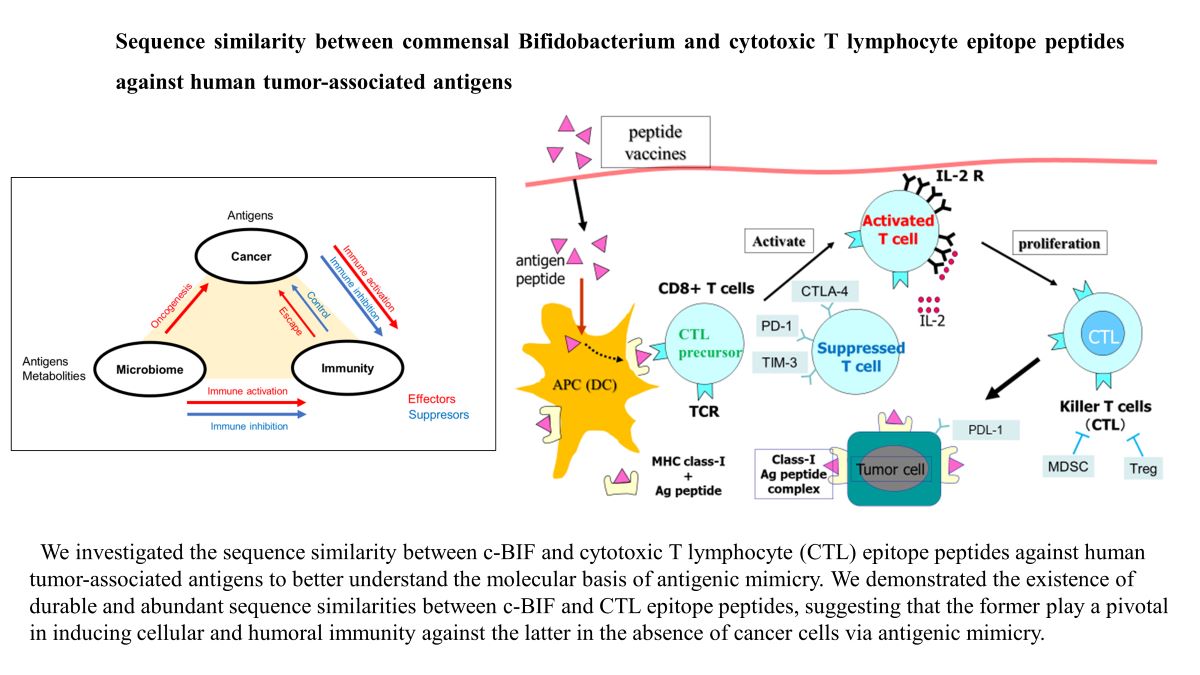
1. Background
Over the past two decades, immune checkpoint inhibitors (1,2), gene-modified T cell therapy (3), and target therapies using various types of monoclonal antibodies (4) have led to remarkable success in cancer therapy and cancer immunoprevention. Further, Sivan et al. reported that commensal Bifidobacterium (c-BIF) not only promoted anti-tumor immunity but also enhanced the efficacy of immune checkpoint inhibitors in a mouse model (5), potentially suggesting a new treatment modality for clinical use. Zitvogel et al. hypothesized that gut microbial proteins might be sufficiently similar to human tumor antigens to be capable of eliciting tumor-specific T lymphocytes and antibodies that can recognize future tumor cells via “antigenic mimicry” (6). Mitsuoka provided an overview of the mechanisms by which intestinal microbiota influence host immunity and cancer prevention (7). We previously reported that the antibodies reactive to each of 31 different CTL epitope peptides not only were detectable in a majority of healthy donors (8), but also inhibited tumor growth in association of activation of dendritic cells and suppression of T regulatory cells at the tumor site in a mouse model (9), suggesting a potential new treatment modality.
Despite this wide body of research, however, the molecular basis of antigenic mimicry in not yet well understood. In the present study, therefore, we investigated the sequence similarity between c-BIF and the 31 CTL epitope peptides against tumor-associated antigens to understand whether c-BIF plays a role in the induction of cellular and humoral immunity in the absence of tumor cells via antigenic mimicry. We also conducted TCR-mediated recognition similarity analysis between commensal clostridium, a pathogenic bacterium, and CTL peptides.
2. Materials and Methods
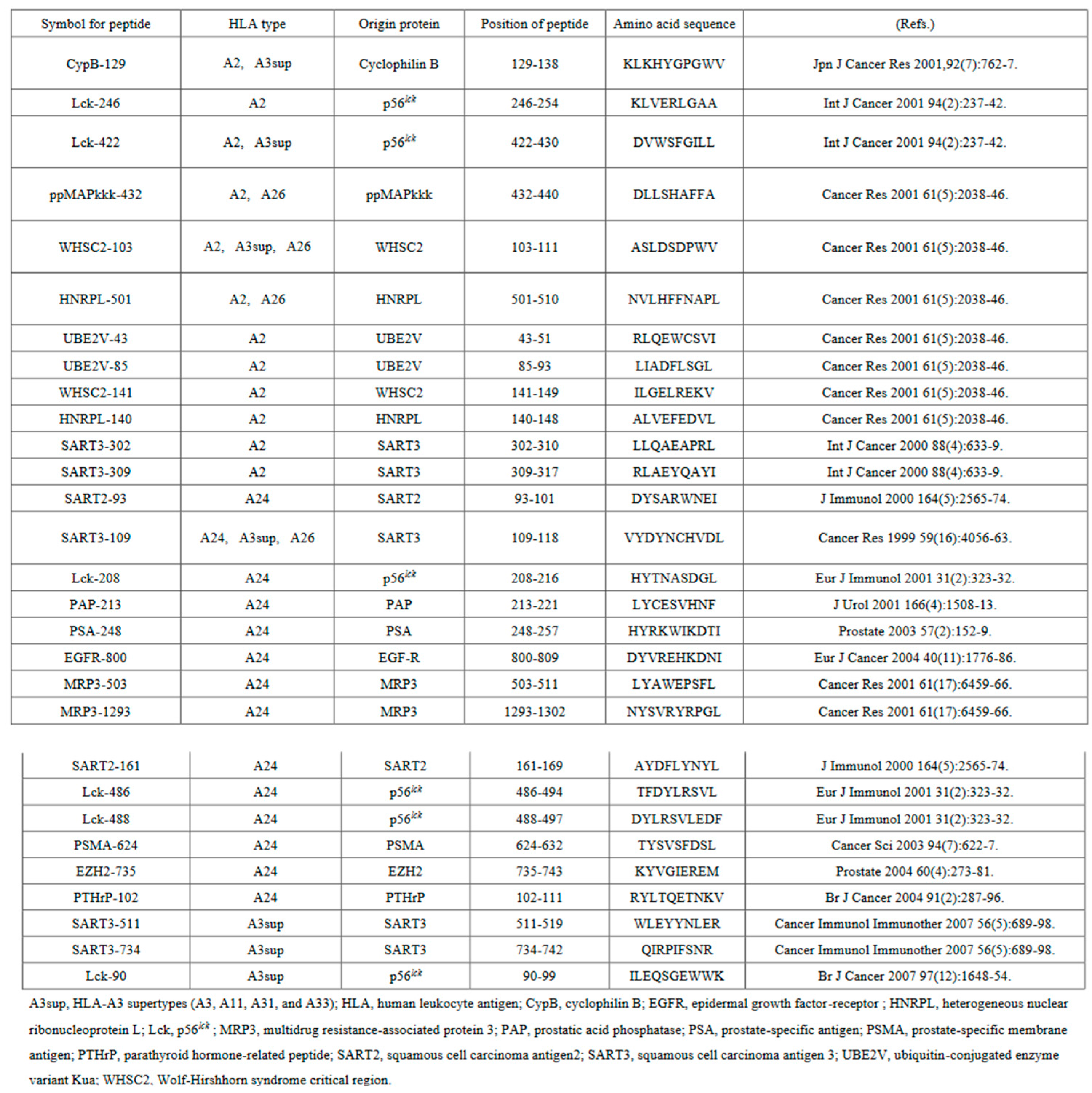
We employed two different similarity analyses: (1) a linear sequence similarity analysis between c-BIF and each of the 31 CTL epitope peptides shown in
Table 1, and (2) a similarity analysis of T cell receptor (TCR)-mediated recognition of c-BIF and each of the 31 CTL epitope peptides on antigen-presenting cells shown in
Table 1. These analyses were conducted based on the information from GenPept.Graphics Next Previous Descriptions, as shown in Supplemental Information 1. We also conducted TCR-mediated recognition similarity analysis between commensal clostridium and CTL peptides based on the information from GenPept.Graphics Next Previous Descriptions shown in Supplemental Information 2.
In order to avoid any possible biases, a sequence was considered to be positive for linear sequence similarity if it consisted of at least 5 identical amino acids between c-BIF and each of 31 CTL epitope peptides.
T lymphocytes recognize complexes of 9 to 10 length peptides and antigen-presenting cells through TCRs. TCRs contain a single binding site for antigens, i.e., the complementarity determining region (CDR) 3 within one of the 4th to 7th positions’ amino acids, and also two different binding sites for major histocompatibility complex (MHC), CDR 1 within one of the positions of 2nd or 3rd amino acid, and CDR2 within one of the positions of 8th to 9th amino acids, respectively, whereas an amino acid at 1st position of CTL epitope peptide is scarcely involved in the binding to TCR, respectively (10). Then, the definition of positive sequence similarity was evaluated between 8 amino acids of the CTL epitope peptide from the 2nd~9th position and the corresponding c-BIF in the case of peptides of 9 amino acids, or both 2nd ~9th and 3rd ~10th amino acids in the case of peptides of 10 amino acids. Positive sequence similarity was defined as follows: (1) a positive sequence consists of at least one identical amino acid from CDR1, CDR2, and CDR3, (2) a positive sequence consists of at least 5 identical amino acids, and (3) the majority (>75%) of amino acids of CTL epitope peptides were identical to those of c-BIF-derived peptide to avoid any possible biases. It also adapted to commensal clostridium.
3. Results
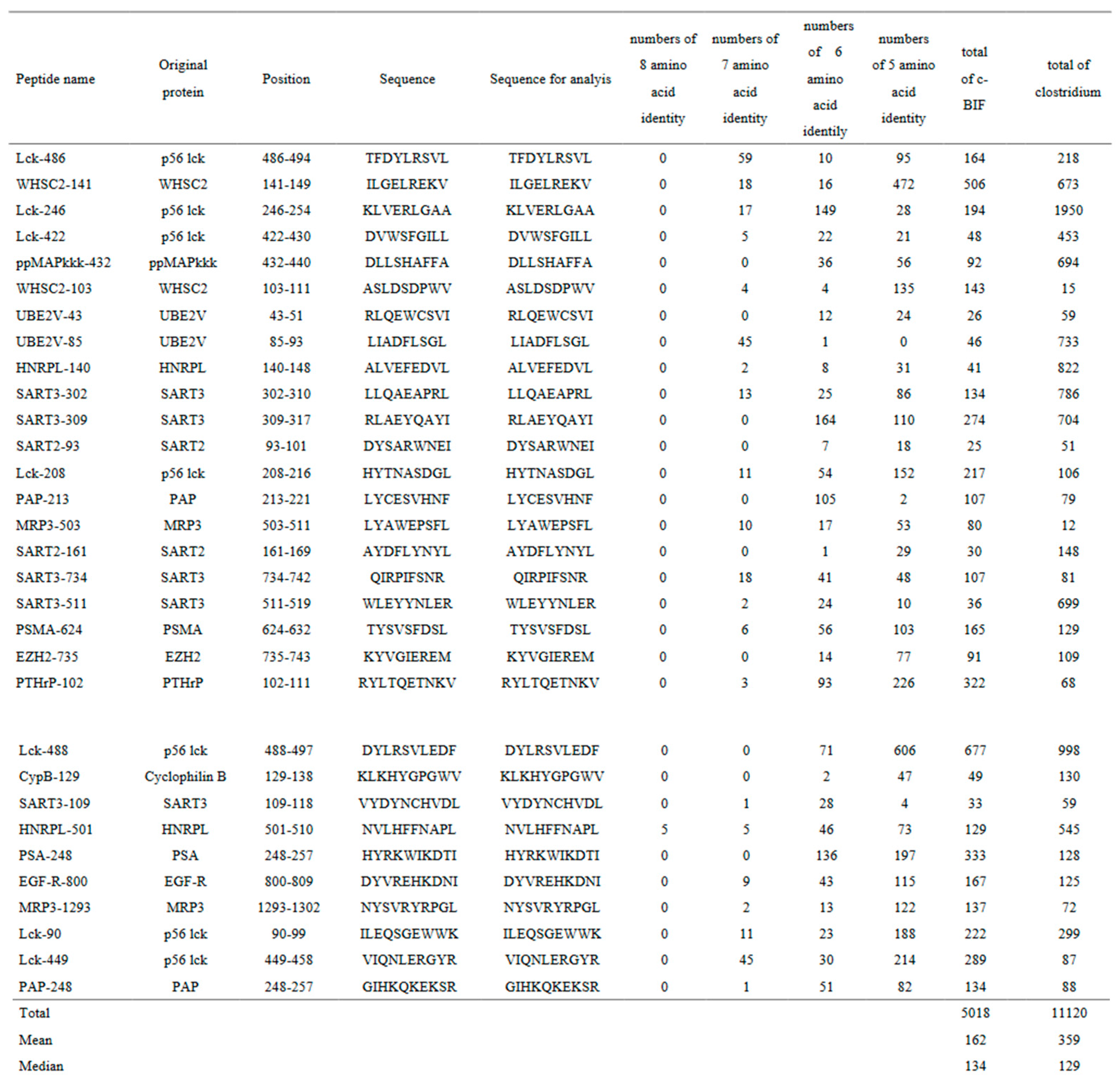
Linear sequence similarity analysis
The linear sequence similarity between c-BIF and each of 31 CTL epitope peptides against tumor-associated antigens was investigated and the results are shown in
Table 2. The analysis revealed 4,002 positive numbers of similarity sites between them with 129 mean per peptide ranging from 84 to 443. Median positive numbers were 107.
Details for the Lck486-94 peptide are shown as a representative case among the 31 peptides, since the Lck486-94 peptide was expressed on both metastatic tumor cells and activated T cells, and then one of the suitable peptides used for the study (11). The Lckprotein, a member of the proto-
oncogene tyrosine-protein kinase family, is essential for T cell activation and also for anchor-independent proliferation of tumor cells. Furthermore, a monoclonal Lck486-94 peptide antibody inhibited tumor growth in a mouse model in association with activation of dendritic cells and suppression of T regulatory cells (9). There are no eight amino acid identity site, 54 seven amino acid identity sites, 13 six amino acid identity sites, and 44 five amino acid identity sites. Collectively, a total of 111 amino acid identity sites existed between c-BIF and the LCK486-94 peptide (
Table 2).
4. Discussion
We employed both a linear sequence analysis and a similarity analysis of TCR-mediated recognition of CTL epitope peptides on antigen-presenting cells. This is mainly because the antibody recognition patterns are different from TCR-mediated recognition. Antibody interacts with antigen in the extracellular space of antigen-presenting cells. Antibodies produced by activated B lymphocytes generally recognize only a small region on the surface of a peptide or protein. In contrast, TCR recognizes the peptide antigen presented in the form of a complex of antigen bound to the MHC molecule. Further, B lymphocyte differentiation into plasma cells to produce a subclass of antibodies requires the assistance of antigen-specific helper T lymphocytes. Accordingly, linear sequence analysis may contribute to a better understanding of antibody-mediated antigenic mimicry, while similarity analysis of T cell receptor (TCR)-mediated recognition may contribute to a better understanding of T lymphocyte-mediated antigenic mimicry. Regardless of those assumption, the results showed the existence of durable and abundant sequence similarities between c-BIF and CTL epitope peptides by either linear sequence analysis or TCR-mediated recognition similarity analysis.
We performed the function of c-BIF peptides showing positive sequence similarity to the Lck486-94 peptide as one representative case among the 31 peptides. Seven amino acids (FDYLRSVL) beside Y (underlined letter) at the 4th position of the Lck486-94 peptide were identical to one site of as large as 57 different sites of 77~84 positions of Bifidobacterium longum (Lacl family transcriptional regulatory). Transcriptional regulators regulate diverse genes and complex regulars in bacteria and eukaryotes, which in turn might be important to better understand how c-BIF plays a role in the induction of cellular and humoral immunity in the absence of tumor cells via antigenic mimicry.
In contrast, commensal clostridium might make almost no contribution to the antigenic mimicry, since there is a very low percentage of commensal clostridium in stool (<one millionth) with little immune activation of tumor immunity (7). Further, more than half of these peptides belonged to hypothetical protein. A few of them function either as the RNA-processing protein RimM, which is essential for processing of 16S rRNA, or the DNA gyrase subunit A, which belongs to type II topoisomerase and negatively supercoils closed circular double-stranded DNA.
Mitsuoka described how intestinal microbiota influenced host immunity and cancer prevention, and mentioned that Bacteroides, eubacterium and c-BIF promote health, including by promoting the maintenance of a normal immune response (7). He also noted that commensal clostridium is mostly involved in many types of diseases as pathogenic bacteria. These results suggest that commensal clostridium rather disturbs the induction of cellular and humoral immunity in the absence of tumor cells via antigenic mimicry.
It might seem reasonable to expect that sequence similarity between c-BIF and human-immunodeficiency virus (HIV)-specific virus via antigenic mimicry also exists, as suggested by Su et al. (12). They showed the existence of memory T lymphocytes specific to HIV and c-BIF. Therefore, the sequence similarity analyses conducted in this study might lead to an improved understanding of whether c-BIF plays a role in the induction of cellular and humoral immunity reactive to HIV in the absence of viral infection via antigenic mimicry.
In this manuscript we have demonstrated that robust and abundant amino acid similarities exist between c-BIF and CTL epitope peptides against tumor-associated antigens, indicating that c-BIF could be a key player capable of inducing cellular and humoral immunity in the absence of cancer cells via antigenic mimicry.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Authors Contributions
K.Itoh provided basic concept of the manuscript. S.Schichijo provided the information from GenPept.Graphics Next Previous Descriptions in Supplemental Information 1 and 2. S. Suekane provided information of intestinal microbiota influence host immunity and cancer prevention. All authors were involved this manuscript preparation with acceptance for the submission.
Funding support
No outside support to be mentioned.
Ethics approval and consent to participate
This manuscript is a research manuscript without information on any individual.
Consent for publication
We agreed this manuscript to be published in British Journal of Cancer.
Data availability
All the data were available upon request.
Acknowledgments
We thanks S.Matueda for information on Lck peptides.
References
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al: Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Eng J Med 366, 2443–2454, 2012. [CrossRef]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu W-J, Topalian SL, Hwu P, et al: Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Eng J Med 366: 2455–2465, 2012. [CrossRef]
- CH June, RS O’Conner, OU Kawalelenet., S Ghassemi MC, Milone DOI: CAR T cell immunotherapy for human cancer. Science 2018, 359. 1361-1365. [CrossRef]
- JP.Manis, DE. Furst, JS.Tirnauer, and AM.Feldweg: https://www.update.com/contents/overview of therapeutic monoclonal antibodies.
- Ayelet Sivan, Leticia Corrales, Nathaniel Hubert, Jason B. Williams, Keston Aquino-Michaels et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015, 350, 1084-1089. [CrossRef]
- L.Zitogel, M.Ayyoub, B.Routy, and G.Kroemer. Microbiome and Anticancer Immunosurveillance. Cell. 165, 2016, 276-287. [CrossRef]
- T.Mitsuoka, Bifidobacteria and their role in human health. J Indust Microbiol 6:263-268 Journal of Industrial Microbiology, Volume 6, Issue 4, 1 December 1990, Pages 263–267. [CrossRef]
- Matsueda S, Komatsu N, Kusumoto K, Koga S, Yamada A, Kuromatsu R, et al. Humoral immune responses to CTL epitope peptides from tumor-associated antigens are widely detectable in humans: a new biomarker for overall survival of patients with malignant diseases. Dev Comp Immunol. 2013;41(1):68-76. [CrossRef]
- Matsueda S, Itoh K, Shichijo S. Antitumor activity of antibody against cytotoxic T lymphocyte epitope peptide of lymphocyte-specific protein tyrosine kinase. Cancer Sci. 2018 Mar;109(3):611-617. [CrossRef]
- C.Szeto, CA.Lobes, AT.Nguyen, and S.Gras, TCR recognition of Peptide-MHC-I: Rule Markers and Breakers. Int. J. Mol. Sci.2021 22, 68. [CrossRef]
- Harashima N. Tanaka K, Sasatomi T, Locoregional Y, Miyagi Y, Yamada A, Tamura M, Yamana Y, Itoh K Shichijo S. Recognition of the Lck tyrosine kinase as a tumor antigen by T cells of metastatic cancer patients. Eur J Immunol 31:323-332, 2001. [CrossRef]
- LF.Su, BA.Kidd, A.Han. JJ.Kotzin, and MM. Davis. 2013. Virus-specific CD4(+)memory-phenotype T cells are abundant 9n unexposed adults. Immunity 38,373-383. [CrossRef]
Table 1.
Information on the 31 peptides CTL epitope peptides against human tumor associated antigens used for.
Table 1.
Information on the 31 peptides CTL epitope peptides against human tumor associated antigens used for.
Table 2.
Linear sequence similarity analysis between commensal BIF and each of the cytotoxic T lymphocyte epitope peptides against human tumor-associated antigen.
Table 2.
Linear sequence similarity analysis between commensal BIF and each of the cytotoxic T lymphocyte epitope peptides against human tumor-associated antigen.
Table 3.
TCR-mediated sequence similarity analysis between commensal BIF and each of the cytotoxic T lymphocyte epitope peptides against human tumor-associated antigen.
Table 3.
TCR-mediated sequence similarity analysis between commensal BIF and each of the cytotoxic T lymphocyte epitope peptides against human tumor-associated antigen.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).