Submitted:
05 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample collection and isolation
2.2. DNA extraction, quantification and quality
2.3. PCR amplification with the 16S rRNA gene and DNA sequencing
2.4. Bioinformatic analysis
3. Results
3.1. Sample collection and isolation
3.2. DNA quantification and quality
3.3. PCR amplification with the 16S rRNA gene and DNA sequencing
3.4. Bioinformatic analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. (2015). Technical Platform on the Measurement and Reduction of Food Loss and Waste. Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/platform-food-loss-waste/en/.
- Maksimov, I.V.; Veselova, S.V.; Nuzhnaya, T.V.; Sarvarova, E.R.; Khairullin, R.M. Plant growth-promoting bacteria in regulation of plant resistance to stress factors. Russ. J. Plant Physiol. 2015, 62, 715–726. [Google Scholar] [CrossRef]
- Gao, H.; Xu, X.; Dai, Y.; He, H. Isolation, Identification and Characterization of Bacillus subtilis CF-3, a Bacterium from Fermented Bean Curd for Controlling Postharvest Diseases of Peach Fruit. Food Sci. Technol. Res. 2016, 22, 377–385. [Google Scholar] [CrossRef]
- Aouadhi, C.; Rouissi, Z.; Kmiha, S.; Mejri, S.; Maaroufi, A. Effect of sporulation conditions on the resistance of Bacillus sporothermodurans spores to nisin and heat. Food Microbiol. 2016, 54, 6–10. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Shurigin, V.V.; Hashem, A.; Abd_Allah, E.F. Endophytic Bacteria Improve Plant Growth, Symbiotic Performance of Chickpea (Cicer arietinum L.) and Induce Suppression of Root Rot Caused by Fusarium solani under Salt Stress. Front. Microbiol. 2017, 8, 1887. [Google Scholar] [CrossRef] [PubMed]
- Lastochkina, O.; Pusenkova, L.; Yuldashev, R.; Babaev, M.; Garipova, S.; Blagova, D.; Khairullin, R.; Aliniaeifard, S. Effects of Bacillus subtilis on some physiological and biochemical parameters of Triticum aestivum L. (wheat) under salinity. Plant Physiol. Biochem. 2017, 121, 80–88. [Google Scholar] [CrossRef]
- Larrea-Izurieta, I.; Borja, C.F.; Arcos-Andrade, A. Aislamiento y caracterización de cepas de Bacillus spp. Con actividad contra Tetranychus urticae Koch en cultivos comerciales de rosas. Revista Colombiana de Biotecnología 2015, 17, 140–148. [Google Scholar] [CrossRef]
- Valdez-Nuñez, R.A.; Ríos-Ruiz, W.F.; Ormeño-Orrillo, E.; Torres-Chávez, E.E.; Torres-Delgado, J. Caracterización genética de bacterias endofíticas de arroz (Oryza sativa L.) con actividad antimicrobiana contra Burkholderia glumae. Revista Argentina de Microbiología 2020, 52, 315–327. [Google Scholar] [CrossRef]
- Huertas, D. (2013). Caracterizacion molecular de cepas de Bacillus thuringiensis con propiedades entomocidas, para vectores transmisores de enfermedades metaxénicas (dengue) [Tesis de doctorado, Universidad Nacional Mayor de San Marcos]. Available online: https://hdl.handle.net/20.500.12672/3513.
- Rodríguez-Tolosaa, R.; Cifuentes-Vega, R.; Hernández-Fernández, J. Caracterización de cepas nativas de Bacillus thuringiensis como método para predecir la susceptibilidad sobre insectos lepidópteros, dípteros y coleópteros plaga de la agricultura. Rev. Mutis 2023, 13, 1–34. [Google Scholar] [CrossRef]
- Clark, D. P., Pazdernik, N. J., & McGehee, M. R. (2019). Chapter 6—Polymerase Chain Reaction. En D. P. Clark, N. J. Pazdernik, & M. R. McGehee (Eds.), Molecular Biology (Third Edition) (Third Edition, pp. 168-198). Academic Cell. [CrossRef]
- Suárez-Contreras, L.Y.; Yañez-Meneses, L.F. 16S rRNA as an applied tool in the molecular characterization of genera and species of bacteria. Respuestas 2020, 25, 127–137. [Google Scholar] [CrossRef]
- Cattani, F.; Barth, V.C.; Nasário, J.S.; Ferreira, C.A.; Oliveira, S.D. Detection and quantification of viable Bacillus cereus group species in milk by propidium monoazide quantitative real-time PCR. J. Dairy Sci. 2016, 99, 2617–2624. [Google Scholar] [CrossRef]
- Tae, H.; Karunasena, E.; Bavarva, J.H.; Garner, H.R. Updating microbial genomic sequences: improving accuracy & innovation. BioData Min. 2014, 7, 25. [Google Scholar] [CrossRef]
- Rahman, M.Z.; Uematsu, S.; Coffey, M.D.; Uzuhashi, S.; Suga, H.; Kageyama, K. Re-evaluation of Japanese Phytophthora isolates based on molecular phylogenetic analyses. Mycoscience 2014, 55, 314–327. [Google Scholar] [CrossRef]
- Lorenz, L.; Lins, B.; Barrett, J.; Montgomery, A.; Trapani, S.; Schindler, A.; Christie, G.E.; Cresawn, S.G.; Temple, L. Genomic characterization of six novel Bacillus pumilus bacteriophages. Virology 2013, 444, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.S.W. Phylogenetic relationships between Bacillus species and related genera inferred from 16s rDNA sequences. Braz. J. Microbiol. 2009, 40, 505–521. [Google Scholar] [CrossRef]
- Mendoza, J. (2022). Caracterizacion molecular de las principales plagas y enfermedades en el cultivo de cacao en la zona nororiental del Perú [Tesis de maestría, Universidad Nacional Toribio Rodríguez de Mendoza]. Available online: https://hdl.handle.net/20.500.14077/2842.
- Chávez-Ambriz, L.A.; Hernández-Morales, A.; Cabrera-Luna, J.A.; Luna-Martínez, L.; Pacheco-Aguilar, J.R. Aislados de Bacillus provenientes de la rizósfera de cactus incrementan la germinación y la floración en Mammillaria spp. (Cactaceae). 2016, 48, 333–341. [Google Scholar] [CrossRef]
- Erban, T.; Nesvorna, M.; Erbanova, M.; Hubert, J. Bacillus thuringiensis var. tenebrionis control of synanthropic mites (Acari: Acaridida) under laboratory conditions. Exp. Appl. Acarol. 2009, 49, 339–346. [Google Scholar] [CrossRef]
- Koilybayeva, M.; Shynykul, Z.; Ustenova, G.; Abzaliyeva, S.; Alimzhanova, M.; Amirkhanova, A.; Turgumbayeva, A.; Mustafina, K.; Yeleken, G.; Raganina, K.; et al. Molecular Characterization of Some Bacillus Species from Vegetables and Evaluation of Their Antimicrobial and Antibiotic Potency. Molecules 2023, 28, 3210. [Google Scholar] [CrossRef]
- Zevallos, M.A.F.; Castañeda, A.; Toledo, O.; Caballero, D.; Cueva, M.; Cedeño, V. Caracterización molecular ómica de una cepa de Bacillus amyloliquefaciens aislada de la microbiota del paiche Arapaima gigas con actividad antagonista contra bacterias patógenas de peces. Revista de Investigaciones Veterinarias del Perú 2019, 3, 90–922. [Google Scholar] [CrossRef]
- Florido, G. M., Rondón, A. J., Pérez, M., Arteaga, F., Bocourt, R., Portilla, Y., Rodríguez, M., Pérez, Y., Beruvides, A., & Laurencio, M. (2017). Methodology for the isolation, identification and selection of Bacillus spp. Strains for the preparation of animal additives Metodología para el aislamiento, identificación y selección de cepas de Bacillus spp. Para la elaboración de aditivos zootécnicos. Cuban Journal of Agricultural Science, 51(2).
- Liu, C.; Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, Y.; Zhang, J.; Liao, X.; Li, C.; Wu, S.; et al. Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiol. 2020, 20, 310. [Google Scholar] [CrossRef]
- Rodríguez-Sahagún, A.; Velasco-Jiménez, A.; Castellanos-Hernández, O.; Acevedo-Hernández, G.; Aarland, R.C. Bacterias rizosféricas con beneficios potenciales en la agricultura. Rev. Terra Latinoam. 2020, 38, 333–345. [Google Scholar] [CrossRef]
- Abdulsalam, R.A.; Ijabadeniyi, O.A.; Cason, E.D.; Sabiu, S. Characterization of Microbial Diversity of Two Tomato Cultivars through Targeted Next-Generation Sequencing 16S rRNA and ITS Techniques. Microorganisms 2023, 11, 2337. [Google Scholar] [CrossRef] [PubMed]
- Galvis, F., & Moreno, L. (2014). Caracterizacion molecular mediante rep-PCR de asilados nativos de Bacillus thuringiensis, obtenidos de muestras de suelo. Agronomía Costarricense, 1(38), 223-229. Available online: https://www.scielo.sa.cr/scielo.php?script=sci_arttext&pid=S0377-94242014000100016.
- Galvis, F.; Carrillo, M. Identificación y Caracterización Molecular de Aislados de Burkholderia glumae: Agente Causante del Añublo Bacterial en el Cultivo de Arroz. Información tecnológica 2015, 26, 33–40. [Google Scholar] [CrossRef]
- Ciuffreda, L.; Rodríguez-Pérez, H.; Flores, C. Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 2021, 19, 1497–1511. [Google Scholar] [CrossRef] [PubMed]
- Kumburu, H.H.; Shayo, M.; van Zwetslaar, M.; Njau, J.; Kuchaka, D.J.; Ignas, I.P.; Wadugu, B.; Kasworm, R.; Masaki, L.J.; Hallgren, M.B.; et al. Nanopore sequencing technology for clinical diagnosis of infectious diseases where laboratory capacity is meager: A case report. Heliyon 2023, 9, e17439. [Google Scholar] [CrossRef]
- Elegbeleye, J.; Buys, E. Molecular characterization and biofilm formation potential of Bacillus subtilis and Bacillus velezensis in extended shelf-life milk processing line. J. Dairy Sci. 2020, 103, 4991–5002. [Google Scholar] [CrossRef]
- Romo-Barrera, C.M.; Castrillón-Rivera, L.E.; Palma-Ramos, A.; Castañeda-Sánchez, J.I.; Luna-Herrera, J. Bacillus licheniformis and Bacillus subtilis, Probiotics That Induce the Formation of Macrophage Extracellular Traps. Microorganisms 2021, 9, 2027. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Ma, J.; Yin, Z.; Liu, K.; Yao, G.; Xu, W.; Fan, L.; Du, B.; Ding, Y.; Wang, C. Comparative genomic analysis of Bacillus paralicheniformis MDJK30 with its closely related species reveals an evolutionary relationship between B. paralicheniformis and B. licheniformis. BMC Genom. 2019, 20, 283. [Google Scholar] [CrossRef]
- Valero, M.; Hernández-Herrero, L.; Fernández, P.; Salmerón, M. Characterization of Bacillus cereus isolates from fresh vegetables and refrigerated minimally processed foods by biochemical and physiological tests. Food Microbiol. 2002, 19, 491–499. [Google Scholar] [CrossRef]
- Ying, T.; Wu, P.; Gao, L.; Wang, C.; Zhang, T.; Liu, S.; Huang, R. Isolation and characterization of a new strain of Bacillus amyloliquefaciens and its effect on strawberry preservation. LWT 2022, 165. [Google Scholar] [CrossRef]
- Rovira, A. G. P. (2014). Aislamiento y caracterización de cepas nativas de Bacillus thuringiensis con miras al desarrollo de un bioinsecticida [Tesis de licenciatura, Universidad ORT]. Available online: https://www.colibri.udelar.edu.uy/jspui/handle/20.500.12008/1593.
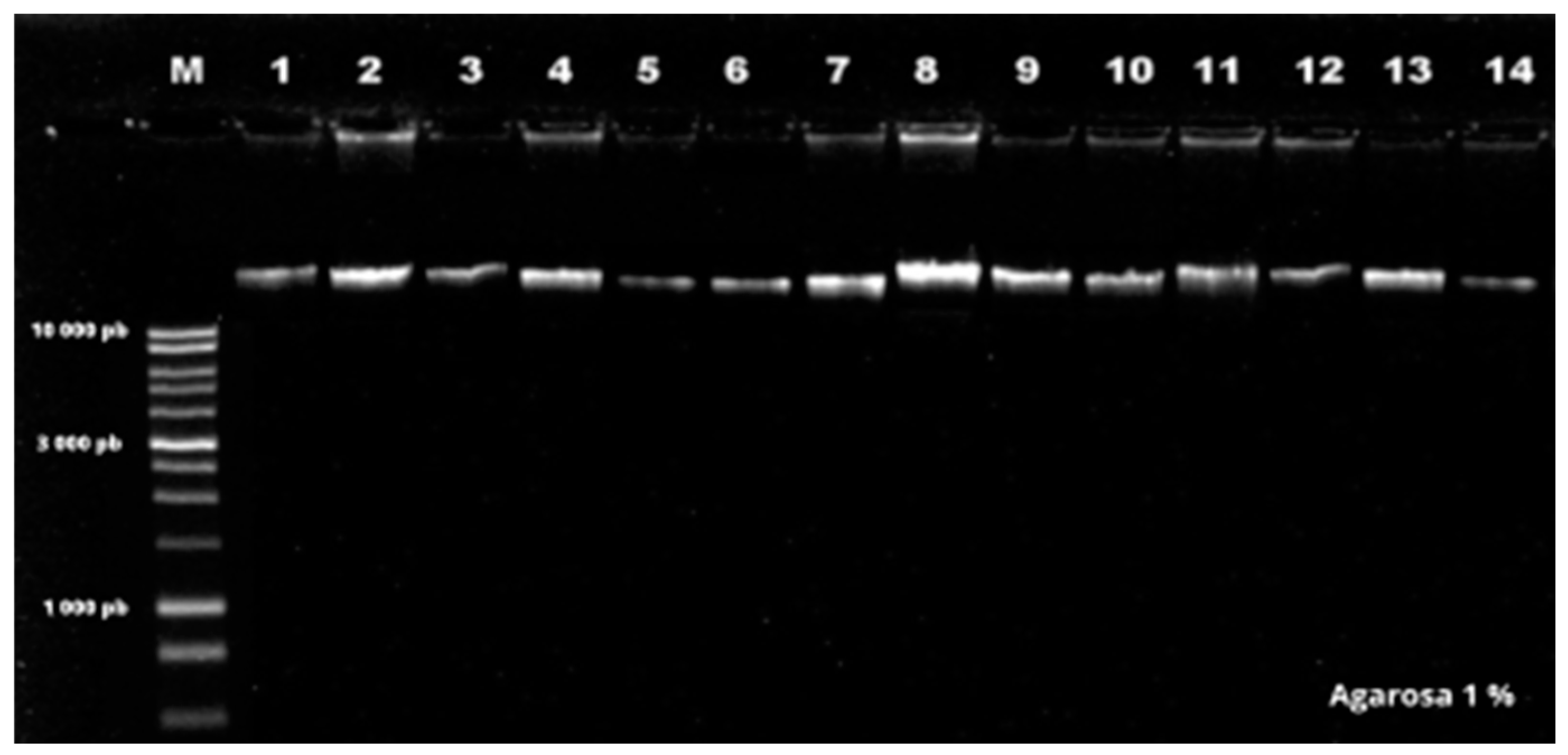
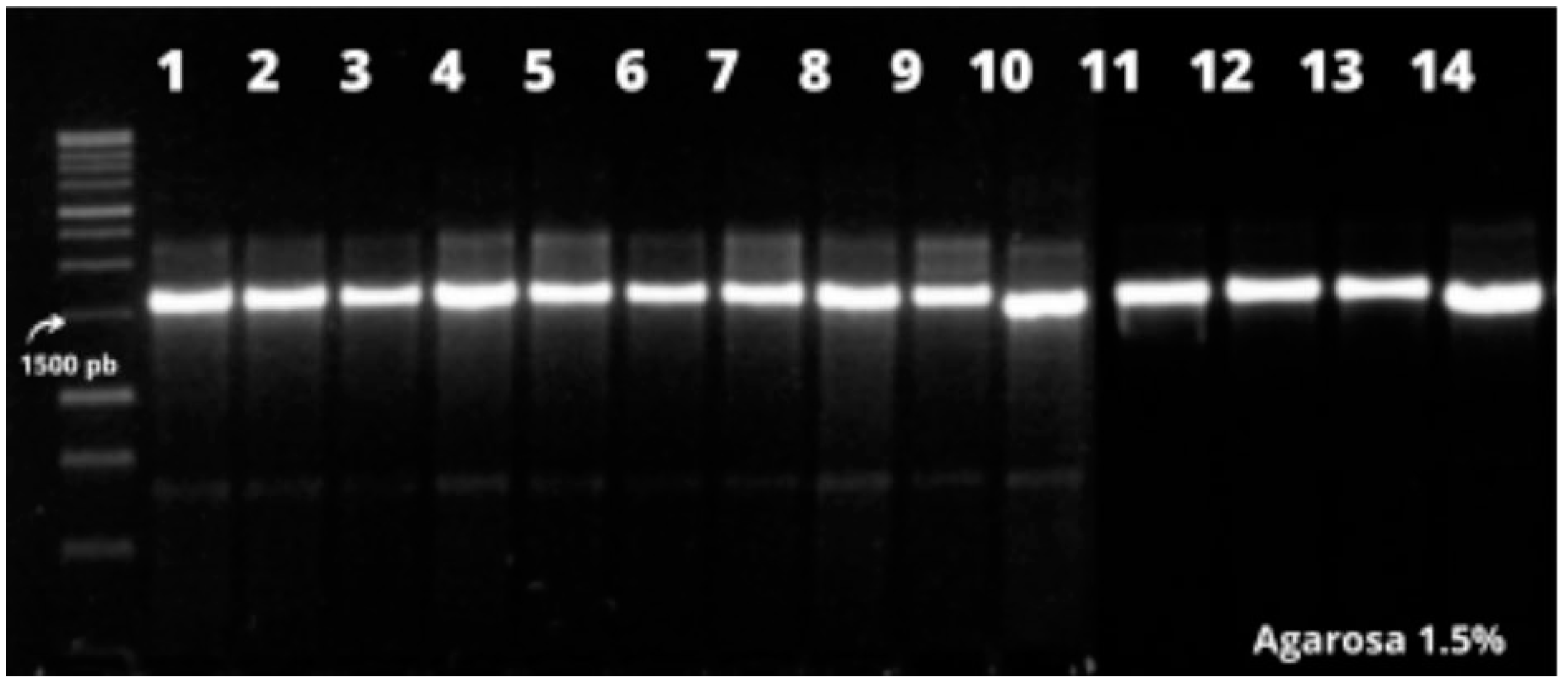
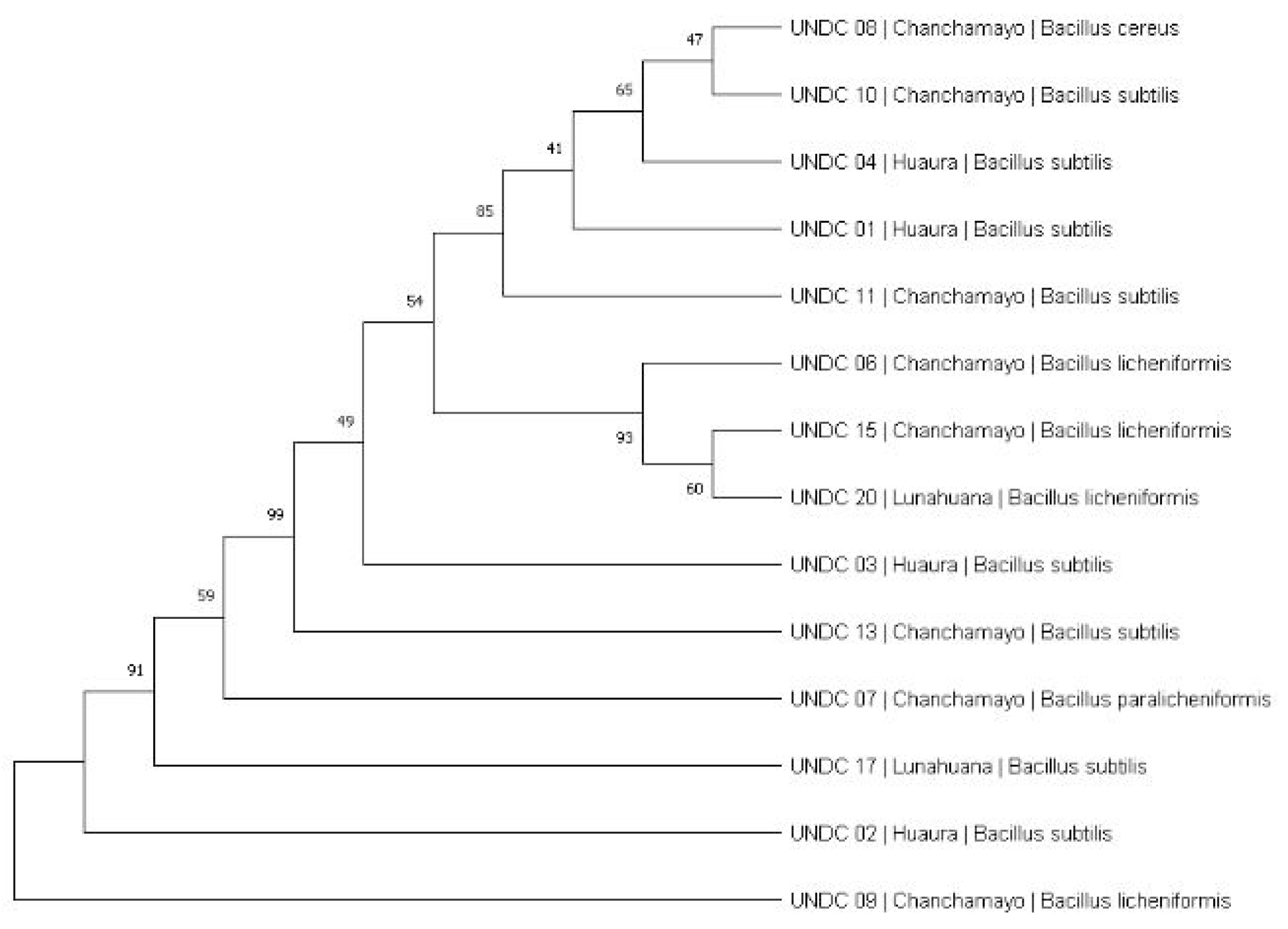
| Code | Species | Location | Identify |
| UNDC_01 | Bacillus subtilis | Huaura | 98,86% |
| UNDC_02 | Bacillus subtilis | Huaura | 95,79% |
| UNDC_03 | Bacillus subtilis | Huaura | 98,09% |
| UNDC_04 | Bacillus subtilis | Huaura | 100,00% |
| UNDC_05 | Bacillus licheniformis | Huaura | 98,18% |
| UNDC_06 | Bacillus paralicheniformis | Huaura | 95,38% |
| UNDC_07 | Bacillus cereus | Huaura | 99,79% |
| UNDC_08 | Bacillus licheniformis | Chanchamayo | 92,05% |
| UNDC_09 | Bacillus subtilis | Chanchamayo | 99,56% |
| UNDC_10 | Bacillus subtilis | Chanchamayo | 98,28% |
| UNDC_11 | Bacillus subtilis | Chanchamayo | 97,91% |
| UNDC_12 | Bacillus licheniformis | Chanchamayo | 97,15% |
| UNDC_13 | Bacillus subtilis | Lunahuaná | 94,05% |
| UNDC_14 | Bacillus licheniformis | Lunahuaná | 98,93% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





