at 60°C for 48 h, followed by ultrathin sectioning. The observations utilizing a Hitachi HT7700 Exalens microscope allowed for detailed examination at high resolution levels.
4.7. Pathogenicity Analysis
In order to assess the pathogenicity of the strains, conidia suspensions (1 × 106·mL-1) were prepared and collected as described previously. These suspensions were then evenly sprayed onto Arabidopsis leaves, followed by subjecting them to a controlled light/dark cycle lasting for 12 hours each day at a temperature of 27°C for 4 d. Additionally, under constant temperature conditions of 27°C and in a completely dark environment, drop 20 μL of conidia suspension onto wounded and unwounded detached Chinese flower cabbage leaves. After 4 days, observe and analyze.
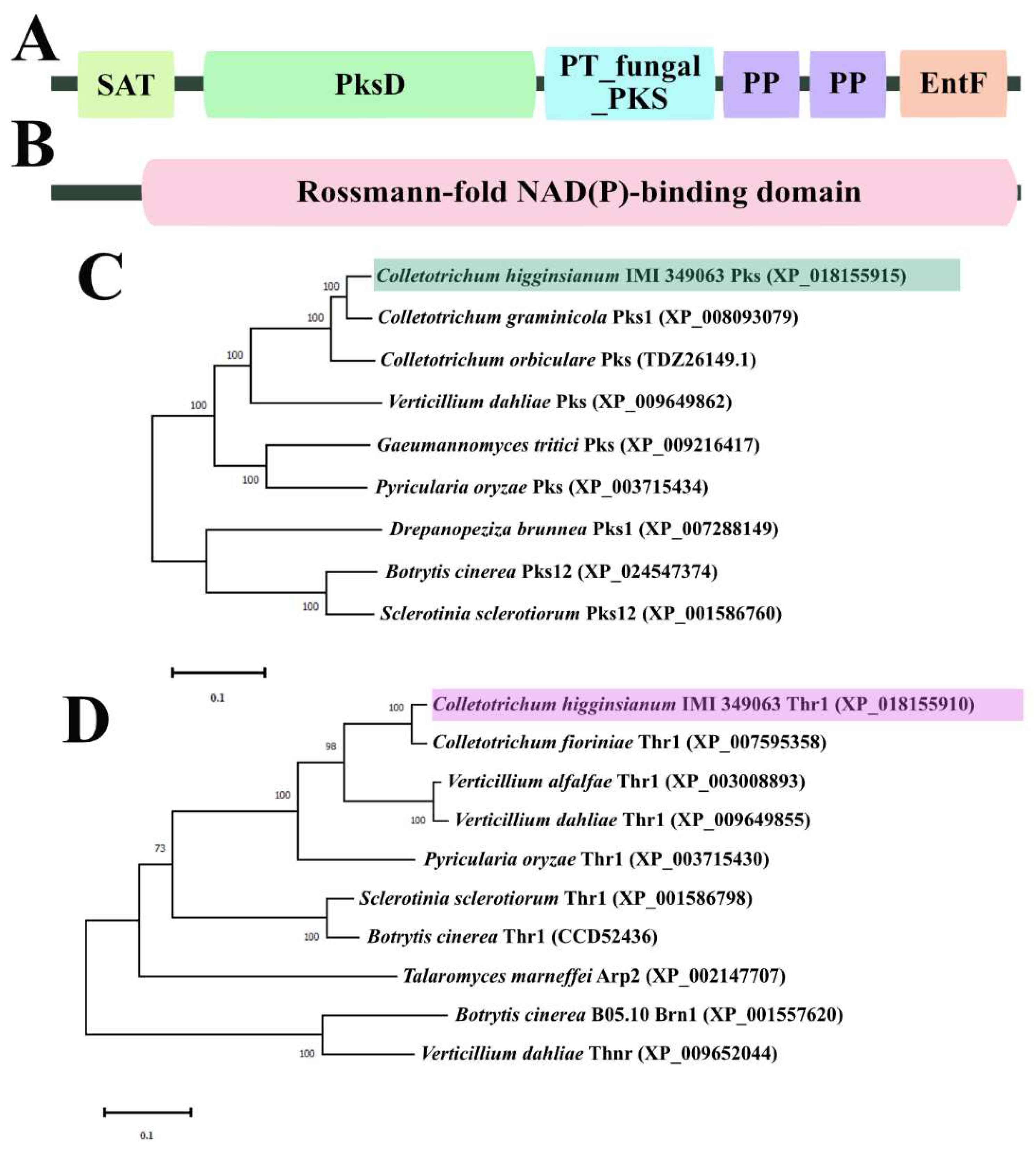
Figure 1.
Bioinformatics analysis of ChPks and ChThr1 in Colletotrichum higginsianum. (A) Pfam domain analysis of ChPks, including SAT, PksD, PT_fungal_PKS, two PP and EntF domain. (B) Pfam domain analysis of ChThr1, including a Rossmann-fold NAD(P)-binding domain. (C) Phylogenetic tree analysis of ChPks and its homologues in C. graminicola, C. orbiculare, V. dahliae, Gaeumannomyces tritici, Pyricularia oryzae, Drepanopeziza brunnea, B. cinerea and Sclerotinia sclerotiorum. (D) Phylogenetic tree analysis of ChThr1 and its homologues in C. fioriniae, V. alfalfae, V. dahliae, P. oryzae, S. sclerotiorum, B. cinerea and Talaromyces marneffei, along with the Thnr homologues in B. cinerea and V. dahliae.
Figure 1.
Bioinformatics analysis of ChPks and ChThr1 in Colletotrichum higginsianum. (A) Pfam domain analysis of ChPks, including SAT, PksD, PT_fungal_PKS, two PP and EntF domain. (B) Pfam domain analysis of ChThr1, including a Rossmann-fold NAD(P)-binding domain. (C) Phylogenetic tree analysis of ChPks and its homologues in C. graminicola, C. orbiculare, V. dahliae, Gaeumannomyces tritici, Pyricularia oryzae, Drepanopeziza brunnea, B. cinerea and Sclerotinia sclerotiorum. (D) Phylogenetic tree analysis of ChThr1 and its homologues in C. fioriniae, V. alfalfae, V. dahliae, P. oryzae, S. sclerotiorum, B. cinerea and Talaromyces marneffei, along with the Thnr homologues in B. cinerea and V. dahliae.
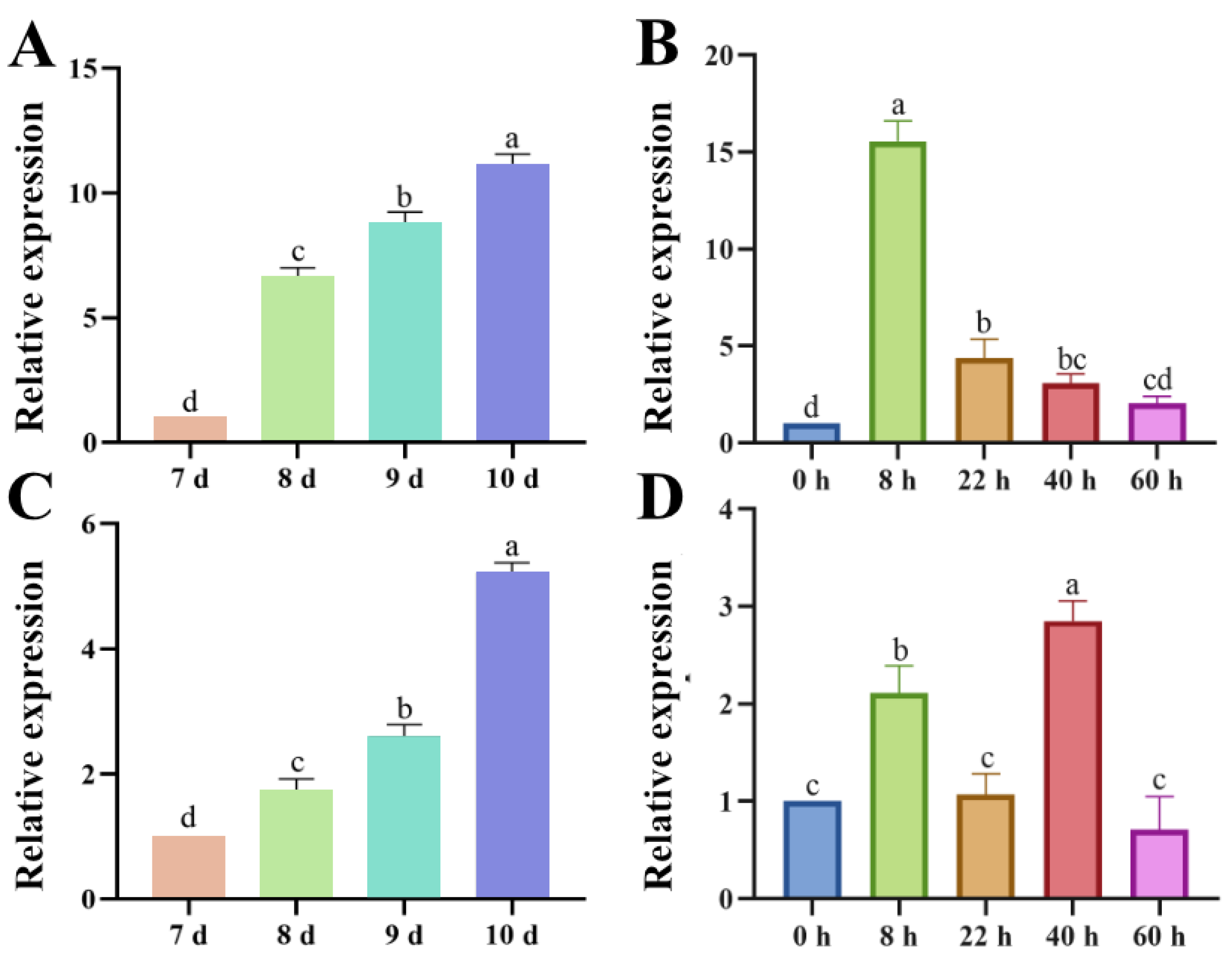
Figure 2.
Expression analysis of genes ChPKS and ChTHR1. (A,B) Expression patterns of gene ChPKS during hyphae and appressorium melanization. (C,D) Expression patterns of gene ChTHR1 during hyphae and appressorium melanization. Error bars represent standard deviations from three replicates, experimental data were analyzed by One-way ANOVA. Different letters indicating significant difference at P<0.05.
Figure 2.
Expression analysis of genes ChPKS and ChTHR1. (A,B) Expression patterns of gene ChPKS during hyphae and appressorium melanization. (C,D) Expression patterns of gene ChTHR1 during hyphae and appressorium melanization. Error bars represent standard deviations from three replicates, experimental data were analyzed by One-way ANOVA. Different letters indicating significant difference at P<0.05.
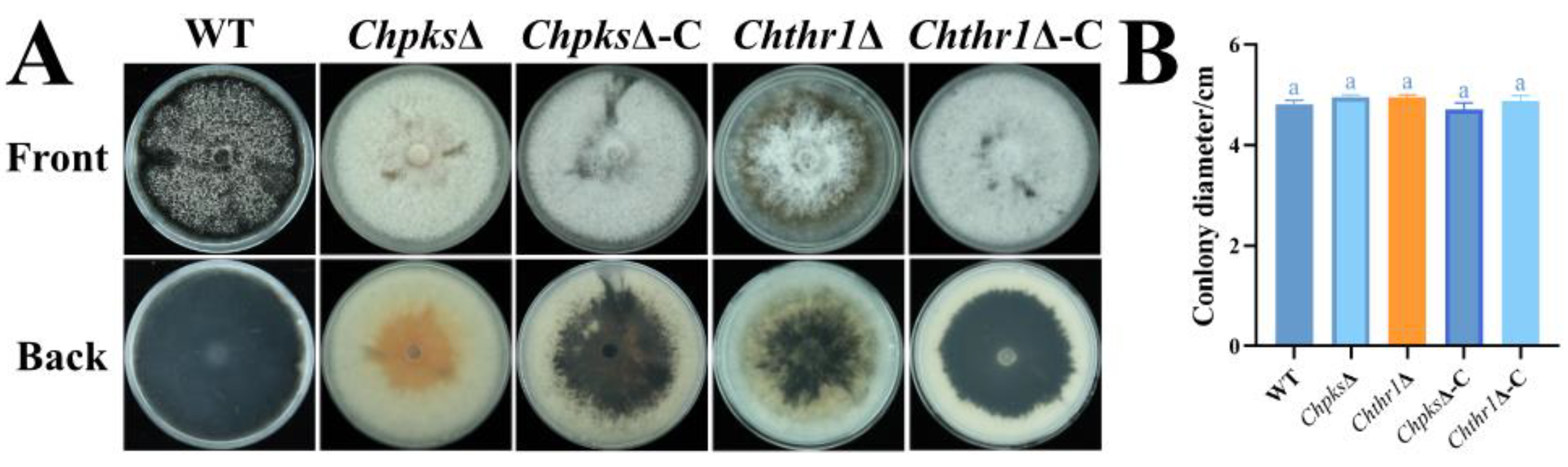
Figure 3.
Defects in hyphae melanization of the ChpksΔ and Chthr1Δ mutants. (A) Colony morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on PDA. (B) Statistical analysis of colony diameters for the indicated strains on PDA. Error bars represent standard deviations from three replicates. The experimental data ware analyzed using One-way ANOVA. Different letters indicate significant difference at P<0.05.
Figure 3.
Defects in hyphae melanization of the ChpksΔ and Chthr1Δ mutants. (A) Colony morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on PDA. (B) Statistical analysis of colony diameters for the indicated strains on PDA. Error bars represent standard deviations from three replicates. The experimental data ware analyzed using One-way ANOVA. Different letters indicate significant difference at P<0.05.
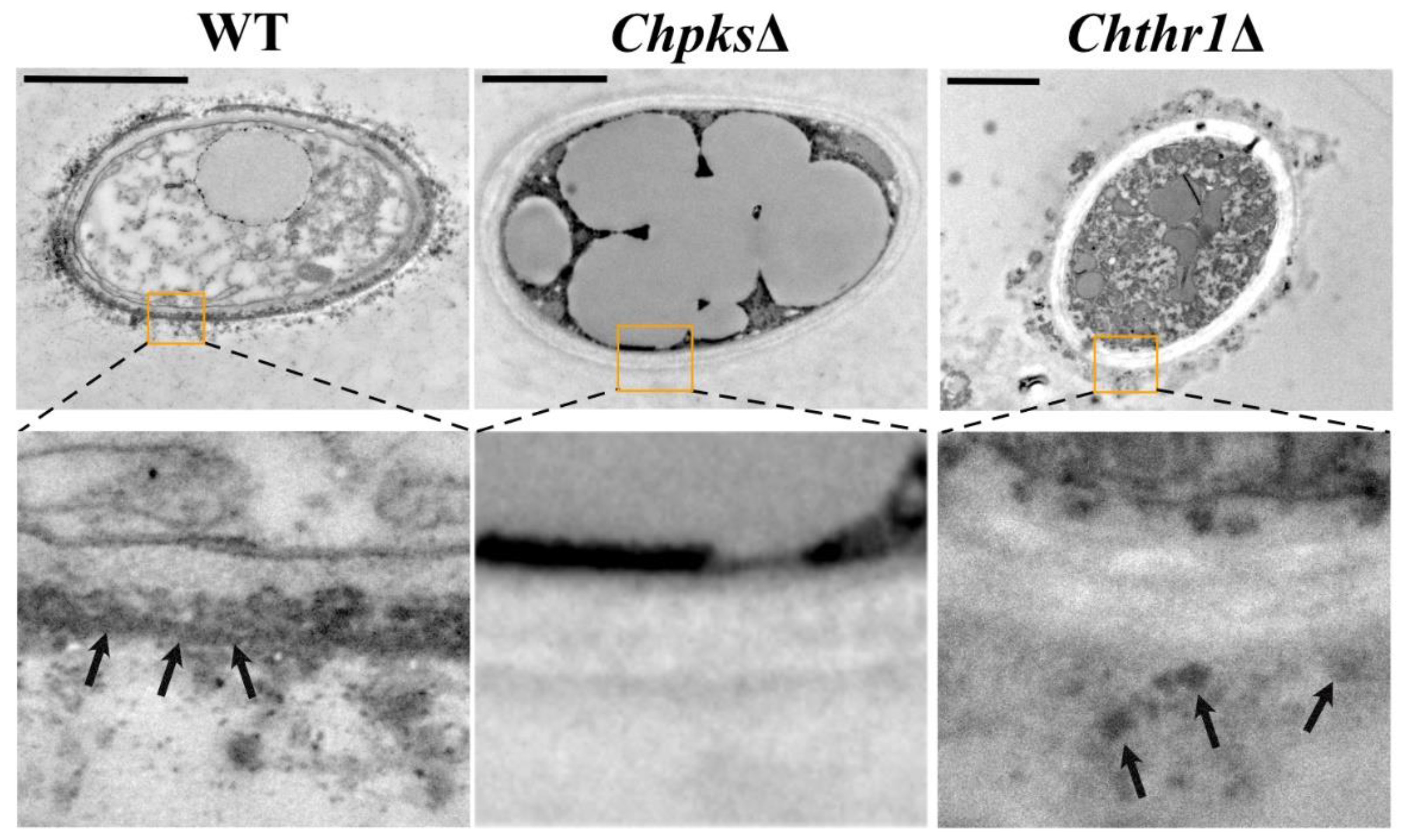
Figure 4.
TEM (transmission electron microscopy) analysis of WT, ChpksΔ and Chthr1Δ strains. The images below each show the region enclosed by the enlarged orange box. Arrows indicate cell wall melanin deposition, and bars represent a scale of 2 μm.
Figure 4.
TEM (transmission electron microscopy) analysis of WT, ChpksΔ and Chthr1Δ strains. The images below each show the region enclosed by the enlarged orange box. Arrows indicate cell wall melanin deposition, and bars represent a scale of 2 μm.
Figure 5.
Sensitivity analysis of the ChpksΔ and Chthr1Δ mutants to cell wall stresses. (A) Colony morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on PDA with 200 μg·mL-1 calcofluor white (CFW), 0.01% SDS, and 200 μg·mL-1 Congo red (CR). (B) Statistical analysis of inhibition rates of these strains under different stress conditions. Error bars represent standard deviations from three replicates. The experimental data were analyzed by One-way ANOVA. Different letters indicate significant difference at P<0.05.
Figure 5.
Sensitivity analysis of the ChpksΔ and Chthr1Δ mutants to cell wall stresses. (A) Colony morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on PDA with 200 μg·mL-1 calcofluor white (CFW), 0.01% SDS, and 200 μg·mL-1 Congo red (CR). (B) Statistical analysis of inhibition rates of these strains under different stress conditions. Error bars represent standard deviations from three replicates. The experimental data were analyzed by One-way ANOVA. Different letters indicate significant difference at P<0.05.
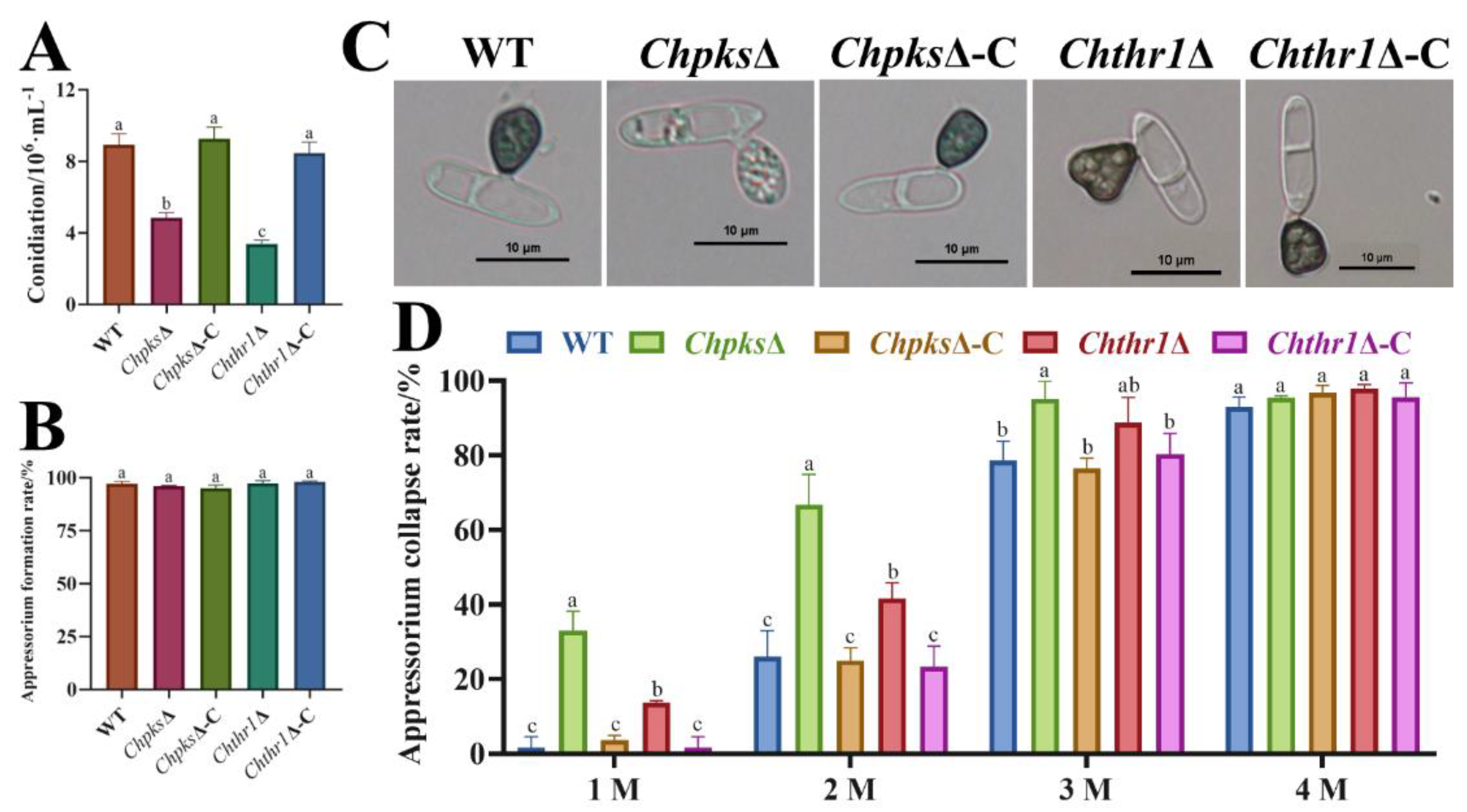
Figure 6.
Conidiation, appressorium formation, morphology, and turgor pressure of the ChpksΔ and Chthr1Δ mutants. (A) Conidiation of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on mathur medium for 5 d. (B) Appressorium formation rates of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (C) Appressorium morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (D) Appressorium collapse rates of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains under glycerol concentration from 1 M to 4 M. Error bars represent standard deviations from three replicates. The experimental data ware analyzed by One-way ANOVA. Different letters indicate significant difference at P<0.05.
Figure 6.
Conidiation, appressorium formation, morphology, and turgor pressure of the ChpksΔ and Chthr1Δ mutants. (A) Conidiation of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains grown on mathur medium for 5 d. (B) Appressorium formation rates of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (C) Appressorium morphology of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (D) Appressorium collapse rates of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains under glycerol concentration from 1 M to 4 M. Error bars represent standard deviations from three replicates. The experimental data ware analyzed by One-way ANOVA. Different letters indicate significant difference at P<0.05.
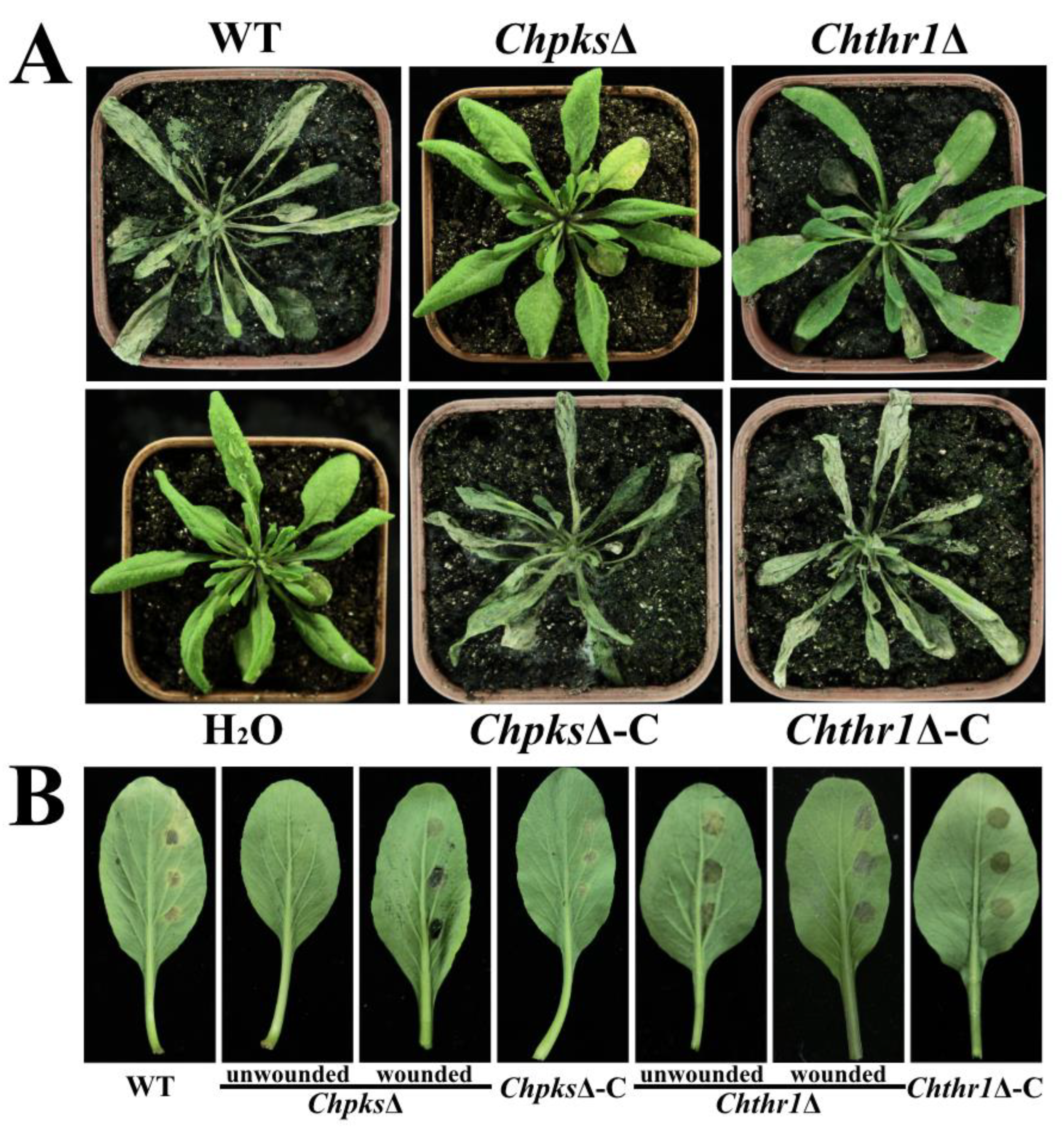
Figure 7.
Analysis of pathogenicity of the ChpksΔ and Chthr1Δ mutants. (A) Symptoms observed on Arabidopsis thaliana plants 5 days post-inoculation with conidial suspensions of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (B) Symptoms observed on detached leaves of Chinese flowering cabbage 4 days post-inoculation with conidial suspensions of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains.
Figure 7.
Analysis of pathogenicity of the ChpksΔ and Chthr1Δ mutants. (A) Symptoms observed on Arabidopsis thaliana plants 5 days post-inoculation with conidial suspensions of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains. (B) Symptoms observed on detached leaves of Chinese flowering cabbage 4 days post-inoculation with conidial suspensions of the WT, ChpksΔ, Chthr1Δ, ChpksΔ-C and Chthr1Δ-C strains.
Table 1.
Primers used in this study.
Table 1.
Primers used in this study.
| Primers |
Sequence (5’~3’) |
| ACTIN |
CCCCAAGTCCAACAGAGAGA |
| CATCAGGTAGTCGGTCAAGTCA |
| PKS-qP |
GTTCAAGTGGTTCTCATGGCTT |
| CTGGCGATGGGGATGTAATTAG |
| THR1-qP |
ACTTTGTCATCTCCAACTCGG |
| ATGGAGGAGGTGAGGAGGAT |
| THNR-qP |
GCACCATCGAGACATTTGT |
| GGTACATGTCGGTCTTGATG |
| PKS-UP |
ACGACGGCCAGTGCCAAGCTTCTGCTTTCAAACTCGTTTCACG |
| GACCTGCAGGCATGCAAGCTCAAGAACAAGATTTCTGGATGTCG |
| PKS-DS |
GACTCTAGAACTAGTGGATCCGGAACTTCATAGCGAAGCATATGA |
| AGCTCGGTACCCGGGGGATCCGGAGTATTCGGGGAAGGAGAAT |
| THR1-UP |
ACGACGGCCAGTGCCAAGCTTTTAAGCCAAAAAACGACTTGATAGA |
| GACCTGCAGGCATGCAAGCTTAGTTGCTATGAAAAGACTGTGGCG |
| THR1-DS |
CCGGGTACCGAGCTCGAATTCGTGCGATGATGCCACAAACTC |
| TATGGAGAAACTCGAGAATTCGACACCGGGAGAGGGGAGG |
| PKSc-F1 |
GAAACTCGAGCTCGAGAATTCATCGGTACCCTATCTGGCTGC |
| CCGGGTACCGAGCTCGAATTCGCTCGCTGGTTACCTCGTTCG |
| PKSc-F2 |
GACTCTAGATCTAGAGTCGACCTGCTTTCAAACTCGTTTCACG |
| CTTGCATGCCTGCAGGTCGACTTACTCGCAAACCGCTTCACG |
| PKSc-F3 |
GACCTGCAGGCATGCAAGCTTGGAACTTCATAGCGAAGCATATGA |
| DATA ACGACGGCCAGTGCCAAGCTTGTAACGACAAAAAAAGCCACAGG |
| THR1c-F1 |
GAAACTCGAGCTCGAGAATTCCTCTAGCCGAAGGTATGAACCG |
| DATA CCGGGTACCGAGCTCGAATTCCCGCCGCCACGGAGGGGC |
| THR1c-F2 |
DATA ACTCTAGATCTAGAGTCGACCGGTGGCGGCGGCATGGTCGCG |
| DATA TCACACCAGATCCGCCTGTGCCGCCACCGGTGACC |
| THR1c-F3 |
CACAGGCGGATCTGGTGTGAGCA |
| GGCATCATCGCACTTACTTGTACAGCTCGTCCATGCC |
| THR1c-F4 |
CAAGTAAGTGCGATGATGCCACAAACTC |
| DATA CTTGCATGCCTGCAGGTCGACCCGACATCCGGGGAACAC |
| PKS |
GCACCGGTACCCAGG |
| GGTAGATGGTGTCGCTGG |
| THR1 |
ATGGCGCCCTCAGCGACTGAGAA |
| CCTAGATATAAGCTATGTTAGGTATCCC |
| HPH1 |
GGGCGTCGGTTTCCACTAT |
| GATATGTCCTGCGGGTAAATAGC |
| NeoR |
GATAGAAGGCGATGCGCT |
| ACCCGGTCATACCTTCTTAAG |
| PKS-probe |
CACAGGACATTACATGACGCA |
| CACCCTTGCTTATAACTTCGCA |
| THR1-probe |
ACATAGCTTATATCTAGGACAGGCG |
| TGTTACTAAACCTAGCTTGGGAAGG |