Submitted:
04 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- A layer structure of MWNNs is designed and optimization is performed through integrated neuro-evolution based heuristic with IPAS to solve the PDM numerically.
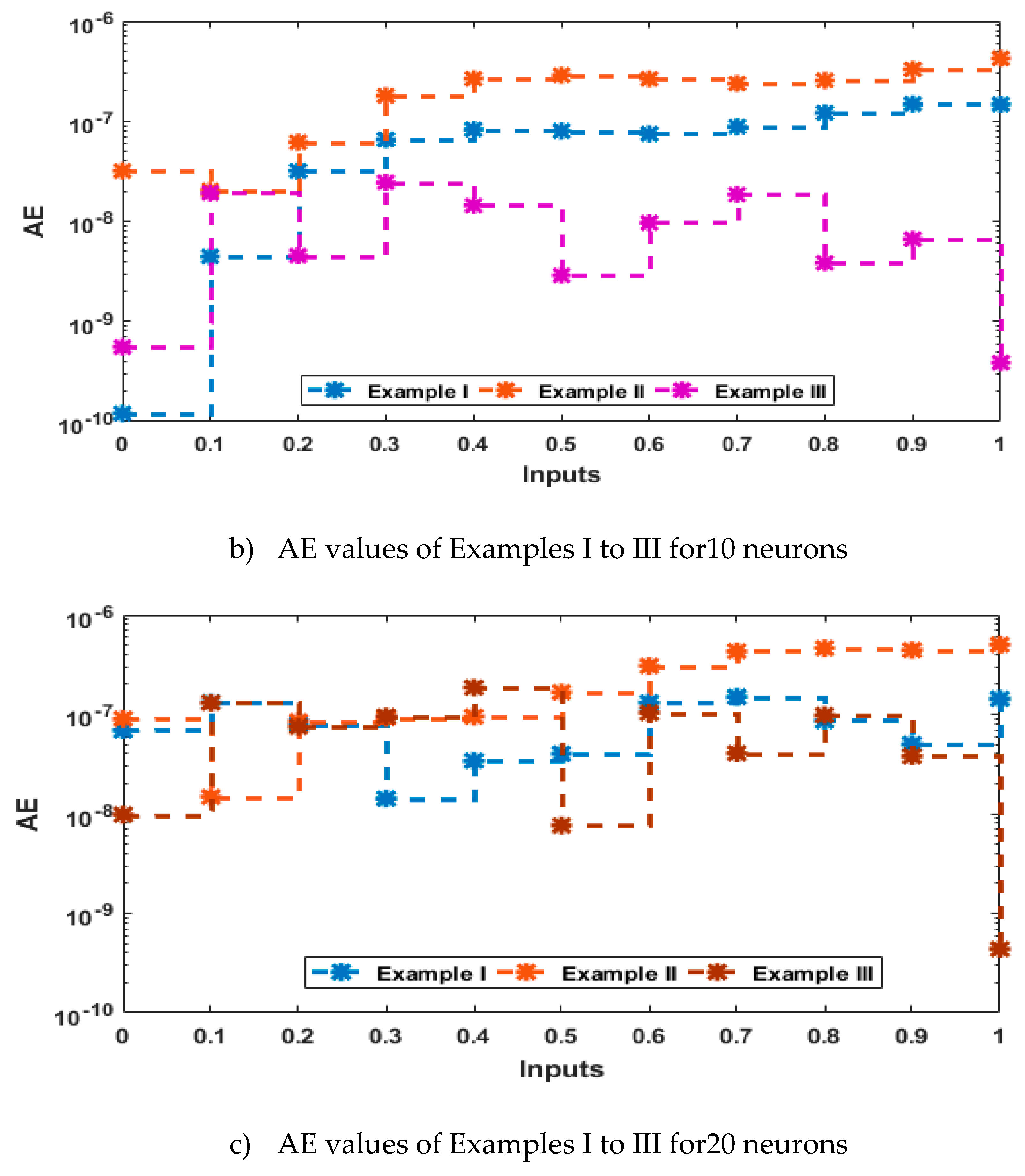
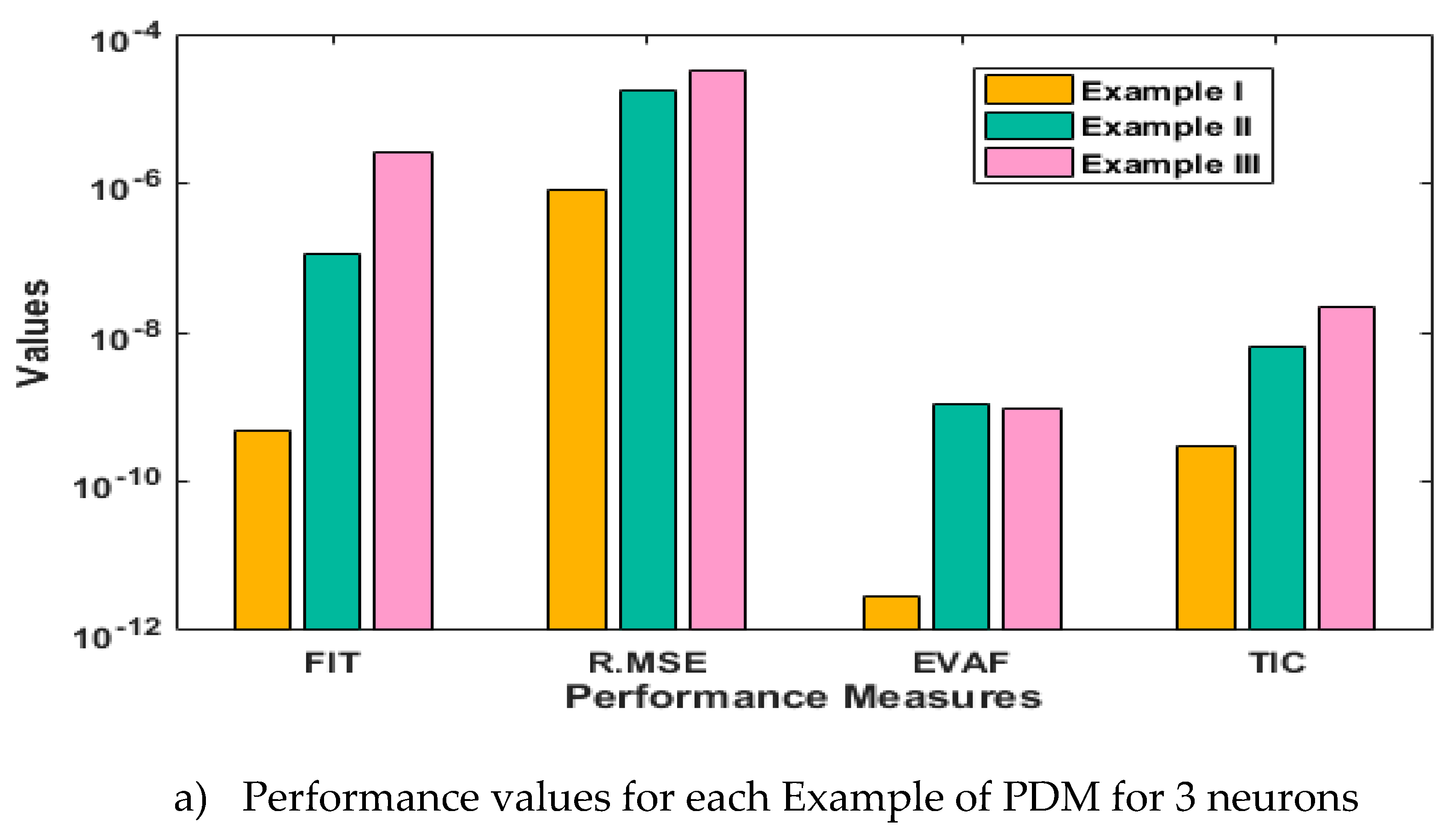
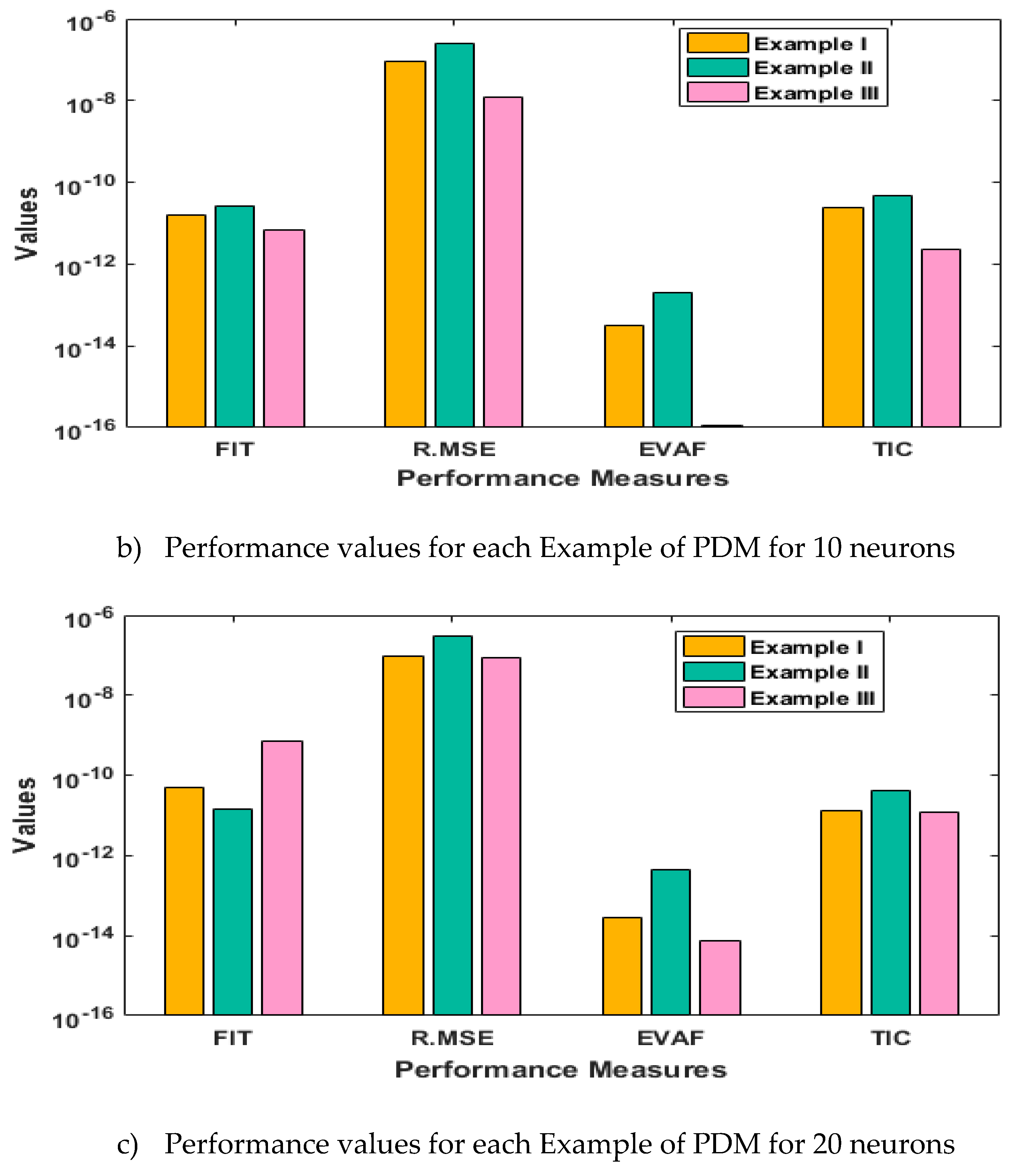
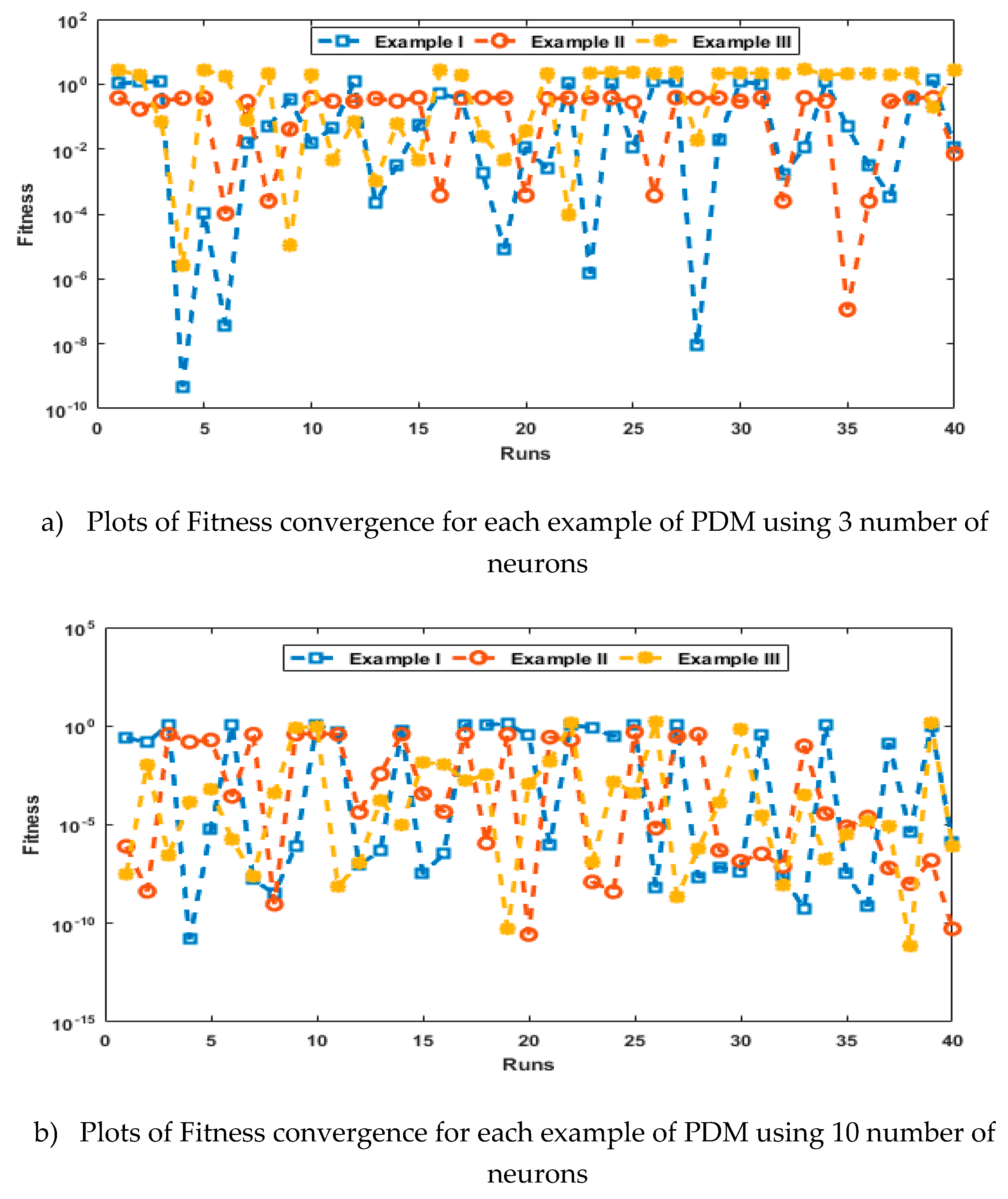
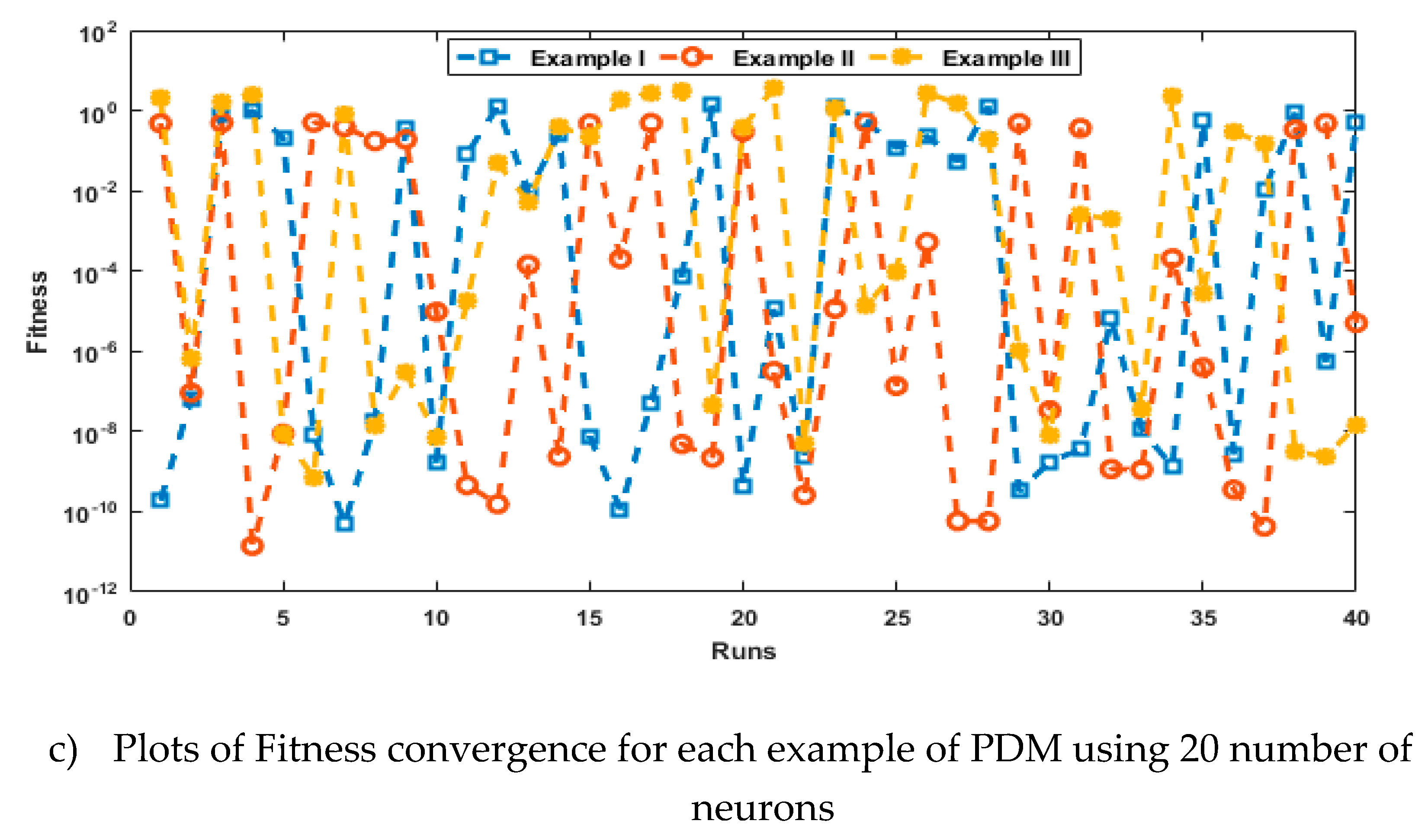
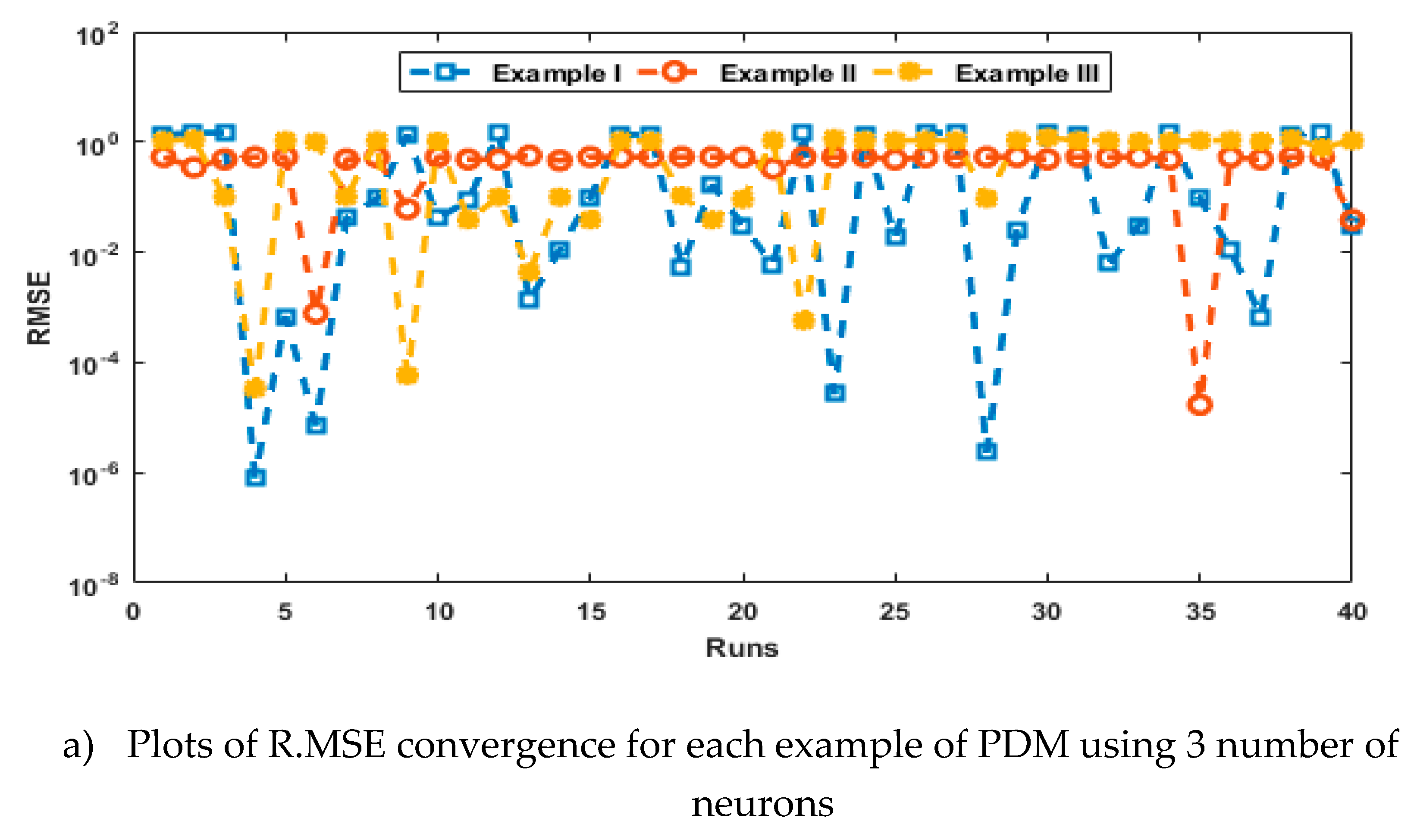
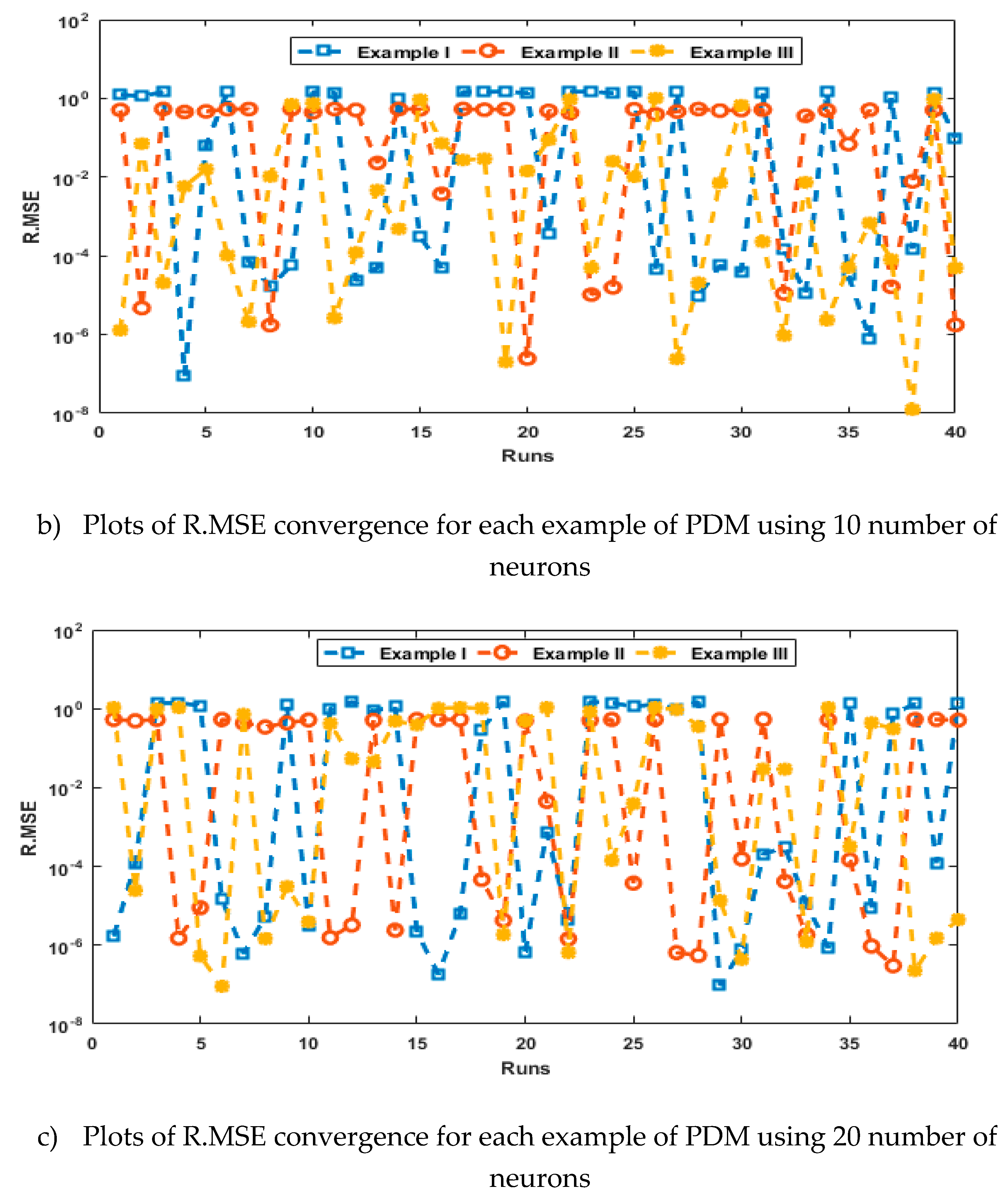
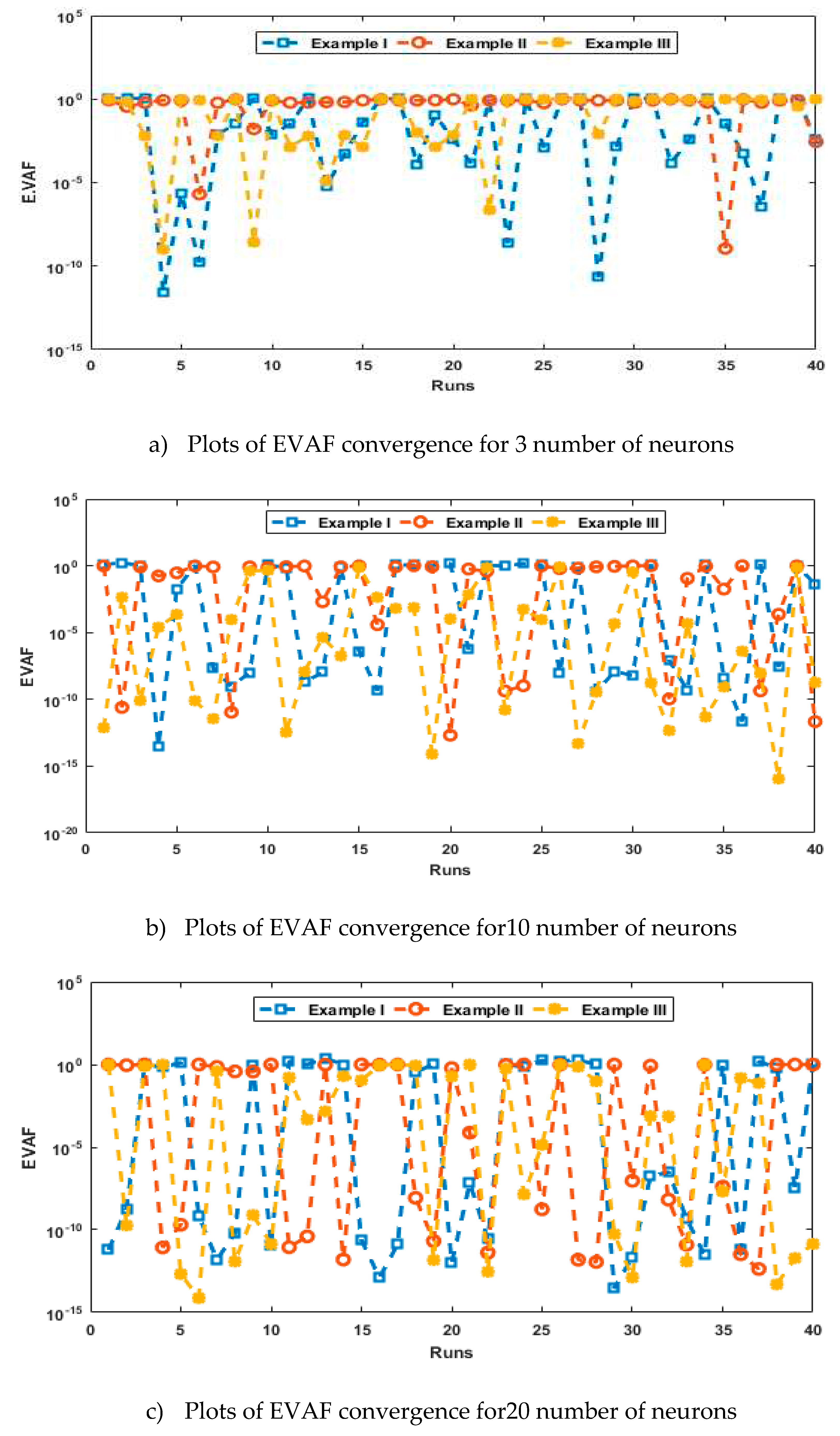
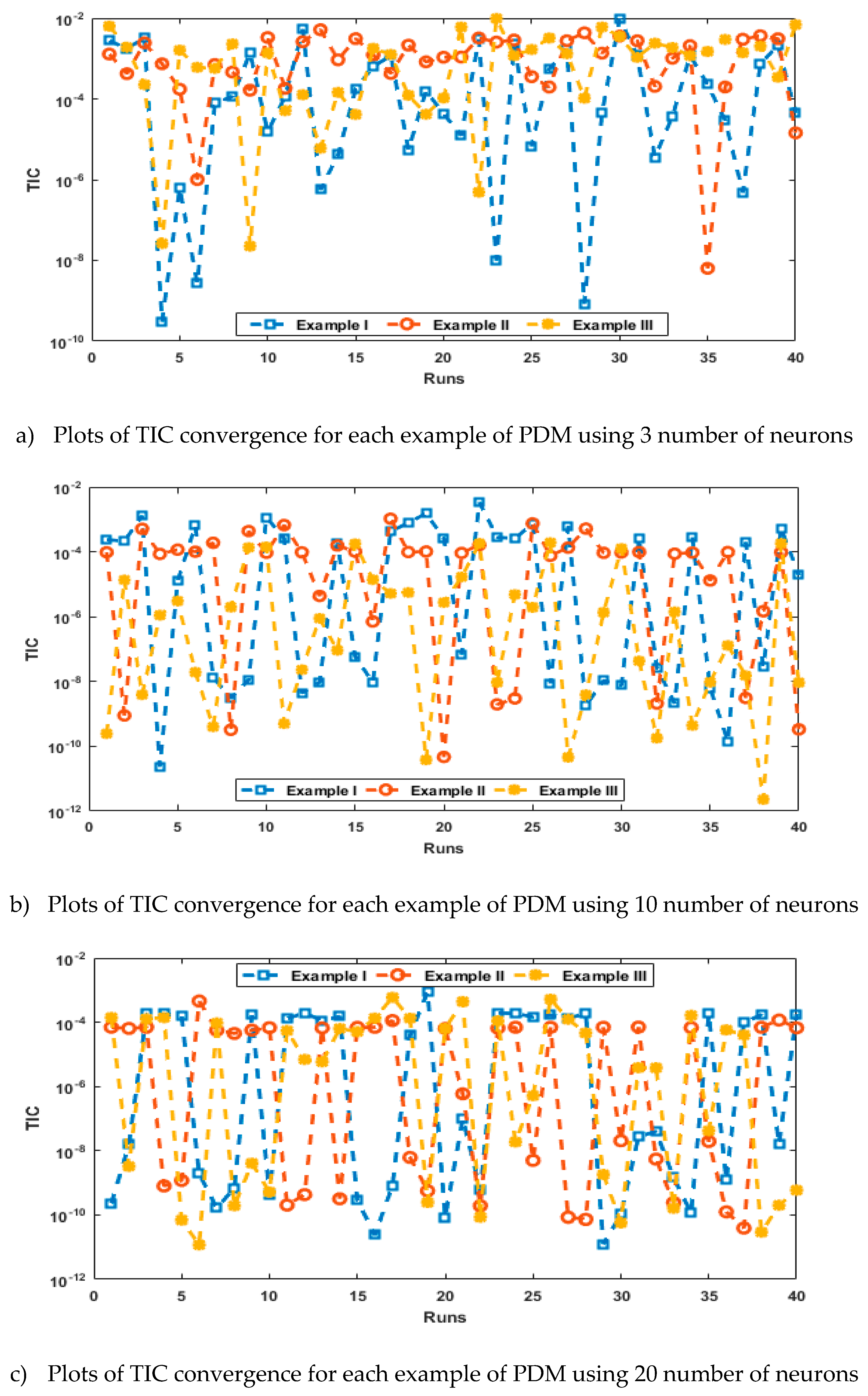
- The analysis with 3, 10 and 20 numbers of neurons is presented to interpret the stability and accuracy of the designed approach for solving the PDM.
- The proposed MWNN-GAIPAS is executed for three different examples based on PDMand comparison is performed with the exact solutions to validate the accurateness of proposed MWNN-GAIPAS.
- Statistics investigations through different performances of fitness, “root mean square error (R.MSE)”, “variance account for (VAF)”, “Theil’s inequality coefficients (TIC)” and semi inter quartile range (S.I.R) further authenticate the MWNN-GAIPAS for solving all examples of the PDM.
- The complexity performance of the MWNN-GAIPAS based on 3, 10 and 20 numbers of neurons using different statistical operators is examined for all the examples of the PDM.
- The proposed MWNN-GAIPAS provides reasonable and accurate results in training span. Furthermore, smooth processes of implementation, constancy, and expendability are other obvious applauses.
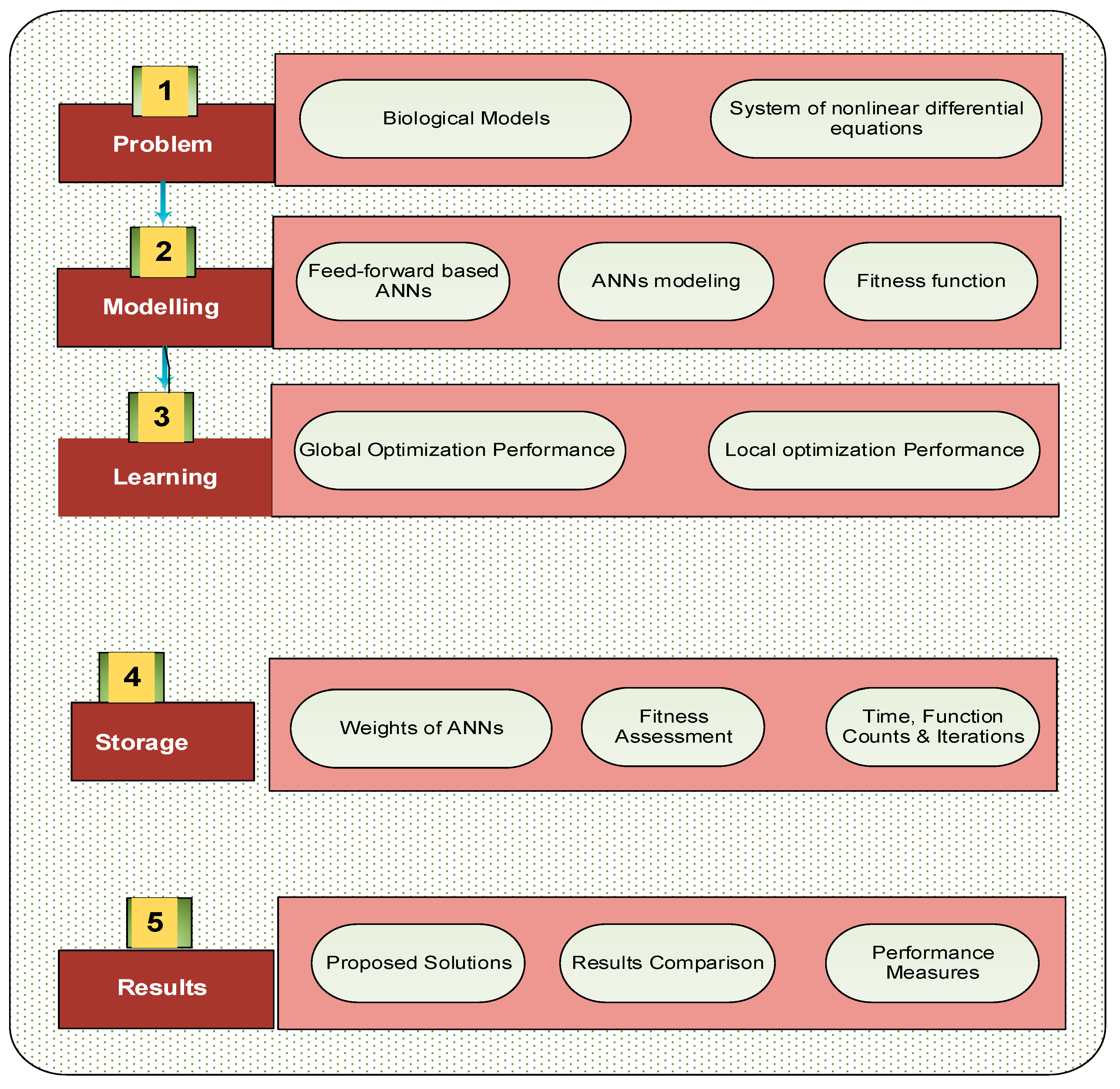
2. Methodology: MWNN-GAIPAS
- An error-based merit function is presented to construct the MWNNs.
- For the optimization of the merit function, the hybrid form of GAIPAS is described for the decision variables of MWNNs.
2.1. MWNN Modeling
2.2. Optimization process: GAIPAS
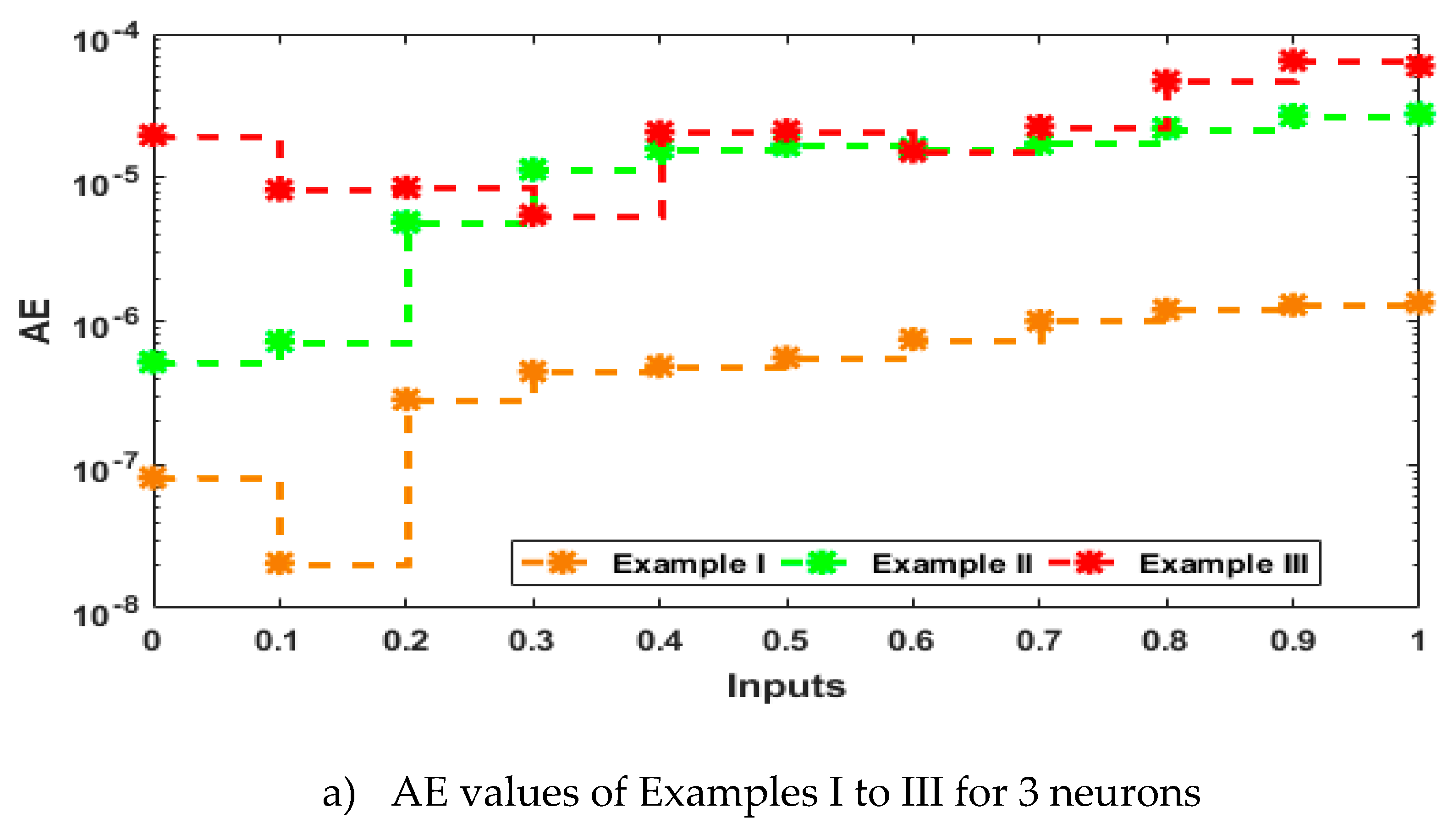
3. Statistical performances
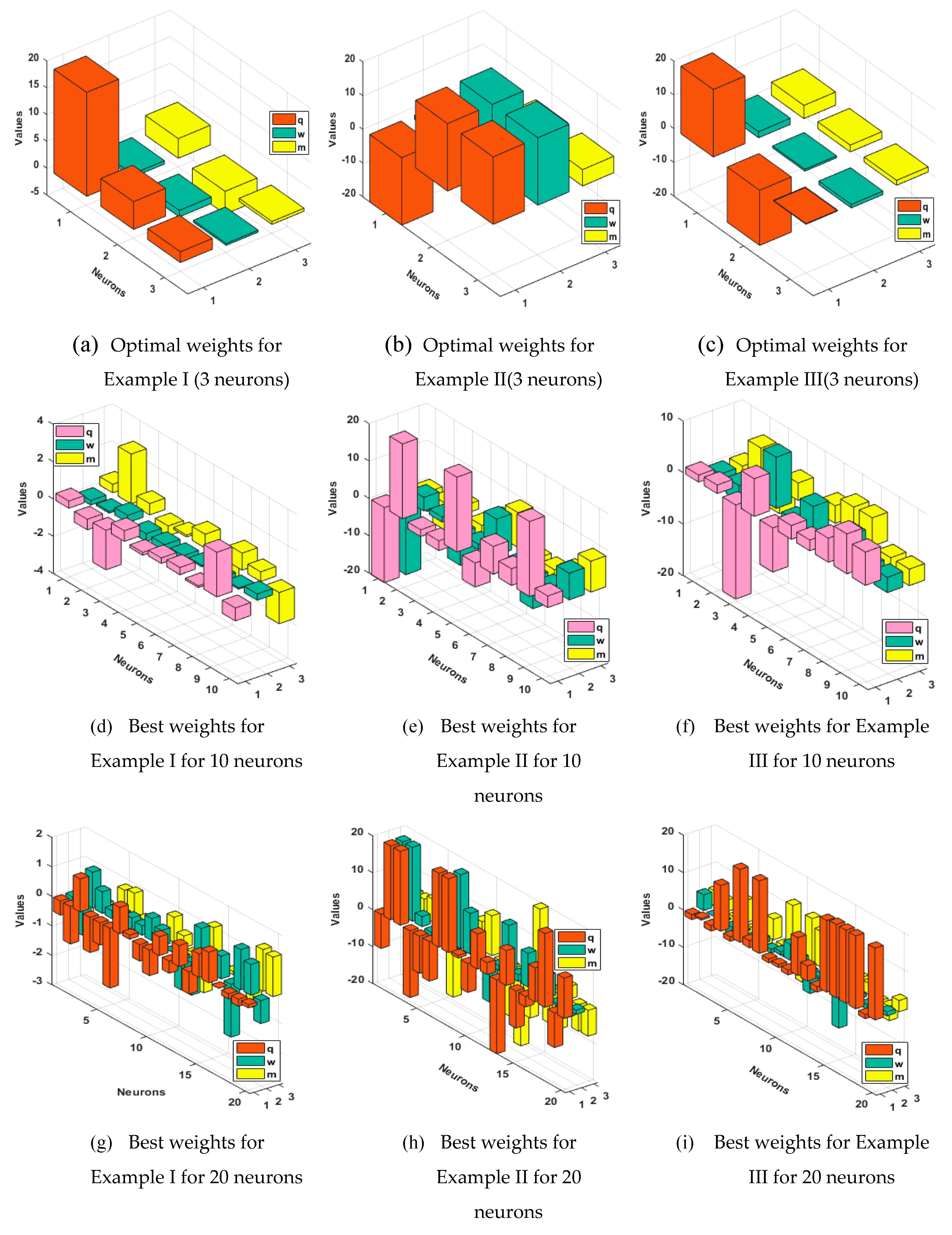
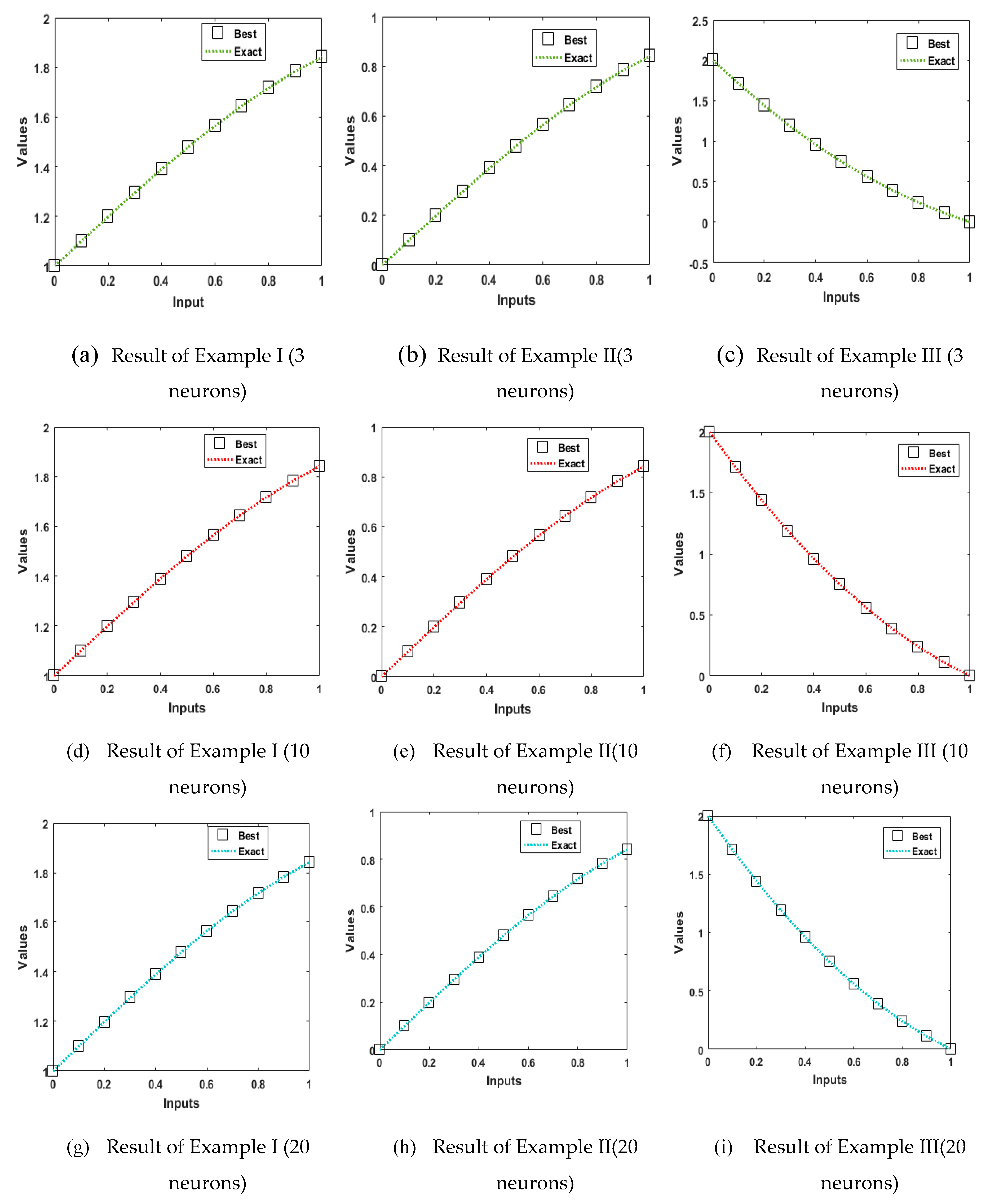
4. Simulations of the results
5. Conclusion
Funding
Conflicts of Interest
References
- Niculescu, S.I., 2001. Delay effects on stability: a robust control approach (Vol. 269). Springer Science & Business Media.
- Zulqurnain Sabir et al., 2021, Design of neuro-swarming heuristic solverfor multi-pantograph singular delay differential equation, Fractals. [CrossRef]
- Guirao, J.L. et al. 2020. Design and numerical solutions of a novel third-order nonlinear Emden–Fowler delay differential model. Mathematical Problems in Engineering, 2020.
- Sabir, Z. et al, 2021. Solving a novel designed second order nonlinear Lane-Emden delay differential model using the heuristic techniques. Applied Soft Computing, p.107105.
- Abdelkawy, M.A. et al., 2020. Numerical investigations of a new singular second-order nonlinear coupled functional Lane–Emden model. Open Physics, 18(1), pp.770-778.
- Bildik, N. and Deniz, S., 2017. A new efficient method for solving delay differential equations and a comparison with other methods. The European Physical Journal Plus, 132(1), pp.1-11.
- Rahimkhani, P., Ordokhani, Y. and Babolian, E., 2017. A new operational matrix based on Bernoulli wavelets for solving fractional delay differential equations. Numerical Algorithms, 74(1), pp.223-245.
- Sabir, Z. et al., 2020. On a new model based on third-order nonlinear multisingular functional differential equations. Mathematical Problems in Engineering, 2020.
- Aziz, I. and Amin, R., 2016. Numerical solution of a class of delay differential and delay partial differential equations via Haar wavelet. Applied Mathematical Modelling, 40(23-24), pp.10286-10299.
- Frazier, M.W., 1999. Background: Complex Numbers and Linear Algebra. An Introduction to Wavelets through Linear Algebra, pp.7-100.
- Tomasiello, S., 2017. An alternative use of fuzzy transform with application to a class of delay differential equations. International Journal of Computer Mathematics, 94(9), pp.1719-1726.
- Vaid, M.K. and Arora, G., 2019. Solution of second order singular perturbed delay differential equation using trigonometric B-spline. International Journal of Mathematical, Engineering and Management Sciences, 4(2), pp.349-360.
- Hashemi, M.S., Atangana, A. and Hajikhah, S., 2020. Solving fractional pantograph delay equations by an effective computational method. Mathematics and Computers in Simulation, 177, pp.295-305.
- Adel, W. and Sabir, Z., 2020. Solving a new design of nonlinear second-order Lane–Emden pantograph delay differential model via Bernoulli collocation method. The European Physical Journal Plus, 135(6), p.427.
- Erdogan, F et al., 2020. A finite difference method on layer-adapted mesh for singularly perturbed delay differential equations. Applied Mathematics and Nonlinear Sciences, 5(1), pp.425-436.
- Seong, H.Y. and Majid, Z.A., 2017. Solving second order delay differential equations using direct two-point block method. Ain Shams Engineering Journal, 8(1), pp.59-66.
- Sabir, Z. et al, 2020,. Design of a novel second order prediction differential model and solved by using Adams and explicit Runge-Kutta numerical methods. Volume 2020 Article ID 9704968 . [CrossRef]
- Sabir, Z., et al., 2020. Integrated neuro-evolution heuristic with sequential quadratic programming for second-order prediction differential models. Numerical Methods for Partial Differential Equations.
- Umar, M., Amin, F., Wahab, H.A. and Baleanu, D., 2019. Unsupervised constrained neural network modeling of boundary value corneal model for eye surgery. Applied Soft Computing, 85, p.105826.
- Raja, M.A.Z. et al, 2019. Numerical solution of doubly singular nonlinear systems using neural networks-based integrated intelligent computing. Neural Computing and Applications, 31(3), pp.793-812.
- Sabir, Z., et al, 2020. Intelligence computing approach for solving second order system of Emden–Fowler model. Journal of Intelligent & Fuzzy Systems, pp.1-16.
- Umar, M. et al., 2020. A stochastic numerical computing heuristic of SIR nonlinear model based on dengue fever. Results in Physics, 19, p.103585.
- Sabir, Z. et al., 2019. Stochastic numerical approach for solving second order nonlinear singular functional differential equation. Applied Mathematics and Computation, 363, p.124605.
- Sabir, Z. et al., 2020. Neuro-swarm intelligent computing to solve the second-order singular functional differential model. The European Physical Journal Plus, 135(6), p.474.
- Umar, M., et al., 2020. Stochastic numerical technique for solving HIV infection model of CD4+ T cells. The European Physical Journal Plus, 135(6), p.403.
- Sabir, Z., et al,. 2018. Neuro-heuristics for nonlinear singular Thomas-Fermi systems. Applied Soft Computing, 65, pp.152-169.
- Umar, M., et al., 2019. Intelligent computing for numerical treatment of nonlinear prey–predator models. Applied Soft Computing, 80, pp.506-524.
- Raja, M.A.Z et al., 2015. Design of stochastic solvers based on genetic algorithms for solving nonlinear equations. Neural Computing and Applications, 26(1), pp.1-23.
- Zulqurnain Sabir et al., Design of neuro-swarming heuristic solverfor multi-pantograph singular delay differential equation, Fractals. [CrossRef]
- Sabir, Z. et al., 2020. FMNEICS: fractional Meyer neuro-evolution-based intelligent computing solver for doubly singular multi-fractional order Lane–Emden system. Computational and Applied Mathematics, 39(4), pp.1-18.
- Raja, M.A.Z. et al., 2018. A new stochastic computing paradigm for the dynamics of nonlinear singular heat conduction model of the human head. The European Physical Journal Plus, 133(9), p.364.
- Sabir, Z. et al., 2020. Design of neuro-swarming-based heuristics to solve the third-order nonlinear multi-singular Emden–Fowler equation. The European Physical Journal Plus, 135(6), p.410.
- Sabir, Z. et al., 2020. Novel design of Morlet wavelet neural network for solving second order Lane–Emden equation. Mathematics and Computers in Simulation, 172, pp.1-14.
- Reddy, G.T., Reddy, M.P.K., Lakshmanna, K., Rajput, D.S., Kaluri, R. and Srivastava, G., 2020. Hybrid genetic algorithm and a fuzzy logic classifier for heart disease diagnosis. Evolutionary Intelligence, 13(2), pp.185-196.
- Mayer, M.J., Szilágyi, A. and Gróf, G., 2020. Environmental and economic multi-objective optimization of a household level hybrid renewable energy system by genetic algorithm. Applied Energy, 269, p.115058.
- Zou, D., Li, S., Kong, X., Ouyang, H. and Li, Z., 2019. Solving the combined heat and power economic dispatch problems by an improved genetic algorithm and a new constraint handling strategy. Applied energy, 237, pp.646-670.
- Sabir, Z. et al., 2020. Heuristic computing technique for numerical solutions of nonlinear fourth order Emden–Fowler equation. Mathematics and Computers in Simulation, 178, pp.534-548.
- Armaghani, D.J., Hasanipanah, M., Mahdiyar, A., Majid, M.Z.A., Amnieh, H.B. and Tahir, M.M., 2018. Airblast prediction through a hybrid genetic algorithm-ANN model. Neural Computing and Applications, 29(9), pp.619-629.
- Umar, M. et al., 2020. A Stochastic Intelligent Computing with Neuro-Evolution Heuristics for Nonlinear SITR System of Novel COVID-19 Dynamics. Symmetry, 12(10), p.1628.
- Sabir, Z. et al., 2020. Design of stochastic numerical solver for the solution of singular three-point second-order boundary value problems. Neural Computing and Applications, pp.1-17.
- Jiang, Y., Wu, P., Zeng, J., Zhang, Y., Zhang, Y. and Wang, S., 2020. Multi-parameter and multi-objective optimisation of articulated monorail vehicle system dynamics using genetic algorithm. Vehicle System Dynamics, 58(1), pp.74-91.
- Bertocchi, C., Chouzenoux, E., Corbineau, M.C., Pesquet, J.C. and Prato, M., 2020. Deep unfolding of a proximal interior point method for image restoration. Inverse Problems, 36(3), p.034005.
- Wright, S.E. and Lim, S., 2020. Solving nested-constraint resource allocation problems with an interior point method. Operations Research Letters, 48(3), pp.297-303.
- Pesteh, S., Moayyed, H. and Miranda, V., 2020. Favorable properties of interior point method and generalized correntropy in power system state estimation. Electric Power Systems Research, 178, p.106035.
- Garreis, S., Surowiec, T.M. and Ulbrich, M., 2021. An interior-point approach for solving risk-averse PDE-constrained optimization problems with coherent risk measures. SIAM Journal on Optimization, 31(1), pp.1-29.
- Asadi, S., Darvay, Z., Lesaja, G., Mahdavi-Amiri, N. and Potra, F., 2020. A Full-Newton Step Interior-Point Method for Monotone Weighted Linear Complementarity Problems. Journal of Optimization Theory and Applications, 186(3), pp.864-878.
- Sanchez, Y.G. et al. 2020. Design of a nonlinear SITR fractal model based on the dynamics of a novel coronavirus (COVID). Fractals, 28, p.2040026.
- Guerrero Sánchez, Y. et al., 2020. Analytical and approximate solutions of a novel nervous stomach mathematical model. Discrete Dynamics in Nature and Society, 2020.
- Sabir, Z., Sakar, M.G., Yeskindirova, M. and Saldir, O., 2020. Numerical investigations to design a novel model based on the fifth order system of Emden–Fowler equations. Theoretical and Applied Mechanics Letters, 10(5), pp.333-342.
- Sajid, T. et al,. 2020. Upshot of radiative rotating Prandtl fluid flow over a slippery surface embedded with variable species diffusivity and multiple convective boundary conditions. Heat Transfer.
- Umar, M. et al., 2020. The 3-D flow of Casson nanofluid over a stretched sheet with chemical reactions, velocity slip, thermal radiation and Brownian motion. Thermal Science, 24(5 Part A), pp.2929-2939.

| Mode | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 | ||
| E-I | Min | 4.14E-9 | 2.07E-8 | 2.85E-7 | 4.46E-7 | 4.86E-7 | 5.58E-7 | 7.48E-7 | 1.00E-6 | 1.20E-6 | 1.30E-6 | 1.34E-6 |
| Mean | 3.75E-1 | 4.13E-1 | 4.54E-1 | 4.98E-1 | 5.41E-1 | 5.83E-1 | 6.22E-1 | 6.59E-1 | 6.92E-1 | 7.23E-1 | 7.49E-1 | |
| SD | 4.46E-1 | 5.1E-1 | 5.57E-1 | 6.7E-1 | 6.53E-1 | 6.95E-1 | 7.35E-1 | 7.72E-1 | 8.05E-1 | 8.34E-1 | 8.60E-1 | |
| Med | 3.93E-2 | 2.13E-2 | 1.21E-2 | 2.48E-2 | 4.61E-2 | 6.65E-2 | 8.59E-2 | 1.05E-1 | 1.23E-1 | 1.41E-1 | 1.59E-1 | |
| S.IR | 4.38E-1 | 4.84E-1 | 5.38E-1 | 5.88E-1 | 6.35E-1 | 6.79E-1 | 7.21E-1 | 7.60E-1 | 7.97E-1 | 8.30E-1 | 8.59E-1 | |
| E-II | Min | 5.20E-7 | 7.23E-7 | 4.89E-6 | 1.14E-5 | 1.59E-5 | 1.68E-5 | 1.60E-5 | 1.72E-5 | 2.20E-5 | 2.71E-5 | 2.77E-5 |
| Mean | 6.72E-2 | 1.31E-1 | 2.03E-1 | 2.76E-1 | 3.50E-1 | 4.22E-1 | 4.91E-1 | 5.57E-1 | 6.18E-1 | 6.74E-1 | 7.23E-1 | |
| SD | 3.19E-2 | 5.19E-2 | 7.16E-2 | 9.52E-2 | 1.19E-1 | 1.43E-1 | 1.66E-1 | 1.89E-1 | 2.10E-1 | 2.29E-1 | 2.46E-1 | |
| Med | 7.70E-2 | 1.57E-1 | 2.33E-1 | 3.6E-1 | 3.89E-1 | 4.79E-1 | 5.65E-1 | 6.44E-1 | 7.17E-1 | 7.83E-1 | 8.41E-1 | |
| S.IR | 1.55E-2 | 2.86E-2 | 1.99E-2 | 1.31E-2 | 1.46E-2 | 2.09E-2 | 2.71E-2 | 3.30E-2 | 3.83E-2 | 4.31E-2 | 4.70E-2 | |
| E-III | Min | 1.98E-5 | 8.21E-6 | 8.50E-6 | 5.50E-6 | 7.92E-6 | 2.12E-5 | 1.52E-5 | 1.19E-5 | 2.95E-5 | 5.74E-5 | 5.59E-5 |
| Mean | 1.25E-1 | 1.08E-1 | 9.21E-1 | 7.77E-1 | 6.43E-1 | 5.19E-1 | 4.03E-1 | 2.95E-1 | 1.90E-1 | 1.28E-1 | 1.50E-1 | |
| SD | 9.57E-1 | 8.20E-1 | 6.93E-1 | 5.72E-1 | 4.60E-1 | 3.60E-1 | 2.75E-1 | 2.15E-1 | 1.98E-1 | 2.18E-1 | 1.06E-1 | |
| Med | 1.91E-1 | 1.65E-1 | 1.40E-1 | 1.17E-1 | 9.51E-1 | 7.42E-1 | 5.37E-1 | 3.29E-1 | 1.37E-1 | 8.53E-2 | 1.65E-1 | |
| S.IR | 9.91E-1 | 8.47E-1 | 7.10E-1 | 5.77E-1 | 4.52E-1 | 3.39E-1 | 2.31E-1 | 1.34E-1 | 5.59E-2 | 6.92E-2 | 7.89E-2 | |
| Mode | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 | ||
| E-I | Min | 1.20E-9 | 4.45E-9 | 3.18E-8 | 6.57E-8 | 8.26E-8 | 7.97E-8 | 7.48E-8 | 8.81E-8 | 1.21E-7 | 1.48E-7 | 1.47E-7 |
| Mean | 4.26E-1 | 4.78E-1 | 5.34E-1 | 5.88E-1 | 6.39E-1 | 6.88E-1 | 7.33E-1 | 7.75E-1 | 8.14E-1 | 8.48E-1 | 8.79E-1 | |
| SD | 4.46E-1 | 4.97E-1 | 5.50E-1 | 6.02E-1 | 6.51E-1 | 6.98E-1 | 7.42E-1 | 7.83E-1 | 8.20E-1 | 8.54E-1 | 8.83E-1 | |
| Med | 2.54E-1 | 2.99E-1 | 3.68E-1 | 4.31E-1 | 4.80E-1 | 5.27E-1 | 5.71E-1 | 6.13E-1 | 6.53E-1 | 6.89E-1 | 7.22E-1 | |
| S.IR | 4.66E-1 | 5.23E-1 | 5.79E-1 | 6.33E-1 | 6.82E-1 | 7.27E-1 | 7.68E-1 | 8.06E-1 | 8.40E-1 | 8.77E-1 | 9.05E-1 | |
| E-II | Min | 1.15E-8 | 2.04E-8 | 6.16E-8 | 1.79E-7 | 2.65E-7 | 2.88E-7 | 2.64E-7 | 2.38E-7 | 2.55E-7 | 3.30E-7 | 4.24E-7 |
| Mean | 4.16E-2 | 8.85E-2 | 1.47E-1 | 2.06E-1 | 2.63E-1 | 3.19E-1 | 3.72E-1 | 4.23E-1 | 4.70E-1 | 5.12E-1 | 5.48E-1 | |
| SD | 6.10E-2 | 7.67E-2 | 1.03E-1 | 1.36E-1 | 1.73E-1 | 2.10E-1 | 2.46E-1 | 2.79E-1 | 3.11E-1 | 3.40E-1 | 3.64E-1 | |
| Med | 2.32E-3 | 7.26E-2 | 1.73E-1 | 2.69E-1 | 3.58E-1 | 4.48E-1 | 5.15E-1 | 5.86E-1 | 6.52E-1 | 7.12E-1 | 7.61E-1 | |
| S.IR | 3.70E-2 | 7.45E-2 | 1.11E-1 | 1.50E-1 | 1.93E-1 | 2.36E-1 | 2.79E-1 | 3.18E-1 | 3.53E-1 | 3.84E-1 | 4.08E-1 | |
| E-III | Min | 5.56E-9 | 1.91E-8 | 4.54E-9 | 2.45E-8 | 1.44E-8 | 2.28E-9 | 9.65E-9 | 1.85E-8 | 3.85E-9 | 6.66E-9 | 3.87E-9 |
| Mean | 2.30E-1 | 2.50E-1 | 2.13E-1 | 1.80E-1 | 1.50E-1 | 1.24E-1 | 9.84E-2 | 7.42E-2 | 5.08E-2 | 2.80E-2 | 3.14E-2 | |
| SD | 5.50E-1 | 5.48E-1 | 4.69E-1 | 3.97E-1 | 3.28E-1 | 2.63E-1 | 2.00E-1 | 1.40E-1 | 8.43E-2 | 4.14E-2 | 4.69E-2 | |
| Med | 7.86E-4 | 7.97E-4 | 4.52E-4 | 6.83E-4 | 1.40E-3 | 2.06E-3 | 3.38E-3 | 4.62E-3 | 5.79E-3 | 7.18E-3 | 8.47E-3 | |
| S.IR | 9.30E-3 | 6.24E-3 | 3.54E-3 | 4.98E-3 | 6.69E-3 | 1.19E-2 | 1.87E-2 | 2.58E-2 | 3.32E-2 | 2.27E-2 | 2.45E-2 | |
| Mode | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 | ||
| E-I | Min | 4.22E-9 | 1.67E-8 | 7.93E-8 | 1.42E-8 | 3.39E-8 | 4.01E-8 | 1.32E-7 | 1.50E-7 | 8.64E-8 | 5.00E-8 | 1.43E-7 |
| Mean | 3.47E-1 | 3.89E-1 | 4.43E-1 | 4.94E-1 | 5.44E-1 | 5.92E-1 | 6.37E-1 | 6.79E-1 | 7.19E-1 | 7.55E-1 | 7.87E-1 | |
| SD | 4.24E-1 | 4.62E-1 | 5.07E-1 | 5.53E-1 | 5.97E-1 | 6.41E-1 | 6.83E-1 | 7.23E-1 | 7.61E-1 | 7.95E-1 | 8.25E-1 | |
| Med | 8.71E-4 | 1.89E-2 | 4.71E-2 | 7.45E-2 | 1.02E-1 | 1.28E-1 | 1.54E-1 | 1.79E-1 | 2.03E-1 | 2.26E-1 | 2.49E-1 | |
| S.IR | 4.23E-1 | 4.62E-1 | 5.14E-1 | 5.62E-1 | 6.04E-1 | 6.48E-1 | 6.92E-1 | 7.32E-1 | 7.69E-1 | 7.99E-1 | 8.23E-1 | |
| E-II | Min | 2.31E-9 | 1.48E-8 | 8.52E-8 | 9.16E-8 | 9.46E-8 | 1.67E-7 | 3.06E-7 | 4.31E-7 | 4.63E-7 | 4.43E-7 | 5.02E-7 |
| Mean | 1.15E-2 | 5.33E-2 | 1.04E-1 | 1.54E-1 | 2.03E-1 | 2.50E-1 | 2.95E-1 | 3.37E-1 | 3.75E-1 | 4.10E-1 | 4.41E-1 | |
| SD | 2.85E-2 | 5.47E-2 | 9.77E-2 | 1.43E-1 | 1.88E-1 | 2.31E-1 | 2.72E-1 | 3.11E-1 | 3.47E-1 | 3.79E-1 | 4.08E-1 | |
| Med | 4.78E-5 | 6.73E-2 | 1.50E-1 | 2.29E-1 | 3.16E-1 | 3.89E-1 | 4.54E-1 | 5.16E-1 | 5.72E-1 | 6.25E-1 | 6.78E-1 | |
| S.IR | 9.75E-4 | 4.99E-2 | 9.93E-2 | 1.48E-1 | 1.95E-1 | 2.40E-1 | 2.82E-1 | 3.22E-1 | 3.59E-1 | 3.92E-1 | 4.21E-1 | |
| E-III | Min | 2.23E-9 | 6.07E-8 | 7.51E-8 | 9.56E-8 | 1.82E-7 | 7.64E-9 | 2.99E-8 | 4.07E-8 | 5.42E-8 | 1.76E-9 | 4.38E-9 |
| Mean | 6.58E-1 | 5.99E-1 | 5.08E-1 | 4.24E-1 | 3.46E-1 | 2.74E-1 | 2.06E-1 | 1.42E-1 | 8.09E-2 | 3.31E-2 | 4.71E-2 | |
| SD | 8.23E-1 | 7.13E-1 | 6.04E-1 | 5.02E-1 | 4.06E-1 | 3.17E-1 | 2.35E-1 | 1.58E-1 | 8.82E-2 | 3.78E-2 | 7.41E-2 | |
| Med | 2.43E-2 | 5.77E-2 | 4.54E-2 | 3.42E-2 | 3.23E-2 | 3.22E-2 | 3.50E-2 | 3.96E-2 | 4.82E-2 | 1.77E-2 | 4.90E-3 | |
| S.IR | 7.97E-1 | 7.09E-1 | 6.07E-1 | 5.11E-1 | 4.21E-1 | 3.36E-1 | 2.47E-1 | 1.64E-1 | 7.70E-2 | 2.73E-2 | 2.96E-2 | |
| Example | (G.FIT) | (G.TIC) | (G.RMSE) | (G.EVAF) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SIR | Mean | SIR | Mean Range | SIR | Mean | SIR | |
| I | 4.269E-05 | 2.163E-01 | 1.963E-05 | 8.540E-05 | 1.493E-01 | 6.500E-01 | 1.776E-01 | 5.001E-01 |
| II | 7.355E-06 | 1.762E-01 | 5.651E-05 | 3.482E-05 | 1.493E-01 | 6.500E-01 | 4.935E-01 | 4.950E-01 |
| III | 3.869E-03 | 6.616E-01 | 6.403E-06 | 5.848E-05 | 1.493E-01 | 6.500E-01 | 1.116E-03 | 3.265E-01 |
| Example | (G.FIT) | (G.TIC) | (G.RMSE) | (G.EVAF) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SIR | Mean | SIR | Mean Range | SIR | Mean | SIR | |
| I | 4.269E-05 | 2.163E-01 | 1.963E-05 | 8.540E-05 | 1.493E-01 | 6.500E-01 | 1.776E-01 | 5.001E-01 |
| II | 7.355E-06 | 1.762E-01 | 5.651E-05 | 3.482E-05 | 1.493E-01 | 6.500E-01 | 4.935E-01 | 4.950E-01 |
| III | 3.869E-03 | 6.616E-01 | 6.403E-06 | 5.848E-05 | 1.493E-01 | 6.500E-01 | 1.116E-03 | 3.265E-01 |
| Example | (G.FIT) | (G.TIC) | (G.RMSE) | (G.EVAF) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SIR | Mean | SIR | Mean Range | SIR | Mean | SIR | |
| I | 4.269E-05 | 2.163E-01 | 1.963E-05 | 8.540E-05 | 1.493E-01 | 6.500E-01 | 1.776E-01 | 5.001E-01 |
| II | 7.355E-06 | 1.762E-01 | 5.651E-05 | 3.482E-05 | 1.493E-01 | 6.500E-01 | 4.935E-01 | 4.950E-01 |
| III | 3.869E-03 | 6.616E-01 | 6.403E-06 | 5.848E-05 | 1.493E-01 | 6.500E-01 | 1.116E-03 | 3.265E-01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





