Submitted:
05 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
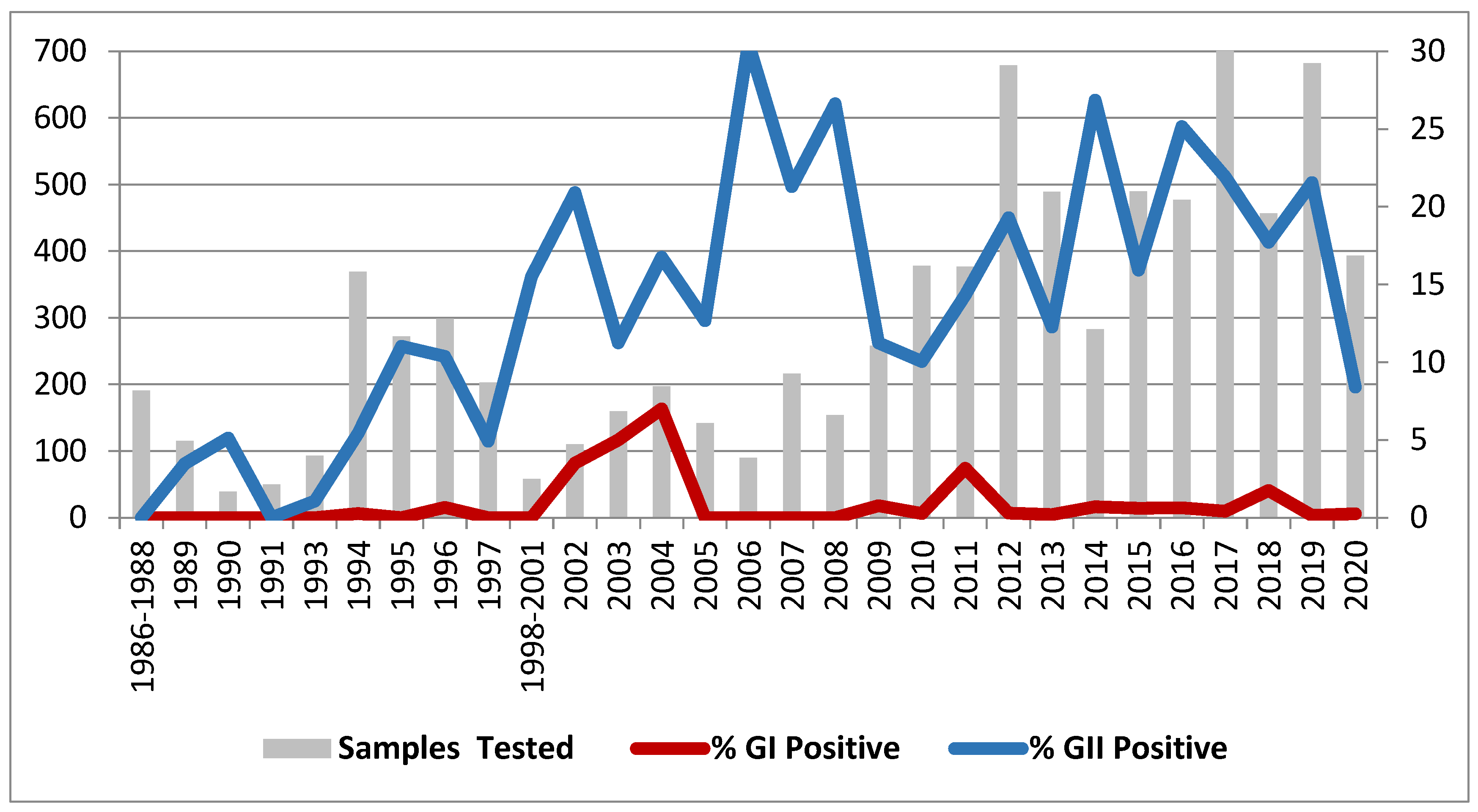
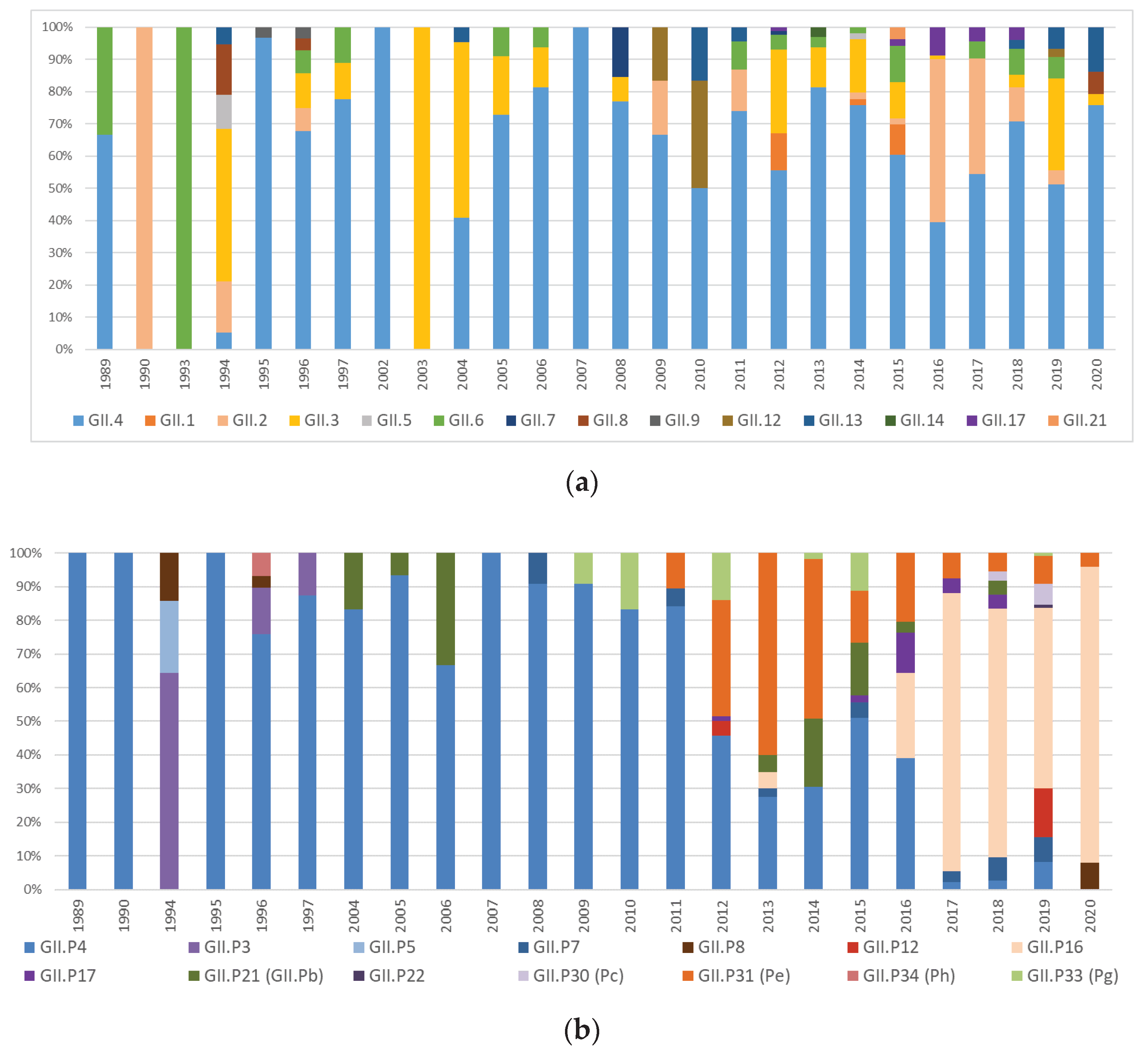
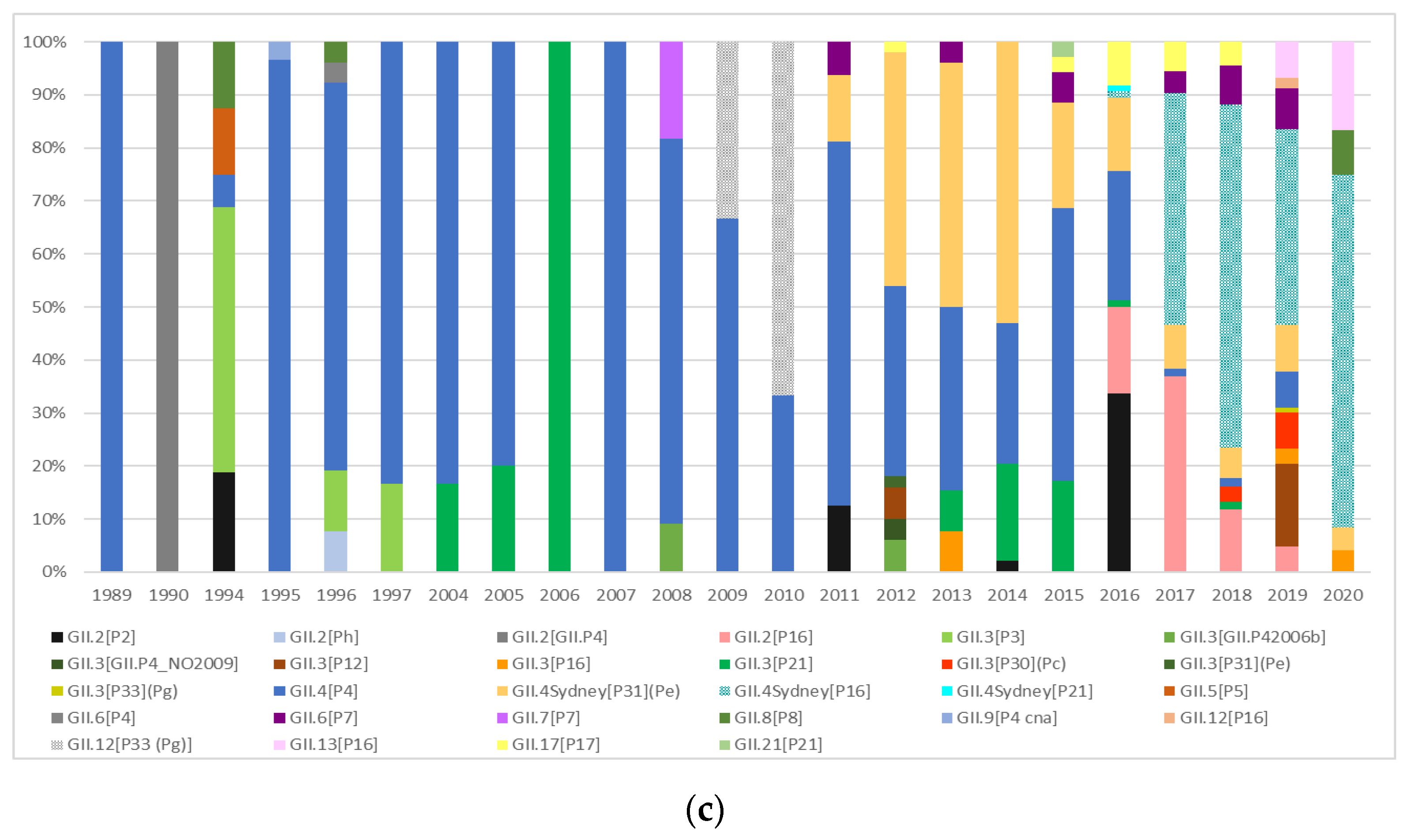
3. Results
3.1. Prevalence and typing of NoVs
3.2. Focus on the early stages of Norovirus circulation
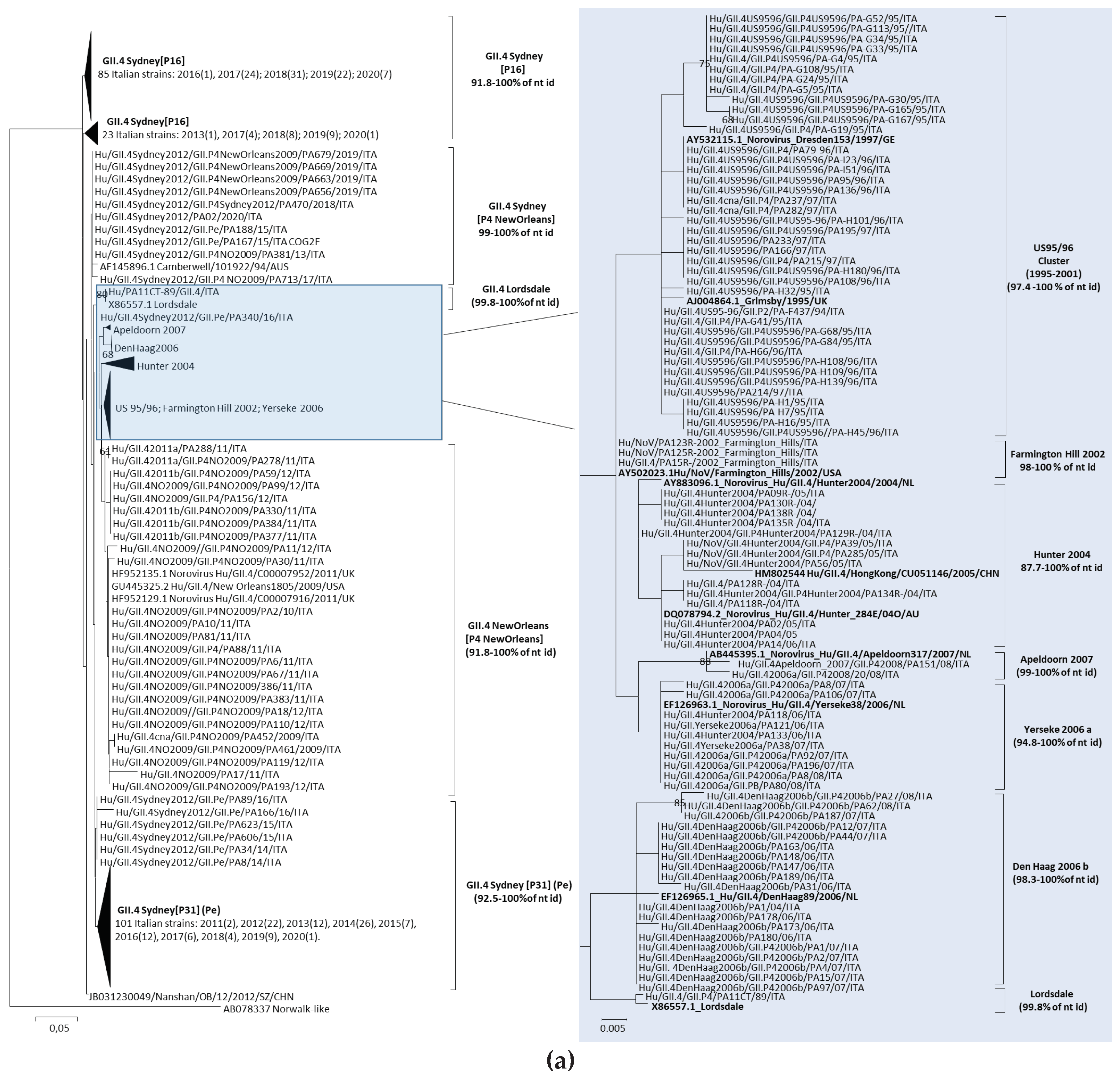
3.3. Genetic evolution and diversification of NoV genotypes
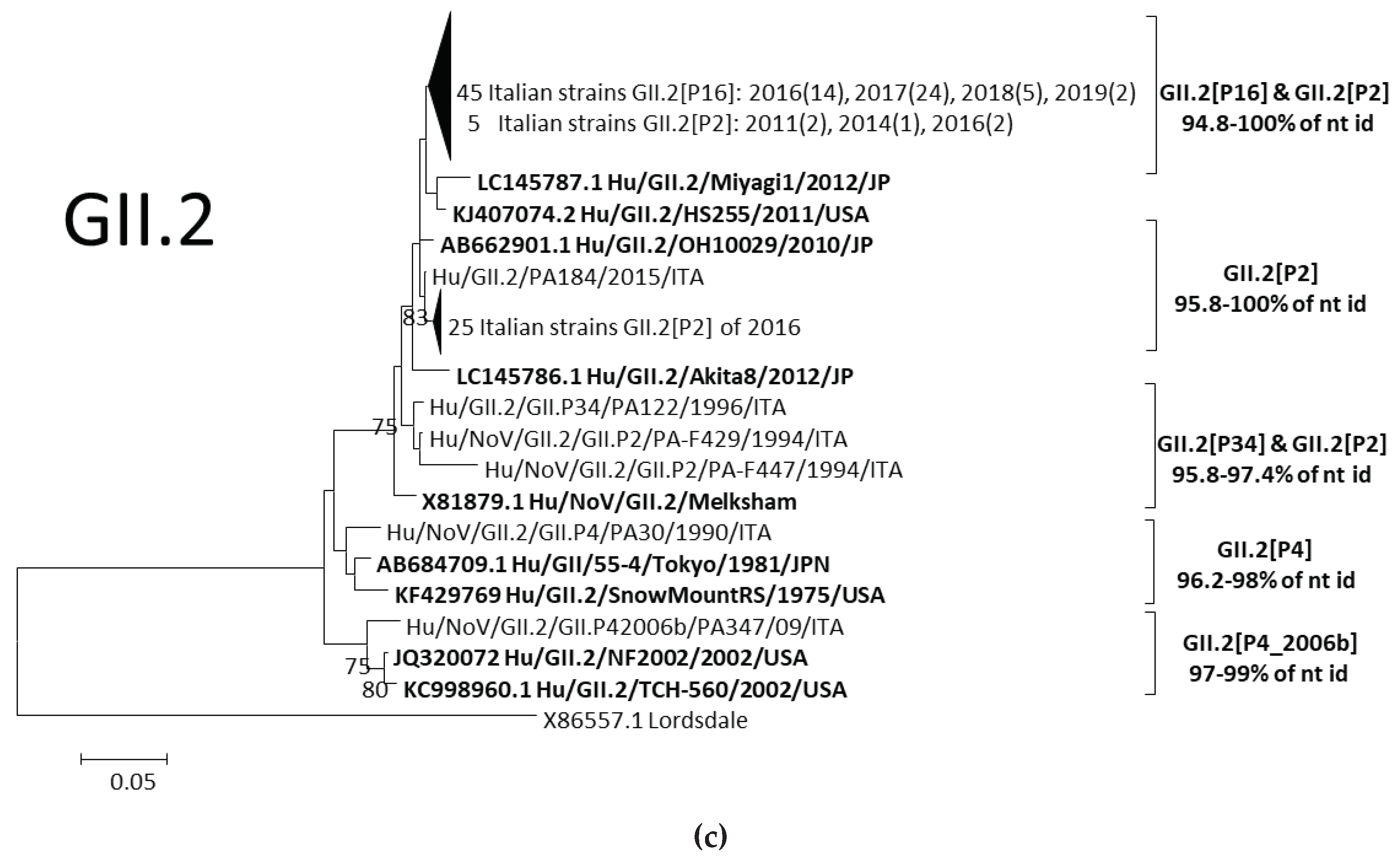
3.3.1. Analyses of GII.3 and GII.2 NoVs
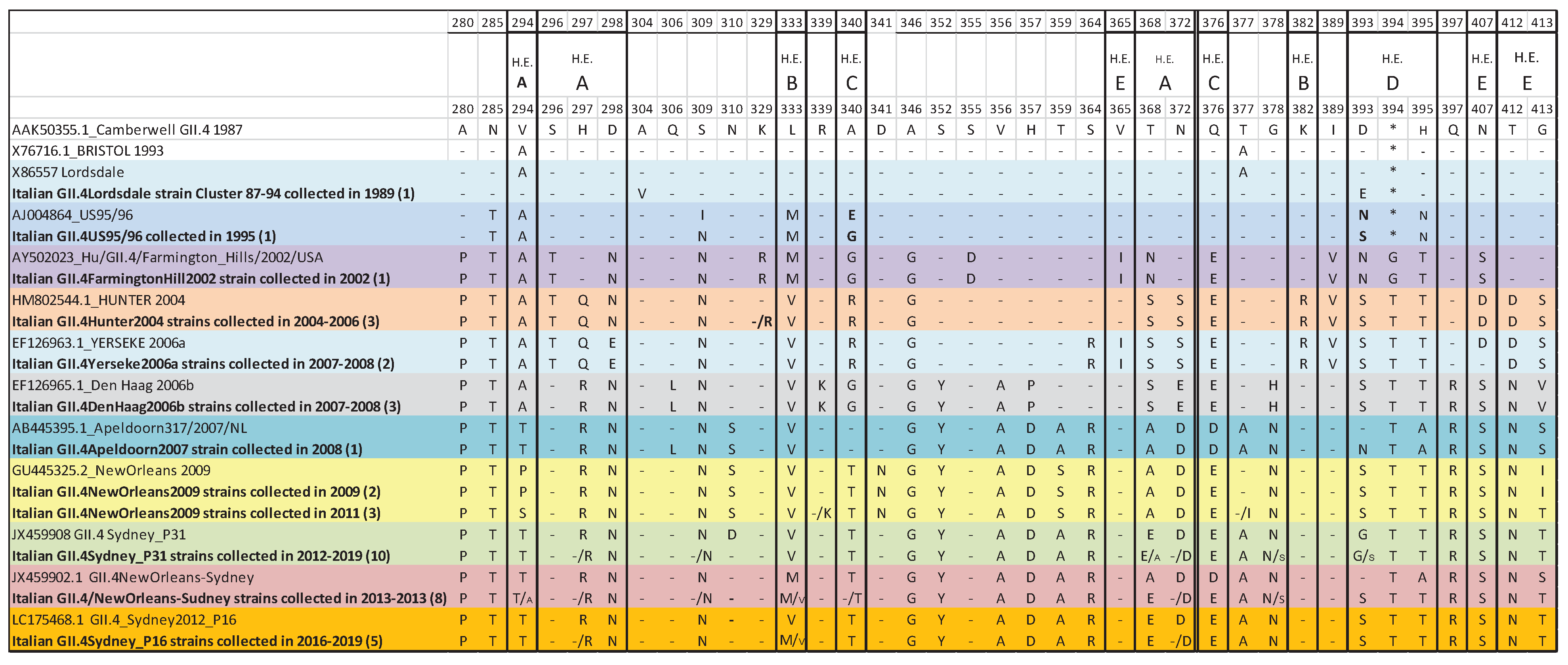
3.3.2. Analysis of the GII.4 NoV variants
3.4. Antigenic variation in the hypervariable P2 domain of GII.4 variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, K.Y. Caliciviridae: The Noroviruses. In Fields virology; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer Health/Lippincott Williams and Wilkins: Philadelphia, 2013; pp. 582–608. [Google Scholar]
- Kapikian AZ; Wyatt RG; Dolin R; Thornhill TS; Kalica AR; Chanock RM Visualization by Immune Electron Microscopy of a 27-Nm Particle Associated with Acute Infectious Nonbacterial Gastroenteritis. J Virol 1972, 10, 1075–1781. [CrossRef] [PubMed]
- Estes, M.K.; Prasad, B.V.; Atmar, R.L. Noroviruses Everywhere: Has Something Changed? Curr Opin Infect Dis 2006, 19, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.M.; Widdowson, M.A.; Glass, R.I.; Akazawa, K.; Vinje, J.; Parashar, U.D. Systematic Literature Review of Role of Noroviruses in Sporadic Gastroenteritis. Emerg Infect Dis 2008, 14, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I.; Squires, R.B.; Karangwa, C.K.; Johnson, J.A.; Lepore, C.J.; Sosnovtsev, S.V.; Green, K.Y. Static and Evolving Norovirus Genotypes: Implications for Epidemiology and Immunity. PLoS Pathog 2017, 13, e1006136. [Google Scholar] [CrossRef]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.-W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated Classification of Norovirus Genogroups and Genotypes. J Gen Virol 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- de Graaf, M.; van Beek, J.; Koopmans, M.P.G. Human Norovirus Transmission and Evolution in a Changing World. Nat Rev Microbiol 2016, 14, 421–433. [Google Scholar] [CrossRef]
- Siebenga, J.J.; Vennema, H.; Zheng, D.-P.; Vinje, J.; Lee, B.E.; Pang, X.-L.; Ho, E.C.M.; Lim, W.; Choudekar, A.; Broor, S.; et al. Norovirus Illness Is a Global Problem: Emergence and Spread of Norovirus GII.4 Variants, 2001-2007. J Infect Dis 2009, 200, 802–812. [Google Scholar] [CrossRef]
- Bok, K.; Abente, E.J.; Realpe-Quintero, M.; Mitra, T.; Sosnovtsev, S.V.; Kapikian, A.Z.; Green, K.Y. Evolutionary Dynamics of GII.4 Noroviruses over a 34-Year Period. J. Virol. 2009, 83, 11890–11901. [Google Scholar] [CrossRef]
- Farahmand, M.; Moghoofei, M.; Dorost, A.; Shoja, Z.; Ghorbani, S.; Kiani, S.J.; Khales, P.; Esteghamati, A.; Sayyahfar, S.; Jafarzadeh, M.; et al. Global Prevalence and Genotype Distribution of Norovirus Infection in Children with Gastroenteritis: A Meta-Analysis on 6 Years of Research from 2015 to 2020. Rev Med Virol 2022, 32, e2237. [Google Scholar] [CrossRef]
- Noel, J.S.; Lee, T.W.; Kurtz, J.B.; Glass, R.I.; Monroe, S.S. Typing of Human Astroviruses from Clinical Isolates by Enzyme Immunoassay and Nucleotide Sequencing. J Clin Microbiol 1995, 33, 797–801. [Google Scholar] [CrossRef]
- White, P.A.; Hansman, G.S.; Li, A.; Dable, J.; Isaacs, M.; Ferson, M.; McIver, C.J.; Rawlinson, W.D. Norwalk-like Virus 95/96-US Strain Is a Major Cause of Gastroenteritis Outbreaks in Australia. J Med Virol 2002, 68, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Lopman, B.; Vennema, H.; Kohli, E.; Pothier, P.; Sanchez, A.; Negredo, A.; Buesa, J.; Schreier, E.; Reacher, M.; Brown, D.; et al. Increase in Viral Gastroenteritis Outbreaks in Europe and Epidemic Spread of New Norovirus Variant. Lancet 2004, 363, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Widdowson, M.-A.; Cramer, E.H.; Hadley, L.; Bresee, J.S.; Beard, R.S.; Bulens, S.N.; Charles, M.; Chege, W.; Isakbaeva, E.; Wright, J.G.; et al. Outbreaks of Acute Gastroenteritis on Cruise Ships and on Land: Identification of a Predominant Circulating Strain of Norovirus--United States, 2002. J Infect Dis 2004, 190, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Tu, E.T.; McIver, C.J.; Rawlinson, W.D.; White, P.A. Emergence of a New Norovirus Genotype II.4 Variant Associated with Global Outbreaks of Gastroenteritis. Journal Clinical Microbiology 2006, 44, 327–333. [Google Scholar] [CrossRef]
- Tu, E.T.-V.; Bull, R.A.; Greening, G.E.; Hewitt, J.; Lyon, M.J.; Marshall, J.A.; McIver, C.J.; Rawlinson, W.D.; White, P.A. Epidemics of Gastroenteritis during 2006 Were Associated with the Spread of Norovirus GII.4 Variants 2006a and 2006b. Clin Infect Dis 2008, 46, 413–420. [Google Scholar] [CrossRef]
- Eden, J.S.; Bull, R.A.; Tu, E.; McIver, C.J.; Lyon, M.J.; Marshall, J.A.; Smith, D.W.; Musto, J.; Rawlinson, W.D.; White, P.A. Norovirus GII.4 Variant 2006b Caused Epidemics of Acute Gastroenteritis in Australia during 2007 and 2008. J Clin Virol 2010, 49, 265–271. [Google Scholar] [CrossRef]
- Yen, C.; Wikswo, M.E.; Lopman, B.A.; Vinje, J.; Parashar, U.D.; Hall, A.J. Impact of an Emergent Norovirus Variant in 2009 on Norovirus Outbreak Activity in the United States. Clin Infect Dis 2011, 53, 568–571. [Google Scholar] [CrossRef]
- Eden, J.S.; Hewitt, J.; Lim, K.L.; Boni, M.F.; Merif, J.; Greening, G.; Ratcliff, R.M.; Holmes, E.C.; Tanaka, M.M.; Rawlinson, W.D.; et al. The Emergence and Evolution of the Novel Epidemic Norovirus GII.4 Variant Sydney 2012. Virology 2014, 450–451, 106–113. [Google Scholar] [CrossRef]
- Eden, J.S.; Tanaka, M.M.; Boni, M.F.; Rawlinson, W.D.; White, P.A. Recombination within the Pandemic Norovirus GII.4 Lineage. J Virol 2013, 87, 6270–6282. [Google Scholar] [CrossRef]
- Siebenga, J.J.; Vennema, H.; Renckens, B.; de Bruin, E.; van der Veer, B.; Siezen, R.J.; Koopmans, M. Epochal Evolution of GGII.4 Norovirus Capsid Proteins from 1995 to 2006. J Virol 2007, 81, 9932–9941. [Google Scholar] [CrossRef]
- White, P.A. Evolution of Norovirus. Clin Microbiol Infect 2014, 20, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Donaldson, E.F.; Lobue, A.D.; Cannon, J.L.; Zheng, D.P.; Vinje, J.; Baric, R.S. Mechanisms of GII.4 Norovirus Persistence in Human Populations. PLoS Med 2008, 5, e31. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Beltramello, M.; Donaldson, E.F.; Corti, D.; Swanstrom, J.; Debbink, K.; Lanzavecchia, A.; Baric, R.S. Immunogenetic Mechanisms Driving Norovirus GII.4 Antigenic Variation. PLoS Pathog 2012, 8, e1002705. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Costantini, V.; Swanstrom, J.; Debbink, K.; Donaldson, E.F.; Vinje, J.; Baric, R.S. Emergence of a Norovirus GII.4 Strain Correlates with Changes in Evolving Blockade Epitopes. J Virol 2013, 87, 2803–2813. [Google Scholar] [CrossRef] [PubMed]
- Debbink, K.; Lindesmith, L.C.; Donaldson, E.F.; Costantini, V.; Beltramello, M.; Corti, D.; Swanstrom, J.; Lanzavecchia, A.; Vinje, J.; Baric, R.S. Emergence of New Pandemic GII.4 Sydney Norovirus Strain Correlates With Escape From Herd Immunity. J Infect Dis 2013, 208, 1877–1887. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, W.B.; Zhang, J.; Hou, J.W.; Tang, F.; Zhang, X.F.; Du, L.F.; Su, J.G.; Li, Q.M. Evolution of the Interactions between GII.4 Noroviruses and Histo-Blood Group Antigens: Insights from Experimental and Computational Studies. PLoS Pathog 2021, 17, e1009745. [Google Scholar] [CrossRef]
- Seto, Y.; Iritani, N.; Kubo, H.; Kaida, A.; Murakami, T.; Haruki, K.; Nishio, O.; Ayata, M.; Ogura, H. Genotyping of Norovirus Strains Detected in Outbreaks between April 2002 and March 2003 in Osaka City, Japan. Microbiol Immunol 2005, 49, 275–283. [Google Scholar] [CrossRef]
- Motomura, K.; Yokoyama, M.; Ode, H.; Nakamura, H.; Mori, H.; Kanda, T.; Oka, T.; Katayama, K.; Noda, M.; Tanaka, T.; et al. Divergent Evolution of Norovirus GII/4 by Genome Recombination from May 2006 to February 2009 in Japan. J Virol 2010, 84, 8085–8097. [Google Scholar] [CrossRef]
- Belliot, G.; Kamel, A.H.; Estienney, M.; Ambert-Balay, K.; Pothier, P. Evidence of Emergence of New GGII.4 Norovirus Variants from Gastroenteritis Outbreak Survey in France during the 2007-to-2008 and 2008-to-2009 Winter Seasons. J Clin Microbiol 2010, 48, 994–998. [Google Scholar] [CrossRef]
- Vinje, J.; Koopmans, M.P. Molecular Detection and Epidemiology of Small Round-Structured Viruses in Outbreaks of Gastroenteritis in the Netherlands. Journal Infection Disease 1996, 174, 610–615. [Google Scholar] [CrossRef]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly Reactive and Highly Sensitive Assay for Norwalk-like Viruses Based on Real-Time Quantitative Reverse Transcription-PCR. Journal Clinical Microbiology 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Kageyama, T.; Fukushi, S.; Hoshino, F.B.; Shinohara, M.; Uchida, K.; Natori, K.; Takeda, N.; Katayama, K. Genogroup-Specific PCR Primers for Detection of Norwalk-like Viruses. Journal Virological Methods 2002, 100, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Vennema, H.; de Bruin, E.; Koopmans, M. Rational Optimization of Generic Primers Used for Norwalk-like Virus Detection by Reverse Transcriptase Polymerase Chain Reaction. J Clin Virol 2002, 25, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Bonura, F.; Urone, N.; Bonura, C.; Mangiaracina, L.; Filizzolo, C.; Sciortino, G.; Sanfilippo, G.L.; Martella, V.; Giammanco, G.M.; De Grazia, S. Recombinant GII.P16 Genotype Challenges RT-PCR-Based Typing in Region A of Norovirus Genome. J Infect 2021, 83, 69–75. [Google Scholar] [CrossRef]
- Vega, E.; Barclay, L.; Gregoricus, N.; Williams, K.; Lee, D.; Vinje, J. Novel Surveillance Network for Norovirus Gastroenteritis Outbreaks, United States. Emerg Infect Dis 17, 1389–1395. [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Boon, D.; Mahar, J.E.; Abente, E.J.; Kirkwood, C.D.; Purcell, R.H.; Kapikian, A.Z.; Green, K.Y.; Bok, K. Comparative Evolution of GII.3 and GII.4 Norovirus over a 31-Year Period. J Virol 2011, 85, 8656–8666. [Google Scholar] [CrossRef]
- Giammanco, G.M.; De Grazia, S.; Tummolo, F.; Bonura, F.; Calderaro, A.; Buonavoglia, A.; Martella, V.; Medici, M.C. Norovirus GII.4/Sydney/2012 in Italy, Winter 2012-2013. Emerg Infect Dis 2013, 19, 1348–1349. [Google Scholar] [CrossRef]
- Mori, K.; Nagano, M.; Kimoto, K.; Somura, Y.; Akiba, T.; Hayashi, Y.; Sadamasu, K.; Kai, A. Detection of Enteric Viruses in Fecal Specimens from Nonbacterial Foodborne Gastroenteritis Outbreaks in Tokyo, Japan between 1966 and 1983. Jpn J Infect Dis 2017, 70, 143–151. [Google Scholar] [CrossRef]
- Siqueira, J.A.M.; Bandeira, R. da S.; Oliveira, D. de S.; Dos Santos, L.F.P.; Gabbay, Y.B. Genotype Diversity and Molecular Evolution of Noroviruses: A 30-Year (1982-2011) Comprehensive Study with Children from Northern Brazil. PLoS One 2017, 12, e0178909. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; White, P.A. Mechanisms of GII.4 Norovirus Evolution. Trends Microbiol 2011, 19, 233–240. [Google Scholar] [CrossRef]
- Kroneman, A.; Verhoef, L.; Harris, J.; Vennema, H.; Duizer, E.; van Duynhoven, Y.; Gray, J.; Iturriza, M.; Böttiger, B.; Falkenhorst, G.; et al. Analysis of Integrated Virological and Epidemiological Reports of Norovirus Outbreaks Collected within the Foodborne Viruses in Europe Network from 1 July 2001 to 30 June 2006. J Clin Microbiol 2008, 46, 2959–2965. [Google Scholar] [CrossRef] [PubMed]
- Motoya, T.; Nagasawa, K.; Matsushima, Y.; Nagata, N.; Ryo, A.; Sekizuka, T.; Yamashita, A.; Kuroda, M.; Morita, Y.; Suzuki, Y.; et al. Molecular Evolution of the VP1 Gene in Human Norovirus GII.4 Variants in 1974-2015. Front Microbiol 2017, 8, 2399. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.L.; Clarke, I.N.; Caul, E.O.; Lambden, P.R. Human Enteric Caliciviruses Have a Unique Genome Structure and Are Distinct from the Norwalk-like Viruses. Arch Virol 1995, 140, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.F.; Lindesmith, L.C.; Lobue, A.D.; Baric, R.S. Norovirus Pathogenesis: Mechanisms of Persistence and Immune Evasion in Human Populations. Immunol Rev 2008, 225, 190–211. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Medici, M.C.; De Grazia, S.; Tummolo, F.; Calderaro, A.; Bonura, F.; Saporito, L.; Terio, V.; Catella, C.; Lanave, G.; et al. Evidence for Recombination between the Pandemic GII.4 Norovirus Strains New Orleans 2009 and Sydney 2012. J Clin Microbiol 2013, 51, 3855–3857. [Google Scholar] [CrossRef] [PubMed]
- De Grazia, S.; Lanave, G.; Giammanco, G.M.; Medici, M.C.; De Conto, F.; Tummolo, F.; Calderaro, A.; Bonura, F.; Urone, N.; Morea, A.; et al. Sentinel Hospital-Based Surveillance for Norovirus Infection in Children with Gastroenteritis between 2015 and 2016 in Italy. PLoS One 2018, 13, e0208184. [Google Scholar] [CrossRef]
- Medici, M.C.; Tummolo, F.; De Grazia, S.; Calderaro, A.; De Conto, F.; Terio, V.; Chironna, M.; Bonura, F.; Pucci, M.; Banyai, K.; et al. Epidemiological Dynamics of Norovirus GII.4 Variant New Orleans 2009. J Gen Virol 2015, 96, 2919–2927. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. The p Domain of Norovirus Capsid Protein Forms a Subviral Particle That Binds to Histo-Blood Group Antigen Receptors. J Virol 2005, 79, 14017–14030. [Google Scholar] [CrossRef]
- Dingle, K.E. Mutation in a Lordsdale Norovirus Epidemic Strain as a Potential Indicator of Transmission Routes. J Clin Microbiol 2004, 42, 3950–3957. [Google Scholar] [CrossRef] [PubMed]
- Vinje, J.; Green, J.; Lewis, D.C.; Gallimore, C.I.; Brown, D.W.; Koopmans, M.P. Genetic Polymorphism across Regions of the Three Open Reading Frames of “Norwalk-like Viruses. ” Arch Virol 2000, 145, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Debbink, K.; Donaldson, E.F.; Lindesmith, L.C.; Baric, R.S. Genetic Mapping of a Highly Variable Norovirus GII.4 Blockade Epitope: Potential Role in Escape from Human Herd Immunity. J Virol 2012, 86, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Lindesmith, L.C.; Donaldson, E.F.; Baric, R.S. Norovirus GII.4 Strain Antigenic Variation. J Virol 85, 231–242. [CrossRef]
- Zhou, H.-L.; Zhen, S.-S.; Wang, J.-X.; Zhang, C.-J.; Qiu, C.; Wang, S.-M.; Jiang, X.; Wang, X.-Y. Burden of Acute Gastroenteritis Caused by Norovirus in China: A Systematic Review. J Infect 2017, 75, 216–224. [Google Scholar] [CrossRef]
- Wangchuk, S.; Matsumoto, T.; Iha, H.; Ahmed, K. Surveillance of Norovirus among Children with Diarrhea in Four Major Hospitals in Bhutan: Replacement of GII.21 by GII.3 as a Dominant Genotype. PLoS One 2017, 12, e0184826. [Google Scholar] [CrossRef]
- Mahar, J.E.; Bok, K.; Green, K.Y.; Kirkwood, C.D. The Importance of Intergenic Recombination in Norovirus GII.3 Evolution. J Virol 2013, 87, 3687–3698. [Google Scholar] [CrossRef]
- Gallimore, C.I.; Cubitt, D.; du Plessis, N.; Gray, J.J. Asymptomatic and Symptomatic Excretion of Noroviruses during a Hospital Outbreak of Gastroenteritis. J Clin Microbiol 2004, 42, 2271–2274. [Google Scholar] [CrossRef]
- Ambert-Balay, K.; Bon, F.; Le Guyader, F.; Pothier, P.; Kohli, E. Characterization of New Recombinant Noroviruses. Journal Clinical Microbiology 2005, 43, 5179–5186. [Google Scholar] [CrossRef]
- Gallimore, C.I.; Cheesbrough, J.S.; Lamden, K.; Bingham, C.; Gray, J.J. Multiple Norovirus Genotypes Characterised from an Oyster-Associated Outbreak of Gastroenteritis. Int J Food Microbiol 2005, 103, 323–330. [Google Scholar] [CrossRef]
- Mahar, J.E.; Kirkwood, C.D. Characterization of Norovirus Strains in Australian Children from 2006 to 2008: Prevalence of Recombinant Strains. J Med Virol 2011, 83, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Medici, M.C.; Tummolo, F.; Martella, V.; Giammanco, G.M.; De Grazia, S.; Arcangeletti, M.C.; De Conto, F.; Chezzi, C.; Calderaro, A. Novel Recombinant GII.P16_GII.13 and GII.P16_GII.3 Norovirus Strains in Italy. Virus Res 2014, 188, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Nahar, S.; Afrad, M.H.; Begum, N.; Al-Mamun, F.; Sarker, A.K.; Das, S.K.; Faruque, A.S.G.; Pourkarim, M.R.; Choudhuri, M.S.K.; Azim, T.; et al. High Prevalence of Noroviruses among Hospitalized Diarrheal Patients in Bangladesh, 2011. J Infect Dev Ctries 2013, 7, 892–896. [Google Scholar] [CrossRef]
- Qin, S.-W.; Chan, T.-C.; Cai, J.; Zhao, N.; Miao, Z.-P.; Chen, Y.-J.; Liu, S.-L. Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016. Int J Environ Res Public Health 2017, 14. [Google Scholar] [CrossRef]
- Hoa Tran, T.N.; Trainor, E.; Nakagomi, T.; Cunliffe, N.A.; Nakagomi, O. Molecular Epidemiology of Noroviruses Associated with Acute Sporadic Gastroenteritis in Children: Global Distribution of Genogroups, Genotypes and GII.4 Variants. J Clin Virol 2013, 56, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.-G.; Shi, C.; Xu, C.; Lin, Q.; Zhang, J.; Yi, Q.-H.; Zhang, J.; Bao, C.-J.; Huo, X.; Zhu, Y.-F.; et al. Outbreaks of Acute Gastroenteritis Associated with a Re-Emerging GII.P16-GII.2 Norovirus in the Spring of 2017 in Jiangsu, China. PLoS One 2017, 12, e0186090. [Google Scholar] [CrossRef] [PubMed]
- Hata, M.; Nakamura, N.; Kobayashi, S.; Onouchi, A.; Saito, T.; Hirose, E.; Adachi, H.; Saito, N.; Ito, M.; Yasui, Y.; et al. Emergence of New Recombinant Noroviruses GII.P16-GII.2 and GII.P16-GII.4 in Aichi, Japan, during the 2016/17 Season. Jpn J Infect Dis 2018, 71, 319–322. [Google Scholar] [CrossRef]
- Medici, M.C.; Tummolo, F.; Martella, V.; De Conto, F.; Arcangeletti, M.C.; Pinardi, F.; Ferraglia, F.; Chezzi, C.; Calderaro, A. Emergence of Novel Recombinant GII.P16_GII.2 and GII. P16_GII.4 Sydney 2012 Norovirus Strains in Italy, Winter 2016/2017. New Microbiol 2018, 41, 71–72. [Google Scholar]
- Giammanco, G.M.; Bonura, F.; Urone, N.; Purpari, G.; Cuccia, M.; Pepe, A.; Li Muli, S.; Cappa, V.; Saglimbene, C.; Mandolfo, G.; et al. Waterborne Norovirus Outbreak at a Seaside Resort Likely Originating from Municipal Water Distribution System Failure. Epidemiol Infect 2018, 146, 879–887. [Google Scholar] [CrossRef]
- Iritani, N.; Kaida, A.; Abe, N.; Sekiguchi, J.-I.; Kubo, H.; Takakura, K.-I.; Goto, K.; Ogura, H.; Seto, Y. Increase of GII.2 Norovirus Infections during the 2009-2010 Season in Osaka City, Japan. J Med Virol 2012, 84, 517–525. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




