1. Introduction
The discovery of the method to make human induced pluripotent stem cells (iPSC) by Yamanaka in 2007 created the opportunity to model human disease and development at a new level (1). However, successful and accurate disease modeling as well as application of human iPSC for human disease is hampered by genetic quality. There have been many improvements of the original cell reprogramming protocol including the use of non-viral methods such as synthetic mRNA transfection for delivery of reprogramming factors to cells (2). Also, replacing c-Myc reprogramming factor with Cyclin D1 that repairs DNA breaks that occur during the reprogramming process to significantly improve genetical stability of iPSC and neural stem cells (NSC) (3). The use of these advanced iPSC and NSC will result in more accurate conclusions when applying as a human disease model in vitro (3).
The peptide products of the proteasome digestion are the source of peptides presented by the major histocompatibility class I molecules (MHC-I) on cells that can be detected by CD8 T lymphocytes. Thus, the expression of different types of proteasome in different cells will likely change the immunopeptidomes bound to MHC-I on the cell surface. The MHC-I, which presents peptides broken down by the proteosome, has been implicated in neuronal plasticity, regulating synaptic density and axonal regeneration in the central and peripheral nervous system during development and in brain diseases (4-5). The mechanisms governing these effects are largely unknown.
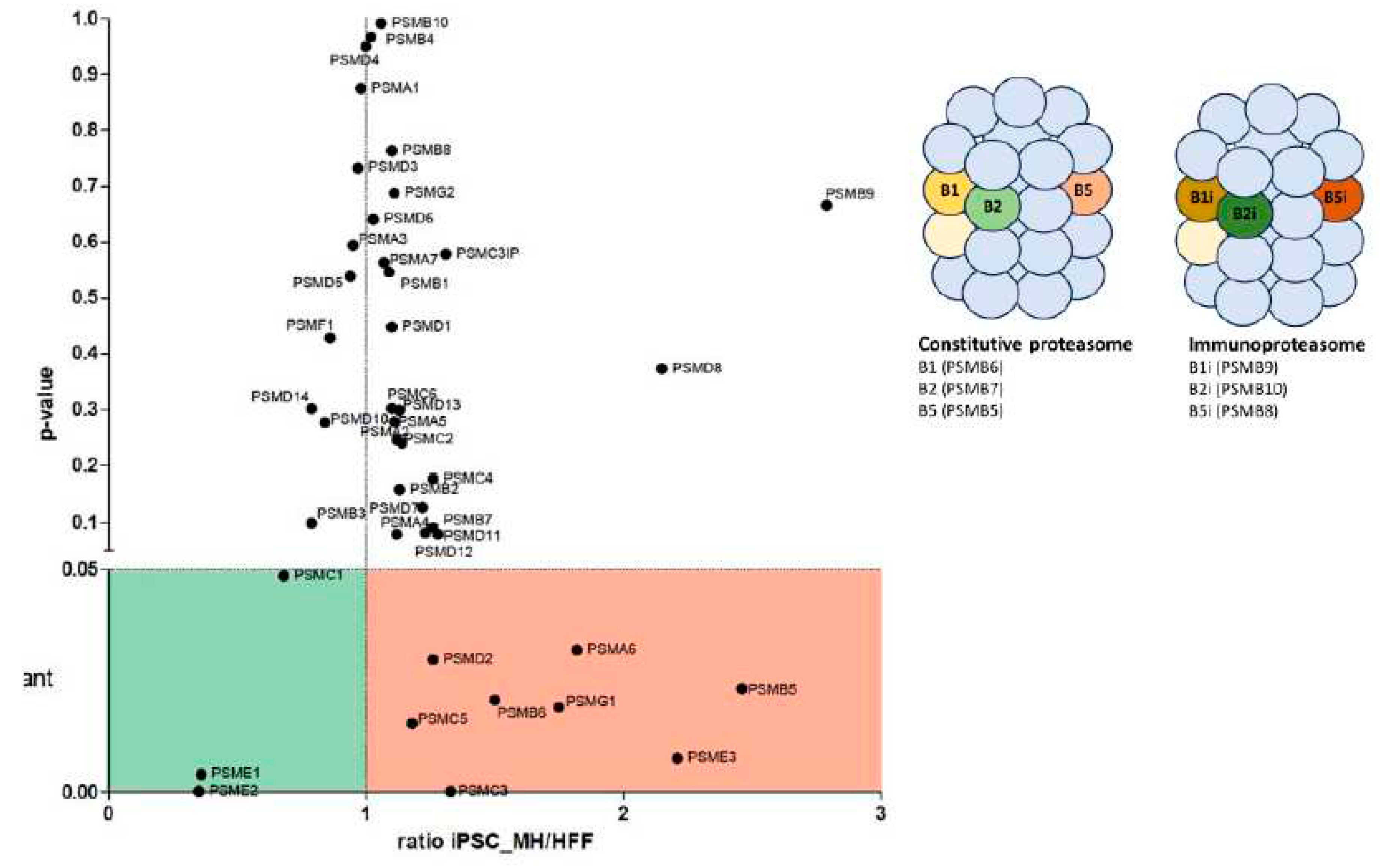
The most abundant proteasome in the cell is the 26S proteasome, which is composed by a catalytic core, the 20S proteasome bound at the ends to a regulatory complex called 19S proteasome. Proteins targeted by the Ubiquitin Proteasome System (UPS) are identified for degradation by the proteasome. The 20S proteasome has a barrel shape with 7 stacking heptameric rings. The outer rings are structural and are composed by 7 α subunits. The 2 inner rings are composed by β subunits, 3 of them have catalytic functions: β1, β2 and β5. Most cells principally express this constitutive or standard proteasome (CP). In addition, other regulatory complexes can replace the 19S complex. In the presence of IFN-γ, these subunits can be replaced by others called β1i, β2i and β5i, forming the immunoproteasome (IP). (6). The assembly of this structure is highly regulated (7). Therefore, an IP structure indicates that the cell is experiencing an inflammatory response.
Dysfunction of the UPS system has been implicated in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) in motor neurons (MN) (8, 9, 10, 11, 12, 13). Mutations in the key UPS regulator UBA1 can cause the juvenile motor neuron disease, spinal muscular atrophy (SMA) (14, 15). In addition, spinal MN appear to present a higher susceptibility than other cell types to UPS stress (16).
The CP is the most abundant proteasome present in most of the body tissues, except in lymphoid tissues where the IP is expressed in higher amount (17). In the nervous system, IP is practically absent in the brain and is more abundant in nerves, but in both the constitutive is the most abundant proteasome type (17).
Here we describe the use of human iPSC cells differentiated to neural stem cells and then motor neurons as a developmental model to better understand the structure of the proteasome in developing motor neurons.
2. Materials and Methods
2.1. Human iPSC differentiation to motor neurons
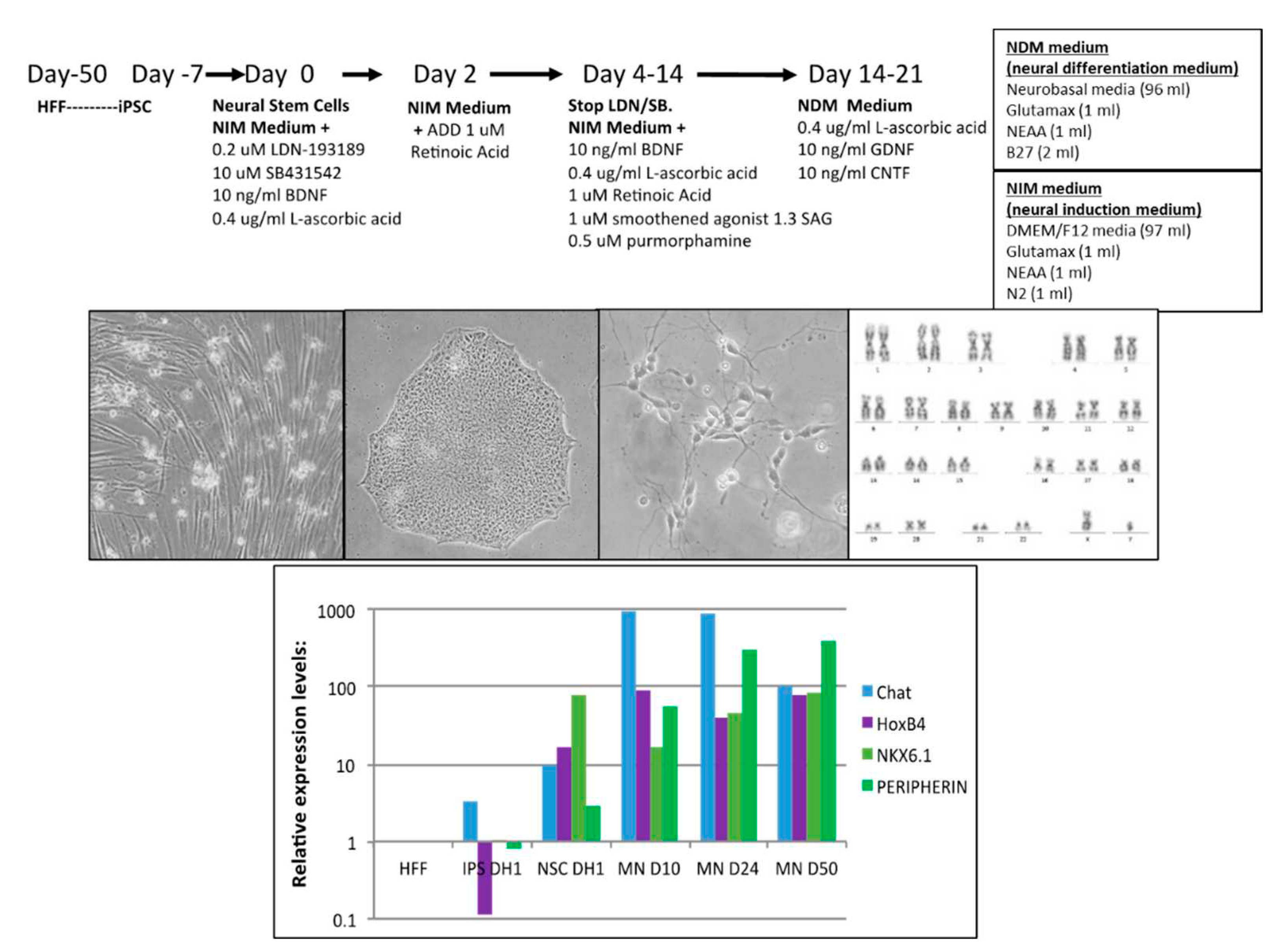
Human iPSC clones were previously generated either by synthetic mRNA transfection methods or retroviral methods and used for this project (3). Briefly, for synthetic mRNA made iPSC, human foreskin fibroblasts (Millipore HFF) were reprogrammed to induced pluripotent stem cells (iPSC) with messenger mRNA transfections and characterized for pluripotency markers (3). Motor Neurons were differentiated from iPSC-derived Neural Stem Cells using the motor neuron induction medium as described in
Figure 1.
HFF, synthetic mRNA made iPSC and NSC were analysed by global proteomics; SP15 and CHB fibroblasts, iPSC1 and iPSC2 (retroviral made) and MN from synthetic mRNA made iPSC included for Western Blot validation of identified proteins (
Table 1).
2.2. Proteomics
Proteomics sample preparation and measurement
Protein concentration of each sample was determined by BCA assay (Pierce). 10µg of each sample were digested by Trypsin and Lys-C endopeptidases applying a modified filter-aided sample preparation protocol as described (Wisniewski et al. Nat Methods 2009). Samples were acidified by trifluoroacetic acid and stored at -20°C. The samples were measured on the Q-Exactive HF mass spectrometer online coupled to an Ultimate 3000 nano-RSLC (Thermo Scientific) in data-independent acquisition (DIA) mode as described previously (Lepper et al. J. Proteome Res. 2018).
Quantitative MS analysis
The DIA LC–MS/MS data set was analysed using Spectronaut Pulsar (Biognosys, Schlieren, Switzerland) as described (Lepper et al. J. Proteome Res. 2018).
Searches were performed using a human spectral meta-library which was generated analysing data-dependent acquisition runs from a variety of human samples in Proteomics Discoverer (Version 2.1, Thermo Scientific). Proteotypic peptide and protein identifications were filtered for a false discovery rate of <1%. Match between runs was enabled with the data filtering function set to q-value percentile mode applying the 20% setting. Resulting peptide and protein quantifications in the individual samples were exported and used for calculations of fold-changes and statistical values.
2.3. Immunofluorescence
Motor neurons grown in chamber slides (ThermoFisher, #177437) were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0,2% Triton X-100 in PBS and blocked in 6% donkey serum for 1 hour. Then, cells were incubated with primary and secondary antibodies. Primary antibodies and dilutions used: anti-Tuj1 (Biolegend, MMS-435P-100, 1:500), anti-Olig2 (R&D systems, AF2418), anti-Nestin (Biolegend, 841801, 1:200) and anti-Sox2 (CalBiochem, sc1002, 1:100). Secondary antibodies used were all Alexa Fluor Series from Invitrogen (dilution 1:200). Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, Roche, 10236276001) and then cover glasses (VWR, ECN631-1575) were mounted on top with Fluoromount-G (Invitrogen, #00-4958-02). Images were taken using a Leica SP5 confocal microscope and were processed using Fiji software.
2.4. Real Time PCR
Total mRNA was isolated using Ambion RNA purification columns kit (#12183018), and 500ng were used to synthesize Cdna using the SensiFAST Cdna synthesis kit (Bioline, BIO65053). One microliter of the reaction was used to quantify gene expression by quantitative PCR as previously described (Aasen et al., 2008). Primers sequences used were Hgapdh Fw: 5’- GCACCGTCAAGGCTGAGAAC -3’, Hgapdh Rv: 5’- AGGGATCTCGCTCCTGGAA -3’, hChat Fw: 5’- AACGAGGACGAGCGTTTG -3’, hChat Rv: 5’- TCAATCATGTCCAGCGAGTC -3’, hHoxB4 Fw: 5’- GTCGTCTACCCCTGGATGC -3’, hHoxB4 Rv: 5’- TTCCTTCTCCAGCTCCAAGA -3’, hNkx6.1 Fw: 5’- ATTCGTTGGGGATGACAGAG -3’, hNkx6.1 Rv: 5’- CCGAGTCCTGCTTCTTCTTG -3’, hPeripherin Fw: 5’- AGACCATTGAGACCCGGAAT -3’ and hPeripherin Rv: 5’- GGCCTAGGGCAGAGTCAAG -3’. Relative quantification was determined according to the ΔΔCT method.
2.5. Karyotype analysis
To stain chromosome G bands, methanol: acetic acid (3:1) fixed cells were dyed with Wright: Sorensen buffer (1:3). Twenty metaphases were assessed per sample and chromosomes were classified using Ikaros software.
2.6. Electrophysiology
For whole-cell patch-clamp experiments, iPSC-derived motor neurons grown in round cover glasses were kept at room temperature in HEPES-based ACSF composed of 135 Mm NaCl, 2 Mm KCl, 2 Mm CaCl2, 1 Mm MgSO4, 10 Mm HEPES, 10 Mm D-glucose; Ph 7.35; 300-310 mOsm/L. Sodium and potassium currents were measured using whole-cell patch-clamp electrophysiology recordings. In several cells, sodium currents were inhibited by tetrodotoxin (TTX). MN firing action potentials were also recorded.
2.7. Western blot
Cell pellets were resuspended in 200μl of Lysis Buffer (LB: 50Mm Tris-HCl Ph 7.4, 150Mm NaCl, 1% Triton X-100, Protease Inhibitor cocktail (completeTM Tablets Mini, Roche)) and homogenized using 30g syringe needles and then, incubated for 1h on ice. All the samples were then centrifuged (13000 g, 10 min, 4 °C) and the supernatants were taken, quantified and stored at -20 °C. The protein concentration of the cell lysates was calculated using the BCA quantification kit (Pierce™ BCA Protein Assay Kit (Thermo Scientific™)) in a colorimetric plate reader (Victor3™ Plate Reader (PerkinElmer ™)). About 10 ug of protein samples were separated by electrophoresis under denaturing conditions (SDS-PAGE) in a 14% polyacrylamide gels. Proteins were transferred to polyvinylidene membranes (Immun-Blot® PVDF Membrane (BioRad ™) previously activated 5 min with methanol for a maximum of 45 min at 100 V. Membranes were subsequently incubated in blocking solution (BS: PBS, 0.1% Tween 20, 5% skimmed milk powder) for 1h with gentle shaking. Then, membranes were washed 3 times with T-PBS (PBS, 0.1% Tween 20) for 5 min with shaking then incubated with the antibody primary at a 1/1000 dilution in T-PBS. Membranes were incubated overnight under shaking conditions at 4°C. After 3 washes with T-PBS, the secondary antibodies were added at a concentration of 1/10000 diluted in T -PBS and membranes were incubated for 1h under shaking conditions at room temperature. Finally, membranes were washed 4 times with T-PBS for 5 min under shaking conditions. Finally, the detection reagent was applied (1: 1 Clarity Western ECL kit Blotting Substrate (BioRad™)) and proteins were detected by chemiluminescence using the Molecular Imager ® VersaDoc ™ (BioRad ™) and QuantityOne software. Fiji software (ImageJ) was used for the densitometry analysis.
The antibodies used for western blot analysis were: anti-β1 (PSMB9) goat monoclonal IgG2a antibody (Santa Cruz BiotechnologyTM); anti-β1i (PSMB6) polyclonal rabbit IgG antibody (Santa Cruz BiotechnologyTM); anti-β2 (PSMB7) mouse monoclonal IgG1 antibody (Santa Cruz BiotechnologyTM); anti-β2i (PSMB10) mouse monoclonal IgG2b antibody (Santa Cruz BiotechnologyTM); anti-β5 (PSMB5) mouse monoclonal IgG2a antibody (Santa Cruz BiotechnologyTM); anti-β5i (PSMB8) mouse monoclonal IgG1 antibody (Santa Cruz BiotechnologyTM); MCP21 mouse polyclonal antibody (anti-α2 subunit); αVinculin (MA5-11690) mouse monoclonal IgG antibody (InvitrogenTM); ECL αMouse IgG, Horseradish Peroxidase-Linked whole antibody from sheep (GE HealthcareTM); Precision ProteinTM StrepTactin-HRP Conjugate 5,000x (BioRadTM); ECL αRabbit IgG, Horseradish Peroxidase-Linked whole antibody from donkey (GE HealthcareTM); αSheep/Goat-Inmunoglobulins Peroxidase (AP360) (The Binding Site LTDTM).
3. Results
3.1. Human iPSC differentiation to motor neurons
Synthetic mRNA generated iPSC clones were successfully differentiated to motor neurons (MN) during 50 days with a defined protocol with good morphology based on phase contrast photos (
Figure 1). Karyotype of MN revealed no major genetic integrity chromosome aberrations (
Figure 1c). RT-PCR characterization captured upregulation of specific MN maturation markers such as Chat, HoxB4, Nkx6.1 and Peripherin at different time points of differentiation (
Figure 1d), and immunofluorescence confirmed positive protein expression of TUJ1, NESTIN, SOX2 and OLIG2 (
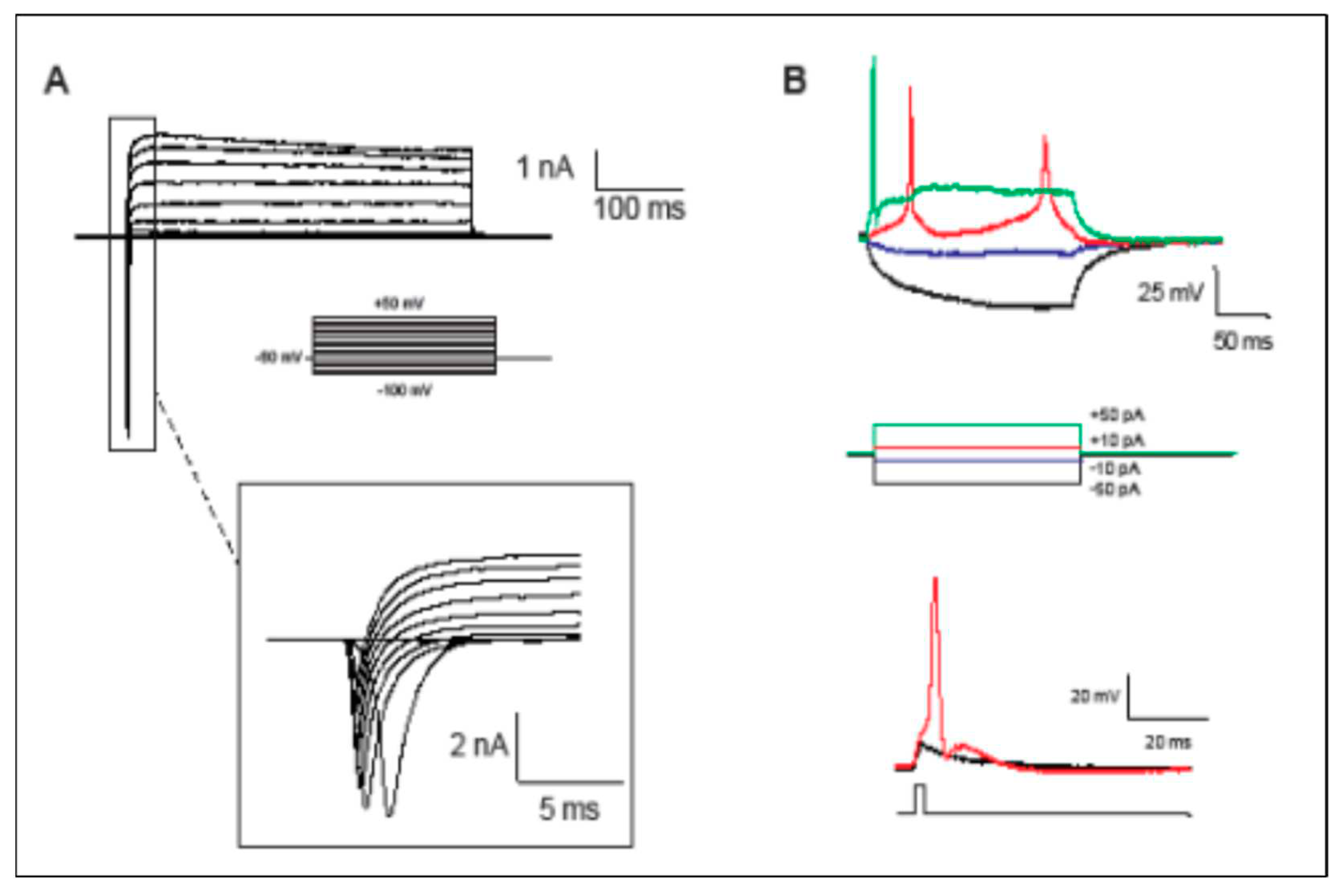
Figure 2), indicative of the successful MN maturation achieved. No significant differences were found between lines. Next, we assessed MN functional activity by patch-clamp electrophysiological recordings (
Figure 3). Neurons demonstrated generation of action potentials (APs) upon electrical stimulation (
Figure 3a and 3b).
3.2. Result of proteomic analysis of HFF, iPSC and NSC
3.2.1. Analysis of HFF vs iPSC.
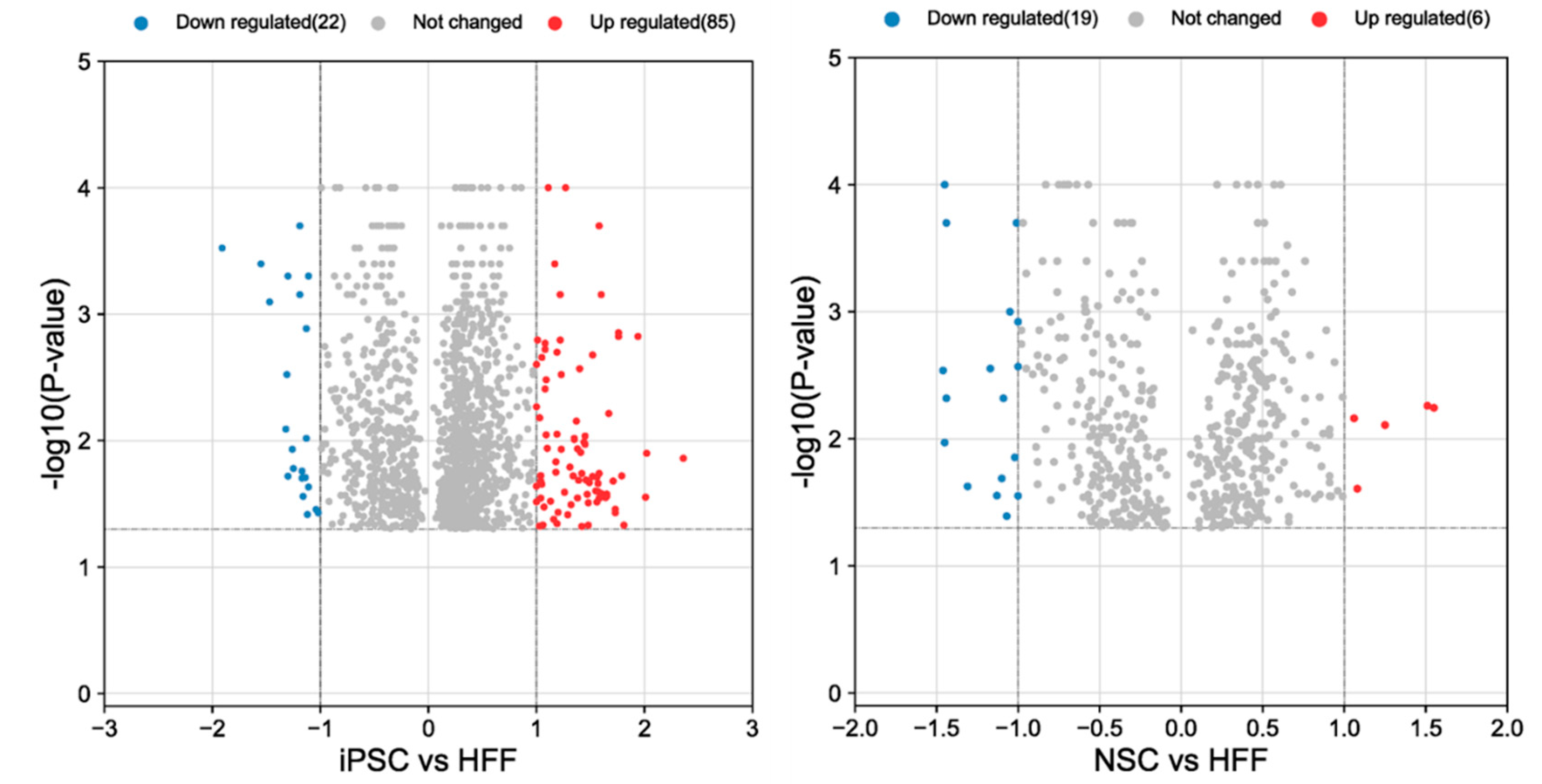
A total of 2192 proteins were identified, and 1600 proteins found to be differentially regulated upon formation of iPSC from HFF. A total of 600 proteins were down regulated (Suppl.
Figure 1, 2 and
Figure 4a) mainly involved in biological processed such as cell proliferation, RNA splicing, protein synthesis, cell signaling, sumo, ubiquitination (ligases, de-ubi), HAT1, HDAC, Methyltransferases, HSP, Hif1a, metabolic changes, RNA polymerase II-related (Suppl.
Figure 1: Excel table of proteomics).
Interestingly, a variation of several UPS proteins was observed. First, we found a strong downregulation of PSME1 (PA28alpha) and PSME2 (PA28beta) upon iPSC differentiation. These are IFNγ-induced proteins that compose a regulatory complex (PA28) that can replace the 19S regulatory complex at the ends of the 20S proteasome to form the immuno-proteasome (IP). In contrast, a significant upregulation of PSME3 (PA28-gamma) and PSMG1 proteins were observed in iPSC. PSME3 protein has been implicated in cancer by inhibiting c-Myc degradation and is also is a target gene of NF-κB during bacterial infections. The PSMG1 protein enables molecular adaptor activity in chaperone-mediated protein complex assembly located in the golgi apparatus, endoplasmic reticulum, and nucleoplasm. Although fibroblasts mainly express CP, the proteomics analysis showed that immunosubunits (β1i (PSMB9), β2i (PSMB10) and β5i (PSMB8) were detected in lower amounts in iPSC in comparison with fibroblasts. The opposite was observed for CP subunits (β1 (PSMB6), β2 (PSMB7) and β5 (PSMB5) (
Figure 5 ). Thus, IFNγ-induced catalytic or regulatory subunits were downregulated in iPSC regarding fibroblasts. Downregulation of these proteins, which are antigen processing related factors, may affect antigen processing, what can produce a change in the HLA-I peptide repertoire presented to CD8 T lymphocytes.
3.2.2. Analysis of iPSC and NSC.
A total of 800 proteins were found with a total of 314 proteins that were down regulated (Suppl Fig 1 Excel file). We found 1.8-fold upregulation of PSMF1, and downregulation of PSMD1, PSMD6 and PSMC6 proteins in NSC compared to HFF (
Figure 4b). Although it was not statistically significant, a trend in which IP subunits were increased and CP decreased in NSC in comparison with iPSC was observed. In addition, PSMF1, a subunit of the Proteasome inhibitor PI31 was statistically increased (
Figure 6). This protein inhibits the activation of the 20S proteasome by PA28.
3.3. Validation of constitutive and immunoproteasome findings
First, we validated the proteomics data by western blot that the CP subunits (β1 (PSMB6), β2 (PSMB7) and β5 (PSMB5) confirming they are upregulated in iPSC compared to the starting HFF (Suppl Fig 3).
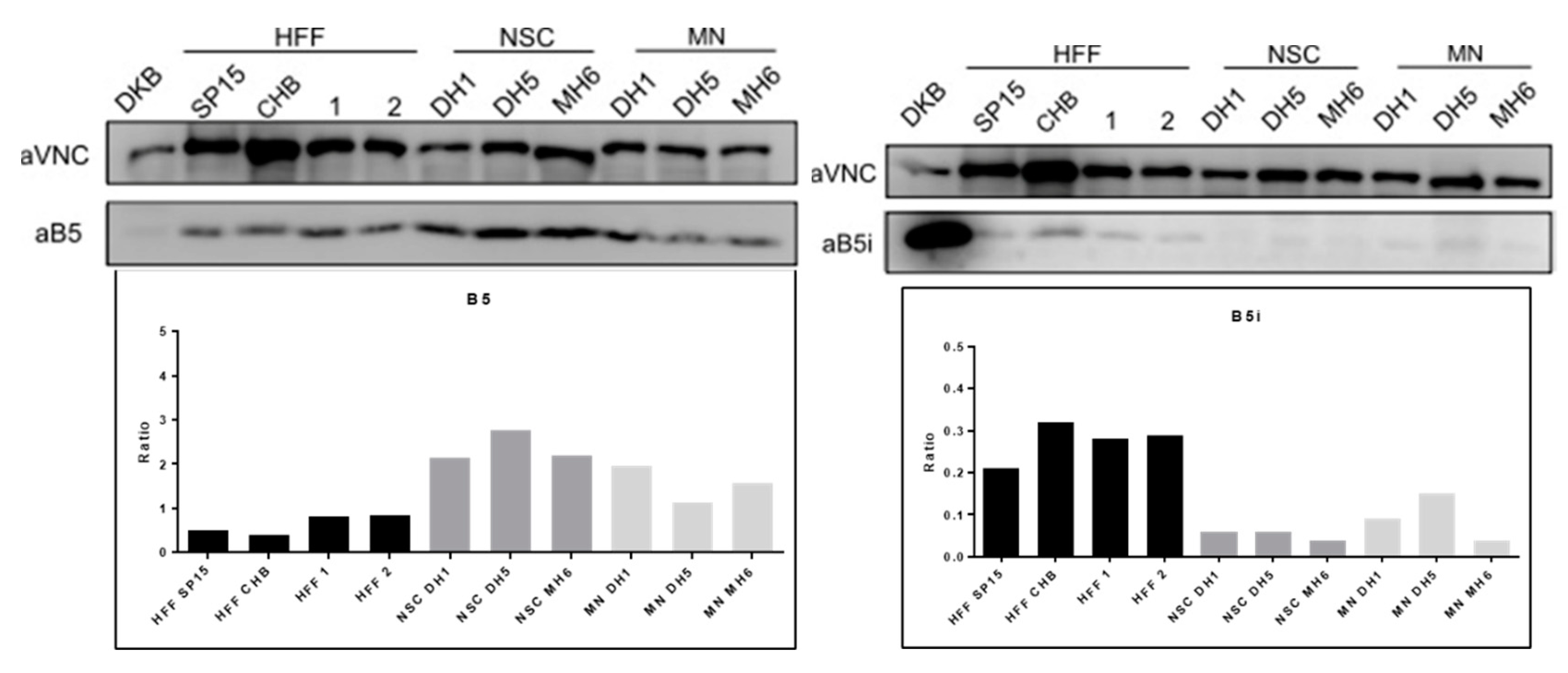
To evaluate the expression levels of CP or IP subunits in fibroblasts, neural stem cells (NSC) and motor neurons (MN), western blot experiments were performed. As expected, the abundance of the CP subunits was high in all cell types, being more expressed in NSC and MN than in fibroblasts, demonstrating that CP subunits (β1 (PSMB6), β2 (PSMB7) and β5 (PSMB5) expression is higher in iPSC, NSC and MN than HFF (Suppl Fig 3 and
Figure 6). On the other hand, IP subunits were present in a lower amount in all cell types. In fact, β1i and β2i could not be detected in fibroblast cells, NSC or MN, and they were only detected in DKB cells, a lymphoblastoid cell line used as control of IP expression. The β5i subunit was detected in fibroblasts, NSC and MN in comparison with DKB cells. The expression level of the β5i subunit was higher in fibroblasts than in NSC or MN. We observe a trend that IP subunits are increased in MN compared to NSC, while CP are decreased, suggesting an IP structural phenotype in MN cells (
Figure 6).
Thus, the data indicate that the undifferentiation of fibroblasts to iPSC reduces the abundance of IP which is progressively recovered during differentiation to MN.
4. Discussion
Proteomics and western blot data validation in iPSC and MN: The western blot data validated the proteomics analysis and consistently demonstrated that the CP subunits were higher expressed in NSC and MN than in fibroblasts, demonstrating that CP subunits β1 (PSMB6), β2 (PSMB7) and β5 (PSMB5) expression is higher in iPSC, NSC and MN than HFF (Suppl Fig 3 and
Figure 6). The data agrees with previous results that showed a reduction of IP subunits in iPSC (18). On the other hand, IP subunits were present in a lower amount in all cell types. In fact, β1i and β2i could not be detected in fibroblast cells, NSC or MN, and they were only detected in DKB cells (data not shown).The β5i subunit was detected in fibroblasts, NSC and MN in comparison with DKB cells. The expression level of the β5i subunit was higher in fibroblasts than in NSC or MN (
Figure 5i). We observe a trend that IP subunits are increased in MN compared to NSC, while CP are decreased, suggesting an IP structural phenotype in MN cells (
Figure 6). Therefore, our data suggest that the catalytic activity and specificity of the proteasomes are associated with MN development. The fact that catalytic proteasome subunits expressed in fibroblasts differ from NSC and MN, indicates that the HLA-I immunopeptidomes presented on the cell surface will likely differ, as CP and IP present different protein cleavage specificities.
The role of the UPS and MHC: The important role of the UPS in pluripotent stem cell survival and motor neuron differentiation has been reported previously (18). This work demonstrated that iPSC are very sensitive to the proteasome inhibitor MG132, and MN were more resilient that iPSC, but more sensitive than fibroblasts. In addition, different mutations in the UPS are related with the development of neurodegenerative diseases, highlighting further the importance in neuron survival and health (9; 10; 11; 12; 13; 14; 15).
MHCI expression in disease: One of the roles of the UPS is to produce small peptides to be presented by MHC-I. Our data suggests that this might be different between patients and transplanted MN because of the structural change of the proteasome. Recent evidence from independent laboratories have reported expression of immune-like MHCI receptors in the central nervous system (4,5). This work provides some insight into the mechanism of the role of Major histocompatibility complex class I proteins in brain development and plasticity (4,5). The expression of MHC-I molecules and the immunoproteasome is highly increased in spinal motor neurons of transgenic mice carrying the mutant SOS1G93A during the progression of the disease (19). Thus, in future studies it may be relevant to evaluate the role of the immunoproteasome in neurologic disorders using iPSC-derived MN.
MHC peptides technique: The MHC-presented small peptide repertoire, also referred as immunopeptidome, cannot be determined from mRNA or protein abundance. The technique used to characterize the immunopeptidome uses mass spectrometry analyses of peptides eluted from MHC complex isolation. However, mass spectrometry is not always sufficient to define the full repertoire of small peptides loaded onto MHC molecules. Moreover, the proinflammatory environment of spinal cord and the dysregulated protein metabolism of motor neurons in ALS may promote the activation of the IP and the membrane presentation of antigenic peptides recognized as non-self by CD8+ T cells, which then activates a cytotoxic autoimmune response (21).
Misfolded proteins in neuronal disease: The role of misfolded proteins in motor neurons is one of the main neuropathological hallmarks of ALS and is regarded as the prime cause of motor neuron degeneration. Misfolded proteins that cannot be degraded by the constitutive proteasome are directed toward the INF-γ activated IP (22). Moreover, apart from misfolded proteins, peripheral interaction of motor neurons with cytotoxic CD8+ T cells may be the direct cause of motor axon injury in SOD1G93A mice. However, how MHCI is triggered in ALS motor neuron remains to be defined. Misfolded proteins in the motor neurons may trigger the inflammation through the release of danger-associated molecular pattern molecules (DAMPs), including ROS, HMGB1, and HSPs, which activate glial and immune cells to produce inflammatory cytokines including IFN-γ (22). IFN-γ may then induce upregulation of the IP in the motor neurons. Recent work highlights a non-detrimental role of MHCI activation in ALS motor neurons and instead hypothesize a protective role (23).
In conclusion, the data reveals a decrease in immunoproteasome associated proteins in NSC and MN derived from iPSC suggesting that the HLA-I immunopeptidomes presented on the cell surface will differ that could have implications in disease modeling and transplant of iPSC-derived MN in the future. Further work is warranted to describe the different small peptide repertoire presented by MHCI in healthy and diseased MN derived from iPSC.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: EXCEL Proteomic data. Figure S2: Heatmap. Figure S3: Western blot of iPSC and HFF for Beta 1,2 and 5 subunits of the CP.
Author Contributions
Conceptualization, M.J.E; I.A Methodology, M.J.E; I.A; B.A.P Software, A.T.H; J.R.O. Validation, I.A.; A.T.H Formal analysis, I.A.; A.T.H; M.J.E; B.A.P; J.R.O. Investigation, I.A.; A.T.H; M.J.E; B.A.P; J.R.O. Resources, I.A; M.J.E Data curation, I.A; M.J.E Writing—original draft preparation, M.J.E Writing—review and editing, I.A.; A.T.H; M.J.E; B.A.P; J.R.O. Visualization, B.A.P, M.J.E Supervision, I.A; M.J.E; B.A.P Project administration, B.A.P; M.J.E Funding acquisition, I.A; M.J.E. All authors have read and agreed to the published version of the manuscript.
Funding
Use of human iPSC and tissue obtained by collaborators from published work funded by approved projects 2012-StG-311736-PD-HUMMODEL, BFU2016-80870-P, Red de Terapia Celular - TerCel RD16/0011/0024 and BFU2011–26596, BFU2014-54467-P and Fundació La Marató de TV3 20172110/FBG309768.
Institutional Review Board Statement
The human fibroblast and iPSC used in this study have been approved from previous publication and the generation and/or use of human iPSC were approved by the Spanish competent authorities (Commission on Guarantees concerning the Donation and Use of Human Tissues and Cells of the Carlos III National Institute of Health (3).
Informed Consent Statement
The human fibroblast and iPSC used in this study have been approved from previous publication and the generation and/or use of human iPSC were approved by the Spanish competent authorities (Commission on Guarantees concerning the Donation and Use of Human Tissues and Cells of the Carlos III National Institute of Health (3).
Data Availability Statement
The full proteomic data is available in an Excel file in the Supplementary Figures.
Acknowledgments
We acknowledge the Silke Meiners laboratory for the Proteomic analysis, performed at the Helmholtz Zentrum München Deutsches Forschungszentrum fuer Gesundheit und Umwelt (GmbH), Ingolstaedter Landstr. 1, 85764 Neuherberg. The Universty of Barcelona, Unit of Physiology for the patch clamp analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [CrossRef]
- Miura K, Okada Y, Aoi T, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743-745. [CrossRef]
- Alvarez-Palomo AB, Requena-Osete J, Delgado-Morales R, et al. A synthetic mRNA cell reprogramming method using CYCLIN D1 promotes DNA repair, generating improved genetically stable human induced pluripotent stem cells. Stem Cells. 2021 Jul;39(7):866-881. [CrossRef] [PubMed]
- Elmer BM, McAllister AK. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci (2012) 35:660–70. [CrossRef]
- Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron (2009) 64:40–5. [CrossRef]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007 Jun 1;316(5829):1349-53. [CrossRef] [PubMed]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009 Feb;10(2):104-15. [CrossRef] [PubMed]
- Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches.. Bioessays. 2008 Nov;30(11-12):1172-84. [CrossRef] [PubMed]
- Yerbury JJ, Ooi L, Dillin A, Saunders DN, Hatters DM, Beart PM, Cashman NR, Wilson MR, Ecroyd H. Walking the tightrope: proteostasis and neurodegenerative disease. J Neurochem. 2016 May;137(4):489-505. [CrossRef] [PubMed]
- Williams KL, Topp S, Yang S, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016 Apr 15;7:11253. [CrossRef] [PubMed]
- Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011 Aug 21;477(7363):211-5. [CrossRef] [PubMed]
- Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010 Dec 9;68(5):857-64. [CrossRef] [PubMed]
- Fecto F, Yan J, Vemula SP et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011 Nov;68(11):1440-6. [CrossRef] [PubMed]
- Dlamini N, Josifova DJ, Paine SM, et al. Clinical and neuropathological features of X-linked spinal muscular atrophy (SMAX2) associated with a novel mutation in the UBA1 gene. Neuromuscul Disord. 2013 May;23(5):391-8. [CrossRef] [PubMed]
- Ramser J, Ahearn ME, Lenski C, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet. 2008 Jan;82(1):188-93. [CrossRef] [PubMed]
- Brockington A, Ning K, Heath PR, et al. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013 Jan;125(1):95-109. [CrossRef] [PubMed]
- Boegel S, Löwer M, Bukur T, et al. HLA and proteasome expression body map. BMC Med Genomics. 2018 Mar 27;11(1):36. [CrossRef] [PubMed]
- Bax M, McKenna J, Do-Ha D, et al. The Ubiquitin Proteasome System Is a Key Regulator of Pluripotent Stem Cell Survival and Motor Neuron Differentiation. Cells. 2019 Jun 13;8(6):581. [CrossRef] [PubMed]
- Bendotti C, Marino M, Cheroni C, et al. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol. 2012 May;97(2):101-26. [CrossRef] [PubMed]
- Thibault P, Perreault C. Immunopeptidomics: Reading the Immune Signal That Defines Self From Nonself. Mol Cell Proteomics. 2022 Jun;21(6):100234. [CrossRef] [PubMed]
- Nardo G, Trolese MC, Bendotti C. Major Histocompatibility Complex I Expression by Motor Neurons and Its Implication in Amyotrophic Lateral Sclerosis. Front Neurol. 2016 Jun 13;7:89. [CrossRef] [PubMed]
- Bendotti C, Marino M, Cheroni C, Fontana E, Crippa V, Poletti A, De Biasi S.Prog Neurobiol. 2012 May;97(2):101-26. [CrossRef] [PubMed]
- Nardo G, Iennaco R, Fusi N, et al. Transcriptomic indices of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis. Brain. 2013 Nov;136(Pt 11):3305-32. [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).