1. Introduction
Lactic acid bacteria (LAB) are a group of Gram-positive, non-spore-forming, cocci or rods, catalase-negative microorganisms [
1] widely used in fermentation, probiotics, and food/beverage manufacturing industries. LAB are Generally Regarded as Safe (GRAS) by the United States Food and Drug Administration (FDA), and boast the Qualified Presumption of Safety (QPS), as assessed by the European Food Safety Authority (EFSA) [
2]. Moreover, LAB have been widely used to improve the taste, texture, and nutritional properties of a wide variety of foods, including vegetables, meat, dairy and cereal substrates [
3], and can extend their shelf life by producing organic acids, carbon dioxide, and antimicrobial peptides [
4].
In the food industry, LAB have become increasingly important, and many studies have been focused on the possible use of bacteriocin-producing species as alternatives to chemical preservatives in foods [
5]. Indeed, certain LAB strains exhibit antimicrobial activity against foodborne pathogens, including bacteria, yeast and filamentous fungi. Furthermore, in recent years, many authors proved that LAB have the ability to neutralize several undesired microbes, including
Clostridium spp.,
Enterococcus faecalis,
Listeria monocytogenes [
6].
Thanks to their fermentation ability, LAB are employed in manufacturing many foods, such as dairy products, sausages, cucumber pickles and olives [
7]. Indeed, LAB can be divided into starter cultures, used to drive biochemical changes of primary relevance for food fermentation, and non-starter cultures, usually deriving from the autochthonous microbiota of the food matrix [
8] and contributing in secondary aspects to the organoleptic characteristics of the final product [
9]. The antimicrobial potential of non-starter LAB isolated from fermented foods has been substantiated by scientific literature and involves the production of organic acids, hydrogen peroxide and bacteriocins.
Besides antimicrobial and fermentation capacities, some LAB also exhibit probiotic features. According to the United Nations of Food and Agriculture Organization and the World Health Organization (FAO/WHO), probiotics are “live microorganisms which, when administered in adequate amounts, confer health benefits on the host” [
10]. It has been confirmed that the consumption of probiotics can reduce cholesterol serum levels, prevent diarrhoea through the enhancement of the intestinal barrier, and decrease irritable bowel syndrome symptoms [
11]. To be applied as a probiotic and to function appropriately, a microbial strain must exhibit several characteristics, such as gastrointestinal tolerance and the ability to colonise the human host. Furthermore, the probiotic candidate should be safe for humans, e.g., with an antibiotic sensitivity phenotype [
12].
Lactiplantibacillus plantarum (
Lpb. plantarum ) is a LAB with a long history of protechnological use in the food sector, in food fermentations and for the design of protective cultures [
13,
14]. This species can adapt to a variety of niches and is widely distributed in the environment, i.e., it can be found in dairy products, in the gastrointestinal tracts of humans and animals, meat, fish, and fermented vegetables [
15]. Increasing evidence also corroborates the probiotic properties of
Lpb. plantarum strains [
16,
17], thus broadening the range of its applications. To date, most studies underline the safety attributes of
Lpb. plantarum [
18,
19], supporting the industrial interest in this species. Recent papers also highlight some
Lpb. plantarum properties that make it intriguing for biomedical purposes, e.g., its capacity to regulate the enteric microbiota, alleviate liver disease [
20], and its anticancer potential [
20]. Moreover, some publications hint at the use of
Lpb. plantarum for the bio-suppression of pathogens in food models [
21,
22].
Fruit and vegetables are important components of a healthy diet, and their consumption helps prevent a wide range of diseases [
23]. Thus, both WHO and FAO recommend the intake of a specific dose of vegetables and fruits
per day. Tomato is highly consumed worldwide; it is economically considered the second-most important fruit or crop [
24]. In 2020, Tunisia ranked tenth in the world in producing tomatoes. The production of tomatoes was estimated at around 1.4 million metric tons in the last four years by Statista Research Department [
25]. Tomato contains a high amount of fiber, oligosaccharides and polysaccharides, which act as prebiotics in the gut [
26]. However, tomatoes can be easily contaminated and get spoilt very fast through contaminated irrigation water during transporting and storage, causing economic losses and serious health issues related to foodborne diseases [
27]. Consequently, there is a need for intervention technologies and techniques to reduce/prevent tomato contamination [
28]. To face food contamination/spoilage, several methodologies have been investigated and applied over the years; however, these methods are not applicable to all foods and can alter the sensory properties of the final product [
29,
30]. Therefore, bio-antimicrobial agents, such as antagonistic bacteria, have emerged as alternatives recently.
Overall, the aims of the present study were to isolate new LAB strains from unexplored Tunisian sources, to evaluate their capability to colonize tomato surfaces and to inhibit the adhesion of pathogenic Escherichia coli O157:H7 CECT 4267 and Listeria monocytogenes CECT 4031 through co-culture on the food matrix. Further, their potential use for the design of probiotic cultures was investigated.
4. Discussion
Probiotics have been traditionally isolated from dairy products, though, in recent years, there has been an increasing trend in exploring novel and alternative sources [
54]. Unconventional niches that are being investigated for discovering new probiotics include traditional fermented foods and beverages, vegetables, fruit juices [
55,
56] and the intestine of insects [
57,
58]. According to scientific research, fermented vegetables are interesting sources of potentially probiotic LAB strains [
7,
59,
60]. Many Tunisian publications reported that fermented vegetables host LAB strains with an antimicrobial and bio-protective effect against germs and moulds [
61,
62,
63], but only a few articles mentioned the possible probiotic properties of strains from Tunisian fermented vegetables.
The present study aimed to isolate LAB strains from unconventional sources and to characterise them for further usage, such as probiotic cultures and/or for food biocontrol. After screening presumptive LAB from very diverse sources, including infant faeces, breast milk, rabbit intestine, fermented olive etc., we focused on six strains exhibiting the highest antagonistic action toward foodborne pathogens. Such selected strains, derived from fermented vegetables (fermented olive and pepper) and locust intestine, were identified as
Lactiplantibacillus plantarum and were further investigated for their antimicrobial, probiotic and biocontrolling properties. Our findings are supported by similar works which reported
Lpb. plantarum as the dominant group in fermented vegetables, e.g. tomatoes, and table olives [
64,
65]. According to recent studies,
Clostridium (
Firmicutes) is one of the most abundant genera found in insects [
58]. Likewise, Garofalo et al. [
66] confirmed the dominance of
Clostridium in locust, mainly represented by
Enterobacteriaceae and
Weissella spp.. Another study reported the presence of
Lactobacillus metriopterae sp. nov. in locust gut [
67]. Unexpectedly, in the current study, LAB isolated from locust, were identified as
Lpb. plantarum with similarity over 98%, and we can mention that these are the first
Lpb. plantarum found and characterized in locust. LAB isolated from insects are being considered as promising probiotics for the benefit of human and animal health due to the survive/persist of their host in hard environments [
58]. Indeed, some very recent researches have looked into the probiotic potential of LAB obtained from insect gut [
57,
68,
69]. A complete genome sequence of
Weissella confusa LM1, found in the gut of the migratory locust, indicated the ability to adapt to different ecological niches [
70]. While there are studies on the microbiological communities (including LAB) in grasshoppers (
Locusta migratoria migratorioides) sold for human consumption [
71], to our knowledge, this is the first report studying the probiotic potentialities of locust-derived LAB.
Nowadays, probiotics with antimicrobial activity are becoming an alternative to traditional drugs due to antibiotics resistance diffusion [
72]. Interestingly, it has been found that all the strains were able to inhibit the indicators growth; a similar study [
73], showed that
Lpb. plantarum exhibits an inhibitory effect against
L. monocytogenes and
E. coli similar to our tested LAB. Moreover, a recent study confirmed the antibacterial potential of
Lpb. plantarum insect strains with a similar range against
E. coli [
57].
The CFSs from the investigated
Lpb. plantarum strains had pH values in the range of 3.5, as reported by previous publications [
74]. After pH neutralisation to 6.0, all CFSs showed minimal to no activity against all the pathogens tested, proving the role of organic acids for antimicrobial effects. It was reported that the increased production of organic acid through carbohydrates fermentation decreases the pH of the medium, which is the major factor suppressing pathogen growth [
75]. In a similar study [
76], CFSs from LAB, including
Lpb. plantarum, showed anti-
E. coli activity with inhibition zone of ranging between 12.89 ± 0.21 to 15.32 ± 0.28 mm. The authors confirmed that the antimicrobial activity was due to the combination of various metabolites, including organic acids. In fact, CFSs of LAB are a complex mixture of metabolic enzymes, secreted proteins, short-chain fatty acids, vitamins, amino acids, peptides, organic acids, and cell components [
77]. It was recently reported that
Lpb. plantarum is able to synthesize various beneficial extracellular metabolites, known as postbiotics [
20], i.e. bioactive soluble compounds or peptides that are produced during LAB growth [
78] that confer health benefits such as infection prevention [
79], antitumor and immunomodulatory effects [
80]. Likewise, previous works have demonstrated the importance of organic acids as bio-preserving agents [
81,
82]. In addition, Mirzaei et al. [
83] reported that the antimicrobial activity of LAB strains disappeared when their CFSs were adjusted to pH 6.5 and treated with catalase. Nevertheless, the bacteriocin production by LAB is highly affected by several factors, including temperature, pH, and incubation time. It was also reported that the optimum secretion of bacteriocin is when the pH ranges between 5.0 and 6.0 [
84]. In the present study, all the tested strains showed antibacterial activity against the target pathogens, most probably due to the organic acids secreted in their CFSs.
The antifungal potential of LAB strains has also been estimated; our results showed that LAB inhibit the growth of
P. expansum,
A. niger,
F. culmorum and
B. cinerea. These findings are similar to the work carried out by other investigators [
85,
86]. A recent Turkish publication [
87] showed that
Lpb. plantarum was active against
P. expansum and
A. niger. Indeed, several studies reported that LAB isolated from vegetables and plants possess a better antifungal activity [
88,
89], while LAB derived from dairy products exhibit antibacterial activity against foodborne pathogens through bacteriocin production capabilities [
90,
91].
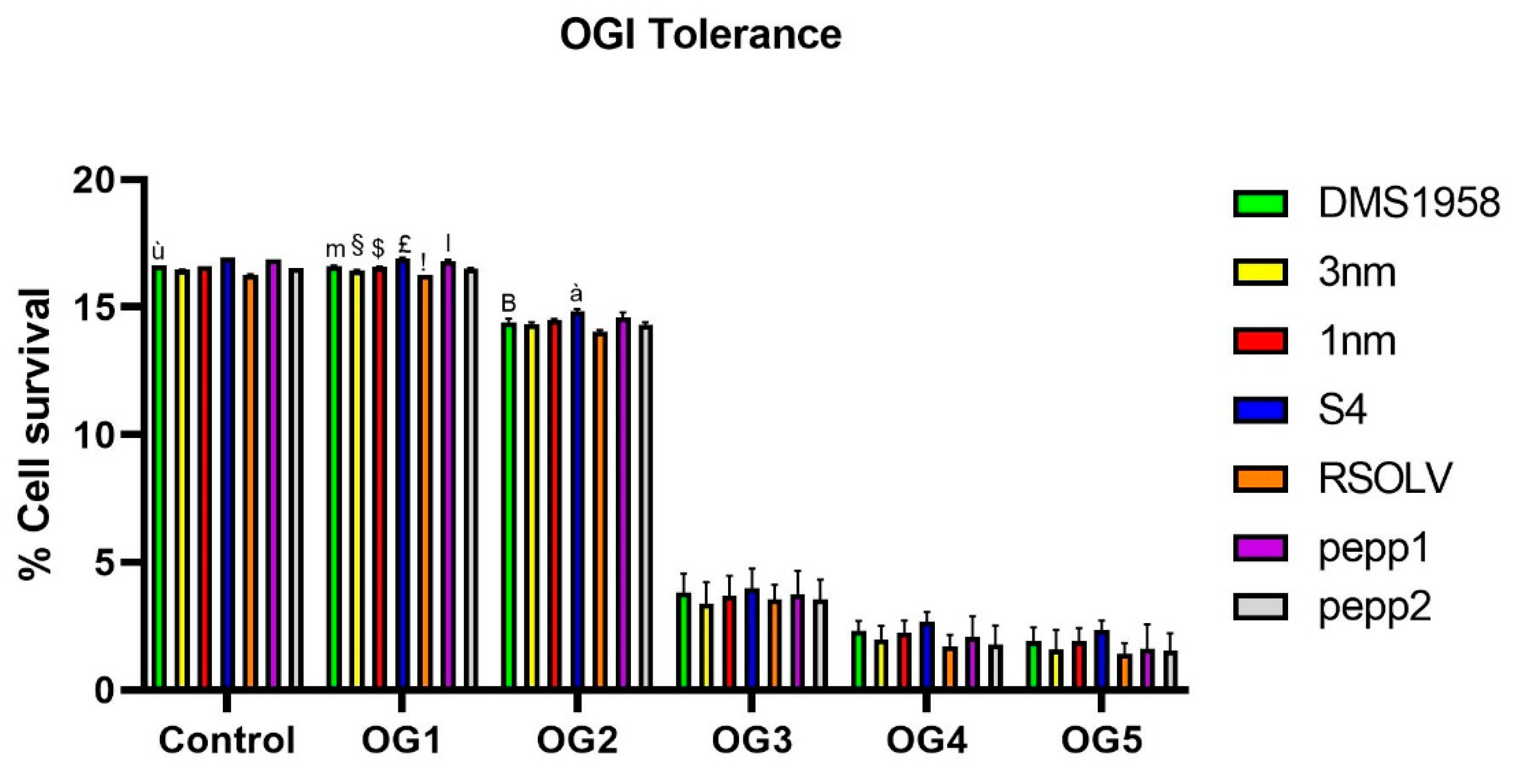
Thereafter, the probiotic traits were investigated. Starting with OGI transit tolerance, all the tested
Lpb. plantarum strains were incubated in successive solutions to mimic the human OGI transit, and their viability was evaluated. In fact, to be applied in the food industry, LAB should possess a good resistance to acidic environment, especially in the preparation of high-acid foods [
92]. All selected
Lpb. plantarum strains were able to survive the simulated OGI transit, with LAB strain of fermented olive S4 exhibiting the highest resistance. Moreover, all the strains showed good resistance to acidic pH through OGI transit, which is congruent with several findings in the literature. Indeed,
Lpb. plantarum were proven to be able to survive in pH varies between 2.5 and 4 [
93,
94]. Besides, in agreement with our findings, several authors demonstrated the ability of
Lpb. plantarum to survive to OGI transit with a rate of 10
3 CFU·mL
−1 [
41,
95].
We further examined the potential probiotics for cell-binding properties. In the current study, all tested LAB exhibited time-dependent auto-aggregation ability, particularly, the fermented olives-derived strains (RSOLV and S4) showed the best auto-aggregation performance. In fact, the auto-aggregation of bacteria has been associated with adherence ability to the intestinal cells, a prerequisite for the colonization of the gastrointestinal tract [
96]. In a similar earlier study [
97],
Lpb. plantarum isolated from fermented vegetables showed auto-aggregation rates very close to our results.
In co-aggregation assays, using the same foodborne pathogenic bacteria as for antibacterial assay, the co-aggregation ability of all tested LAB increased over time and it was high for
Lbp. plantarum strains from fermented green pepper, indicating a potential in preventing and/or excluding colonization of pathogens in the gastrointestinal tract. The adhesion ability to cells was strain-specific as it varied even within the same species. Our findings are supported by recent work on LAB strains obtaining similar results [
44,
53], and are in agreement with the studies of Ben Taheur et al [
61], reporting a lower rate of co-aggregation of probiotic LAB with
E.coli.
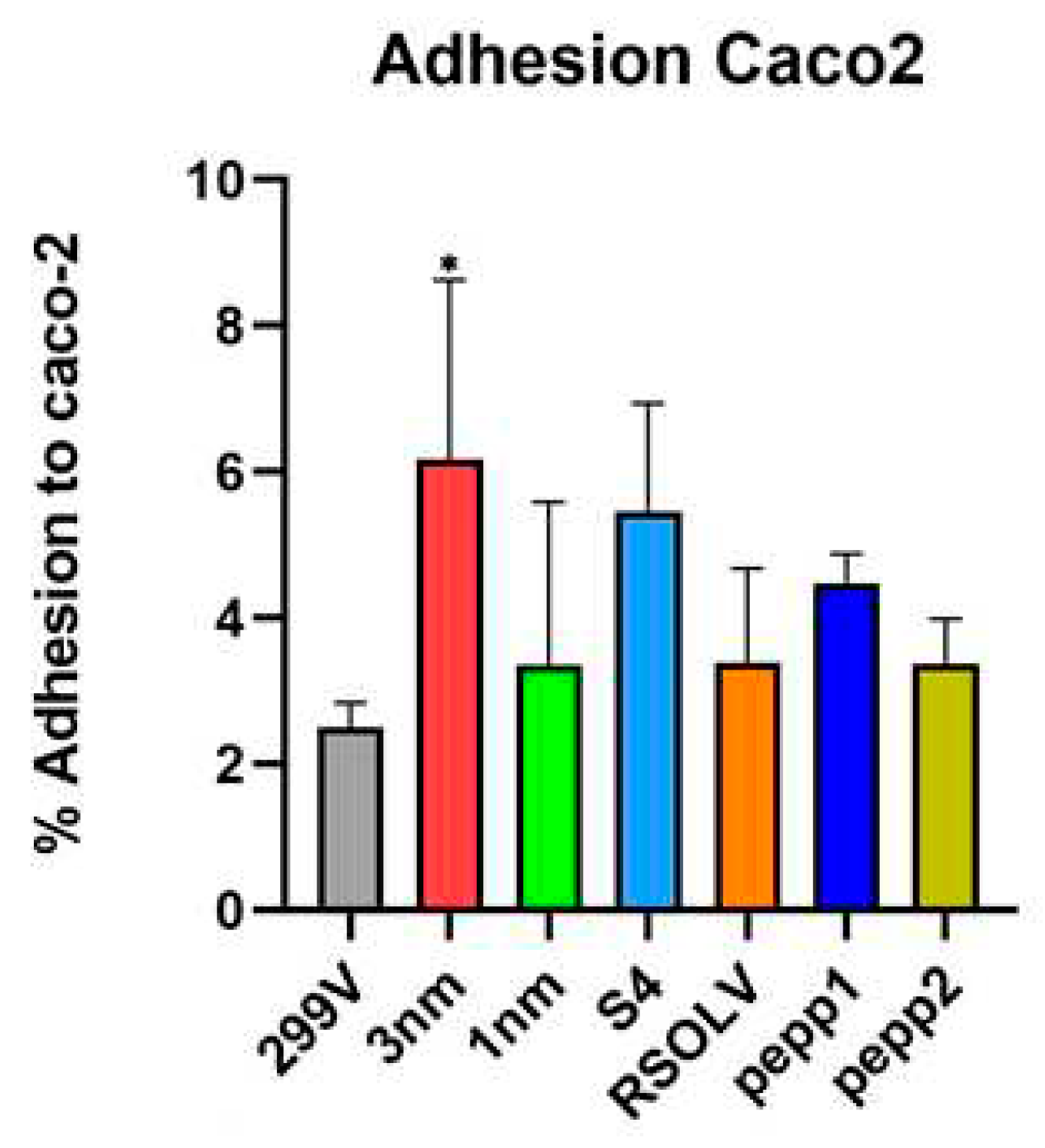
Regarding the adhesion capability, the
Lpb. plantarum strains were first evaluated for biofilm production ability on polystyrenes surface, then for adhesion to human enterocyte-like cells and, finally, to tomatoes as a food model. In fact, bacterial adhesion to epithelial cell is considered one of the most accurate features for selection criteria for probiotic strains [
98]. Results showed that all selected strains were able to form biofilm on abiotic surface, especially S4 and 1nm in a longer time, with a value similar to previous studies [
99], demonstrating that
Lpb. plantarum strains provide various levels of biofilm production capability from “no biofilm” to “good biofilm producer”, possibly depending on the amount of exopolysaccharides produced [
100]. Furthermore, knowledge about the surface conditions and the bacterial properties influencing adhesion is still insufficient [
101]. Concerning the adhesion to human cells, our findings demonstrated that all the tested
Lpb. plantarum strains survived and attached to Caco-2 cells better than the probiotic model strain used as control; our results are comparable to values previously obtained for
Lpb. plantarum [
102]. We also found that the CFSs of the tested
Lpb. plantarum strains were able to decrease colorectal adenocarcinoma cells viability, proving their anti-cancer and anti-proliferative potential; these findings are in line with current studies reporting the cytotoxicity of
Lpb. plantarum metabolites on cancer cells [
20,
50].
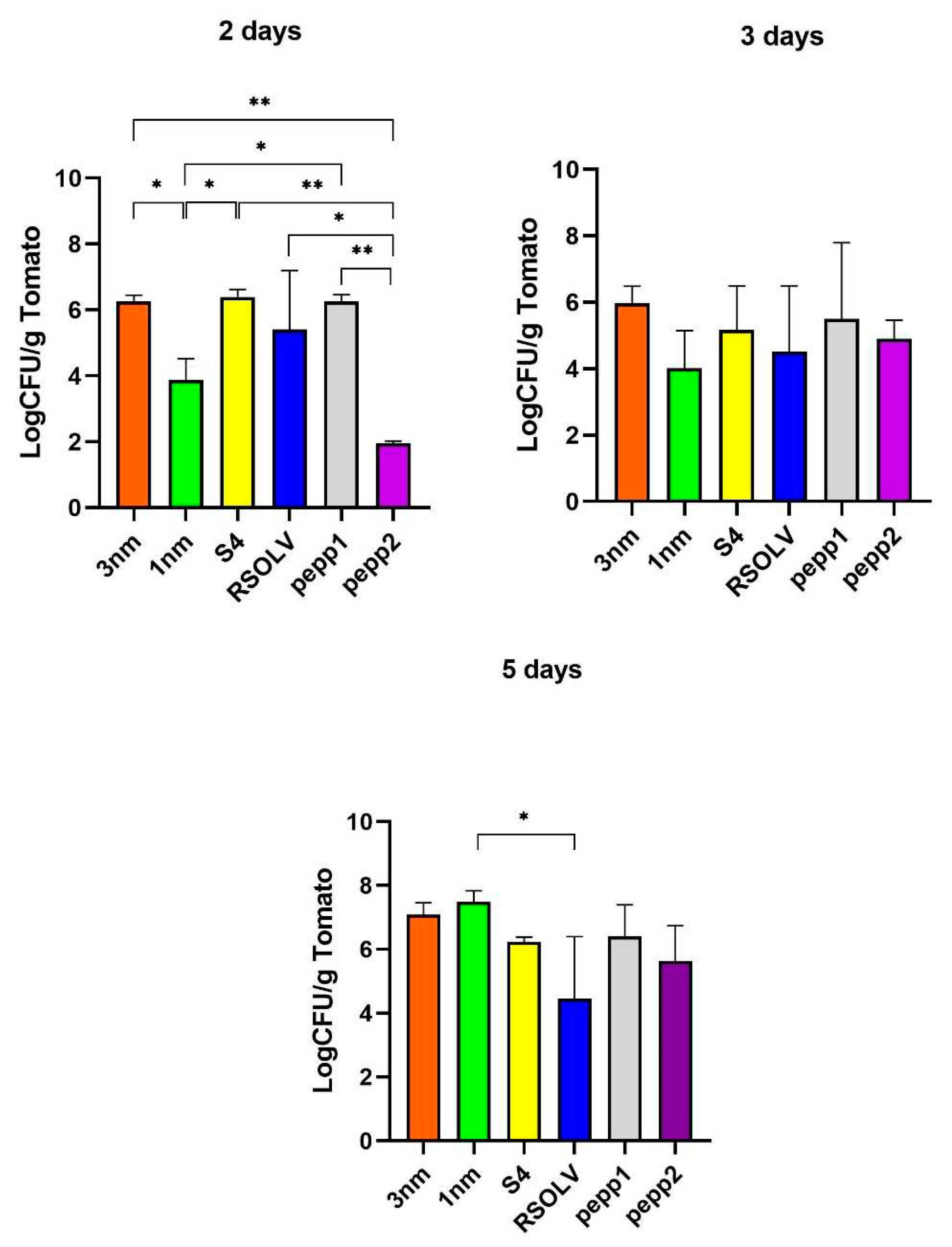
In the present study, we suggest the use of
Lpb. plantarum strains as probiotics and bio-controlling agents on tomatoes. Non-dairy, plant-based food matrices such as fruits, vegetables and legumes have been used successfully in producing probiotic products [
103]. The investigated
Lpb. plantarum strains showed strong attachment
in vivo to tomatoes. Arellano-Ayala et al. [
53] reported similar adhesion percentages of LAB to tomato fruits. The selection of tomatoes as a model to elucidate the adhesion ability of the probiotic strains reflect the importance of this vegetable in the human diet. Latest researches investigated LAB's efficiency as protective cultures tool to control
L. monocytogenes in ready-to-eat and dairy-ripened products [
104]. Moreover, Yin et al., confirmed the possible use of LAB as biocontrol agents for limiting/inhibiting pathogens contamination on leafy greens [
105]. Here we investigated the ability of
Lpb. plantarum strains to attach to tomatoes in co-culture with pathogens. The enumeration of microorganisms was done in different selective mediums; the biocontrol assay on tomatoes confirmed the ability of all
Lpb. plantarum strains, principally 1 nm and S4, to antagonise the tested pathogens, with a maintained count of CFU after 5 days, and with a significant difference compared to the control.
In conclusion, this is one of the few studies that have investigated the probiotic potential of Tunisian vegetables- and locust intestine- derived LAB and their biocontrol capacities on a food model matrix. Six selected Lpb. plantarum strains were analysed for their antagonism potential, demonstrating that they could be good candidates as food-protective cultures and with an interesting probiotic profile. Nevertheless, further studies are needed to deepen their characterisation and to guarantee their use as probiotic strains in biocontrol.
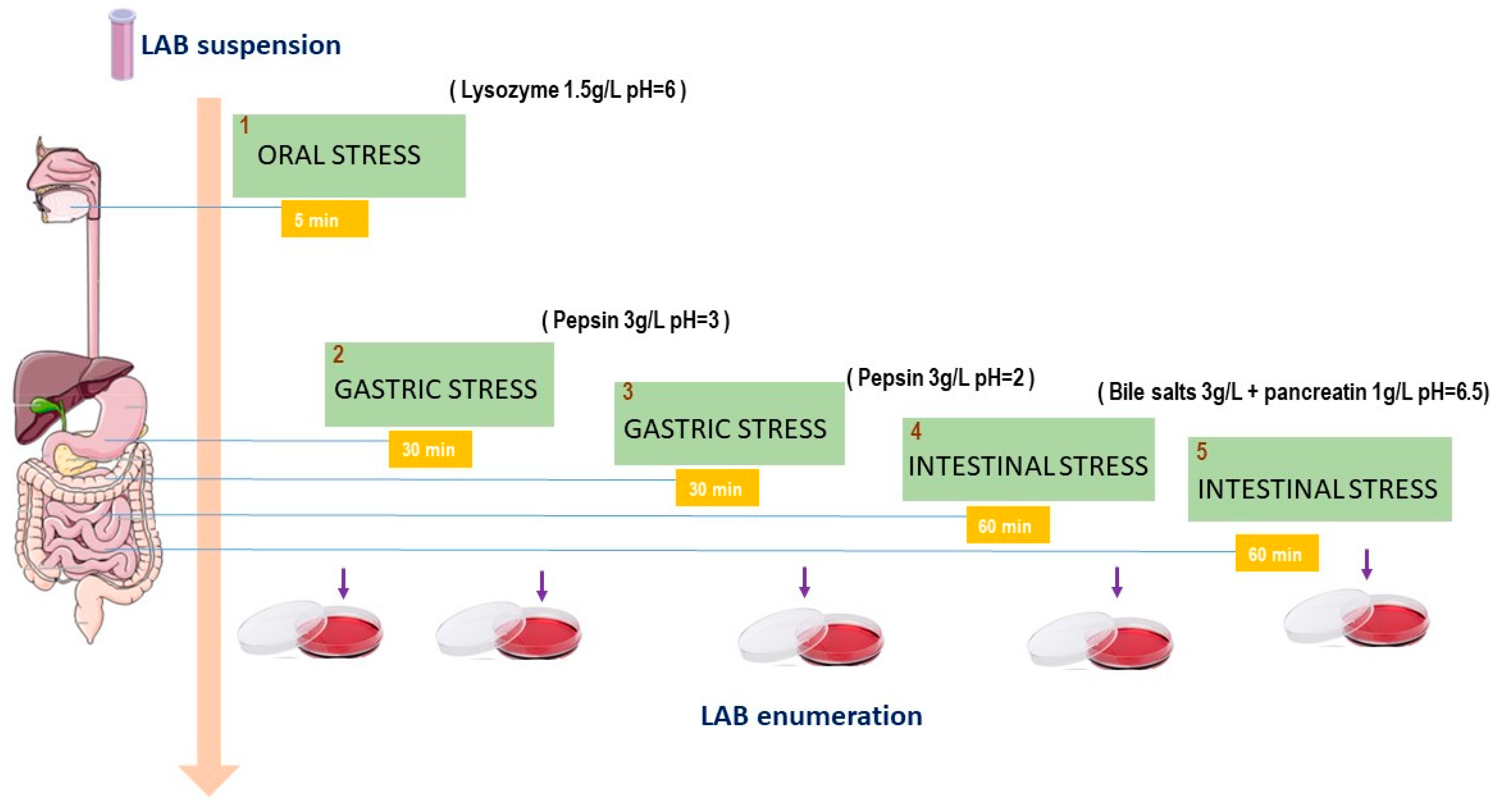
Figure 1.
Schematic diagram of the oro-gastrointestinal transit (OGI) assay used to test the tolerance of Lpb. plantarum strains in vitro. Bacterial cultures were first washed twice with sterile buffer, then incubated in lysozyme solution to mimic the saliva stress for 5 min; later, the pellet was incubated in a solution containing pepsin with different pH values, arriving at the intestinal stress in which cells were inoculated in bile salts and pancreatin for 60 min and then, incubated for 1h at 37 °C in a sterile electrolyte solution.
Figure 1.
Schematic diagram of the oro-gastrointestinal transit (OGI) assay used to test the tolerance of Lpb. plantarum strains in vitro. Bacterial cultures were first washed twice with sterile buffer, then incubated in lysozyme solution to mimic the saliva stress for 5 min; later, the pellet was incubated in a solution containing pepsin with different pH values, arriving at the intestinal stress in which cells were inoculated in bile salts and pancreatin for 60 min and then, incubated for 1h at 37 °C in a sterile electrolyte solution.
Figure 2.
Schematic representation of adhesion in food model assay. First, tomatoes were washed with sterile distilled water and decontaminated with UV light for 15 mins; then, the scar was created using a sterile syringe and Lpb. plantarum cultures were inoculated in spots. Inoculated tomatoes were kept for 2 h to dry under the laminar flow hood and incubated at room temperature. Sterile PBS buffer pH=7.0 was used as a control.
Figure 2.
Schematic representation of adhesion in food model assay. First, tomatoes were washed with sterile distilled water and decontaminated with UV light for 15 mins; then, the scar was created using a sterile syringe and Lpb. plantarum cultures were inoculated in spots. Inoculated tomatoes were kept for 2 h to dry under the laminar flow hood and incubated at room temperature. Sterile PBS buffer pH=7.0 was used as a control.
Figure 3.
Oro-gastrointestinal tolerance in vitro showed by Lpb. plantarum strains. Strains were incubated in MRS (control), then exposed successively to lysozyme (OG1), pepsin pH 3 (OG2), pepsin pH 2 (OG3) and bile salts and pancreatic enzymes (OG4 ). Finally, bacterial cultures were incubated in an intestinal electrolyte solution pH 6.5 (OG5). The assay was performed in triplicate. ANOVA was used for significant differences at each time point (control, OG1, OG2, OG3, OG4) of the OGI transit between strains. ù p < 0.0001 vs all strains; m p<0.05 vs OG1 of strains 3nm, S4, RSOLV, pepp1 and pepp2; § p<0.05 vs OG1 of strains 1nm, S4, RSOLV and pepp1; $ p<0.0001 vs OG1 of strains S4, RSOLV, pepp1; £p<0.0001 vs OG1 of strains RSOLV, pepp2; !p<0.0001 vs OG1 of strains pepp1 and pepp2; l p<0.0001 vs OG1 pepp2; B p<0.05 vs OG2 of strains S4 and RSOLV; à p<0.05 vs OG2 of strains 3nm, 1nm, RSOLV and pepp2.
Figure 3.
Oro-gastrointestinal tolerance in vitro showed by Lpb. plantarum strains. Strains were incubated in MRS (control), then exposed successively to lysozyme (OG1), pepsin pH 3 (OG2), pepsin pH 2 (OG3) and bile salts and pancreatic enzymes (OG4 ). Finally, bacterial cultures were incubated in an intestinal electrolyte solution pH 6.5 (OG5). The assay was performed in triplicate. ANOVA was used for significant differences at each time point (control, OG1, OG2, OG3, OG4) of the OGI transit between strains. ù p < 0.0001 vs all strains; m p<0.05 vs OG1 of strains 3nm, S4, RSOLV, pepp1 and pepp2; § p<0.05 vs OG1 of strains 1nm, S4, RSOLV and pepp1; $ p<0.0001 vs OG1 of strains S4, RSOLV, pepp1; £p<0.0001 vs OG1 of strains RSOLV, pepp2; !p<0.0001 vs OG1 of strains pepp1 and pepp2; l p<0.0001 vs OG1 pepp2; B p<0.05 vs OG2 of strains S4 and RSOLV; à p<0.05 vs OG2 of strains 3nm, 1nm, RSOLV and pepp2.
Figure 4.
Biofilm formation (expressed as absorbance at 595 nm) of the Lpb. plantarum strains after 24h and 72h of incubation at 37°C using Resazurin (RZ) (A) or Crystal Violet (CV) (B). Error bars indicate the standard deviation of triplicate experiments.ANOVA was used between strains and to control at each time (24h, 48h and 72h).b p<0.05 vs RZ 24h of all strains except pepp2; T p<0.05 vs RZ 24 of strains 3nm, 1nm and pepp2; H p<0.05 vs RZ 24h of strains V583, RSOLV and pepp1; G p<0.05 vs RZ 72h of strains V583, RSOLV and pepp1; & p < 0.0001 vs CV 24h of all strains; é p<0.0001 vs CV 24h of strains S4 and pepp2; à p<0.05 vs CV 24h of strains S4, RSOLV and pepp2; è p<0.0001 vs CV 24h of strains RSOLV and pepp1; $ p<0.001 vs CV 24h of strain pepp2; * p<0.0001 vs CV 24h of pepp1; @ p<0.0001 vs CV 48h of all strains except S4; “ p<0.0001 vs CV 48h all strains except pepp1; µ p<0.0001 vs CV 48h of all strains; = p<0.0001 vs CV 48h of all strains; L p<0.0001 vs CV 48h of all strains; m p<0.0001 vs CV 48h of all strains; O p<0.0001 vs CV 48h of all strains; p p<0.0001 vs CV 72h of all strains; q p<0.0001 vs CV 72h of all strains except pepp1 and pepp2; n p<0.05 vs CV 72h of all strains; v p<0.0001 vs CV 72h of all strains; I p<0.0001 vs CV 72h of all strains; w p<0.0001 vs CV 72h of 1nm, S4 and RSOLV; Z p<0.05 vs CV 72h of strains 1nm, S4 and RSOLV.
Figure 4.
Biofilm formation (expressed as absorbance at 595 nm) of the Lpb. plantarum strains after 24h and 72h of incubation at 37°C using Resazurin (RZ) (A) or Crystal Violet (CV) (B). Error bars indicate the standard deviation of triplicate experiments.ANOVA was used between strains and to control at each time (24h, 48h and 72h).b p<0.05 vs RZ 24h of all strains except pepp2; T p<0.05 vs RZ 24 of strains 3nm, 1nm and pepp2; H p<0.05 vs RZ 24h of strains V583, RSOLV and pepp1; G p<0.05 vs RZ 72h of strains V583, RSOLV and pepp1; & p < 0.0001 vs CV 24h of all strains; é p<0.0001 vs CV 24h of strains S4 and pepp2; à p<0.05 vs CV 24h of strains S4, RSOLV and pepp2; è p<0.0001 vs CV 24h of strains RSOLV and pepp1; $ p<0.001 vs CV 24h of strain pepp2; * p<0.0001 vs CV 24h of pepp1; @ p<0.0001 vs CV 48h of all strains except S4; “ p<0.0001 vs CV 48h all strains except pepp1; µ p<0.0001 vs CV 48h of all strains; = p<0.0001 vs CV 48h of all strains; L p<0.0001 vs CV 48h of all strains; m p<0.0001 vs CV 48h of all strains; O p<0.0001 vs CV 48h of all strains; p p<0.0001 vs CV 72h of all strains; q p<0.0001 vs CV 72h of all strains except pepp1 and pepp2; n p<0.05 vs CV 72h of all strains; v p<0.0001 vs CV 72h of all strains; I p<0.0001 vs CV 72h of all strains; w p<0.0001 vs CV 72h of 1nm, S4 and RSOLV; Z p<0.05 vs CV 72h of strains 1nm, S4 and RSOLV.
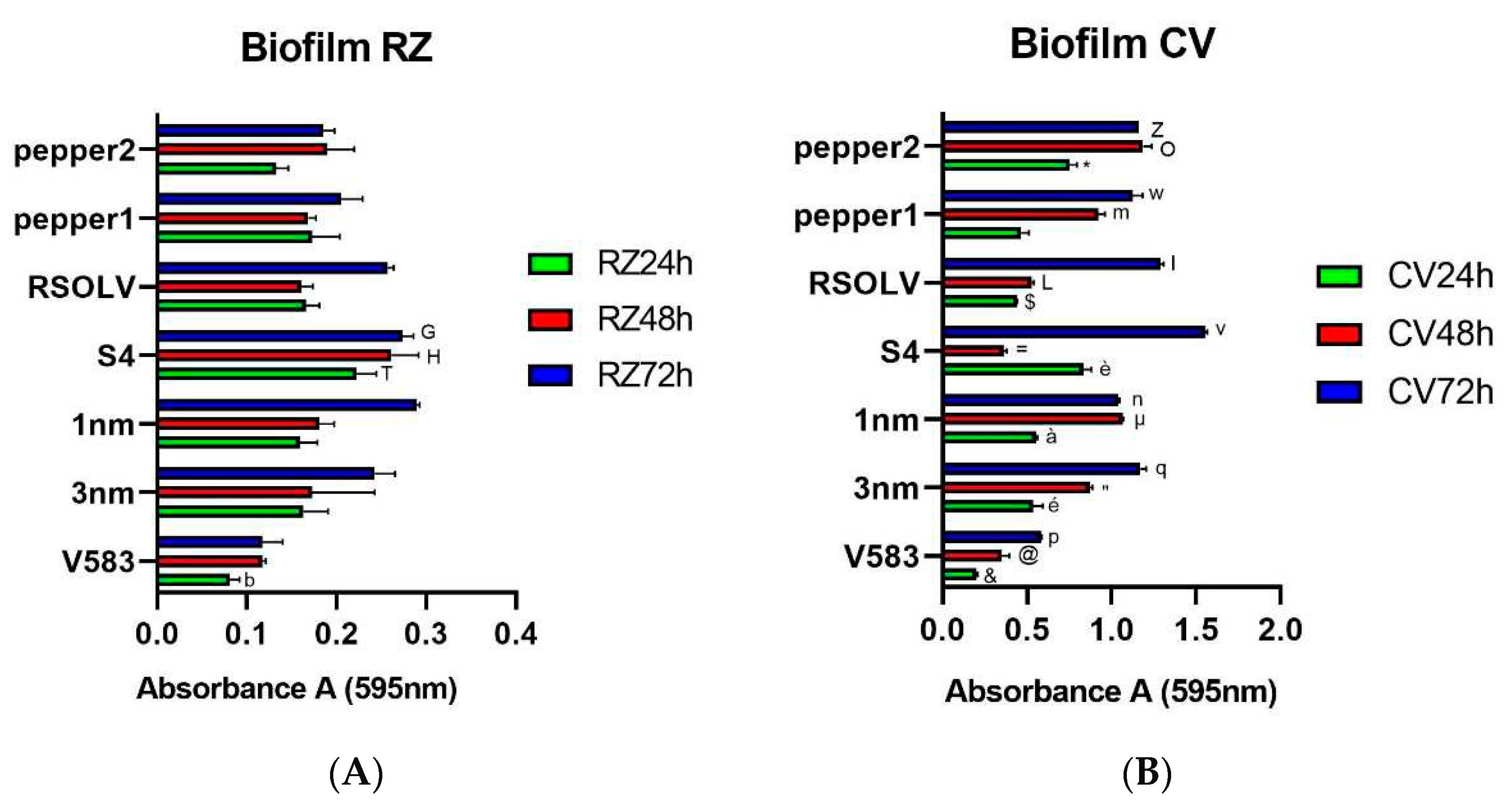
Figure 5.
The effect of different concentration of CFS of the Lpb. plantarum strains on the proliferation of LS cells after 24h (A) and 72h (B) incubation at 37°C. Error bars indicate the standard deviation of triplicate experiments. One-way ANOVA test at each concentration comparing strains among them and to the control (MRS). * p < 0.05 vs all 3nm and all MRS doses; ù p<0.05 vs all 1nm and all MRS doses; µ p<0.05 vs all S4 and all doses; $ p<0.05 vs all RSOLV and all MRS doses; à p<0.005 vs all pepp1 and all MRS doses; ç p<0.05 vs all pepp2 and all MRS doses. + p<0.0001 vs 1mg/ml MRS;& p<0.0001 vs 2mg/ml MRS; “ p<0.0001 vs 4mg/ml of all strains; £ p<0.05 vs 4mg/ml of the strains 3nm, S4, RSOLV, pepp1; / p<0.05 vs 4mg/ml of the strains 1nm;< p<0.05 vs 4mg/ml of S4, RSOLV, pepp1; ; p<0.0001 vs 8mg/ml of all strains; // p<0.05 vs 8mg/ml of strains 3nm, S4, RSOLV and pepp1; ^ p<0.0001 vs 10mg/ml of all strains; ) p<0.05 vs 10 mg/ml of the strains 1nm, S4, RSOLV; ( p<0.05 vs 10mg/ml of the strains pepp1 and pepp2; mp<0.05 vs 10mg/ml of the strains RSOLV and pepp1; np<0.05 vs 10mg/ml of the strains pepp1 and pepp2; l p<0.05 vs 2mg/ml of all strains; - p<0.05 vs 2mg/ml of the strains S4 and pepp1; v p<0.05 vs 4mg/ml of all strains; p p<0.05 vs 4mg/ml of the strains 1nm, S4, RSOLV; h p<0.05 vs 4mg/ml of the strains RSOLV, pepp1 and pepp2; XX p<0.05 vs 4mg/ml of all strains; Y p<0.05 vs 4mg/ml of the strain pepp1; U p<0.001 vs 8mg/ml of all strains; F p<0.05 vs 8mg/ml of all strains; R p<0.05 vs 8mg/ml of all strains; O p<0.05 vs 4mg/ml of the strains 3nm,1nm,pepp1 and pepp2; G p<0.05 vs 8mg/ml of all strains; Q p<0.05 vs 8mg/ml of all strains; Wp<0.001 vs 10mg/ml of all strains; X p<0.05 vs 10mg/ml pepp2.
Figure 5.
The effect of different concentration of CFS of the Lpb. plantarum strains on the proliferation of LS cells after 24h (A) and 72h (B) incubation at 37°C. Error bars indicate the standard deviation of triplicate experiments. One-way ANOVA test at each concentration comparing strains among them and to the control (MRS). * p < 0.05 vs all 3nm and all MRS doses; ù p<0.05 vs all 1nm and all MRS doses; µ p<0.05 vs all S4 and all doses; $ p<0.05 vs all RSOLV and all MRS doses; à p<0.005 vs all pepp1 and all MRS doses; ç p<0.05 vs all pepp2 and all MRS doses. + p<0.0001 vs 1mg/ml MRS;& p<0.0001 vs 2mg/ml MRS; “ p<0.0001 vs 4mg/ml of all strains; £ p<0.05 vs 4mg/ml of the strains 3nm, S4, RSOLV, pepp1; / p<0.05 vs 4mg/ml of the strains 1nm;< p<0.05 vs 4mg/ml of S4, RSOLV, pepp1; ; p<0.0001 vs 8mg/ml of all strains; // p<0.05 vs 8mg/ml of strains 3nm, S4, RSOLV and pepp1; ^ p<0.0001 vs 10mg/ml of all strains; ) p<0.05 vs 10 mg/ml of the strains 1nm, S4, RSOLV; ( p<0.05 vs 10mg/ml of the strains pepp1 and pepp2; mp<0.05 vs 10mg/ml of the strains RSOLV and pepp1; np<0.05 vs 10mg/ml of the strains pepp1 and pepp2; l p<0.05 vs 2mg/ml of all strains; - p<0.05 vs 2mg/ml of the strains S4 and pepp1; v p<0.05 vs 4mg/ml of all strains; p p<0.05 vs 4mg/ml of the strains 1nm, S4, RSOLV; h p<0.05 vs 4mg/ml of the strains RSOLV, pepp1 and pepp2; XX p<0.05 vs 4mg/ml of all strains; Y p<0.05 vs 4mg/ml of the strain pepp1; U p<0.001 vs 8mg/ml of all strains; F p<0.05 vs 8mg/ml of all strains; R p<0.05 vs 8mg/ml of all strains; O p<0.05 vs 4mg/ml of the strains 3nm,1nm,pepp1 and pepp2; G p<0.05 vs 8mg/ml of all strains; Q p<0.05 vs 8mg/ml of all strains; Wp<0.001 vs 10mg/ml of all strains; X p<0.05 vs 10mg/ml pepp2.

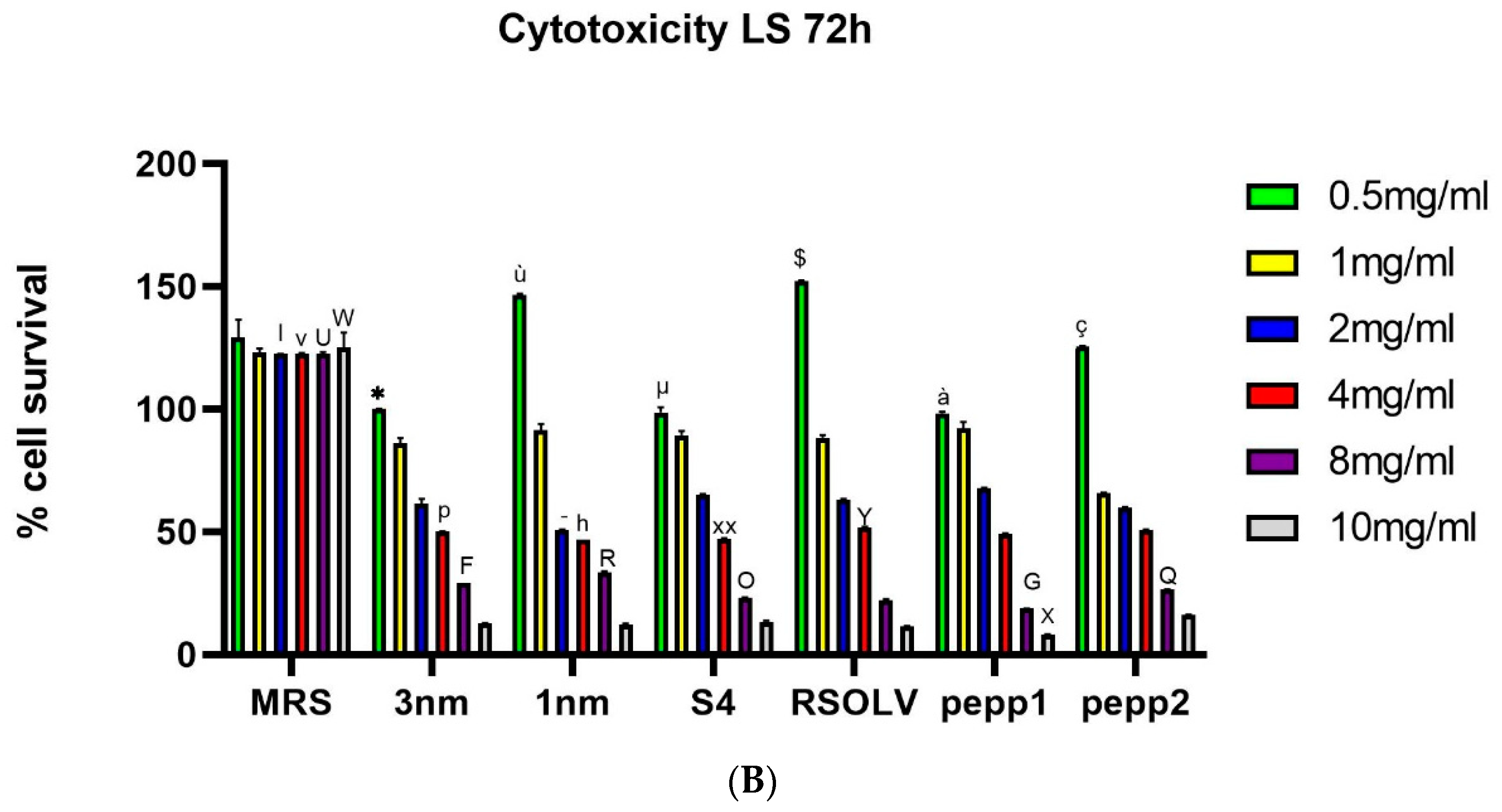
Figure 6.
Adhesion percentage of Lpb. plantarum strains to caco-2 monolayers after 1 h of co-incubation at 37 °C. Lpb. plantarum V299 was used as control. Error bars indicate the standard deviation of triplicate experiments.*: p < 0.05 vs Lpb. plantarum 299V.
Figure 6.
Adhesion percentage of Lpb. plantarum strains to caco-2 monolayers after 1 h of co-incubation at 37 °C. Lpb. plantarum V299 was used as control. Error bars indicate the standard deviation of triplicate experiments.*: p < 0.05 vs Lpb. plantarum 299V.
Figure 7.
Adhesion percentage/capacity of Lpb. plantarum strains on Tomato after 2, 3 and 5 days. Error bars indicate the standard deviation of Triplicate experiments. One-way ANOVA test for strains at each incubation time (2days, 3days and 5 days), Tukey’s multiple comparisons test, * p < 0.05; ** p<0.0001.
Figure 7.
Adhesion percentage/capacity of Lpb. plantarum strains on Tomato after 2, 3 and 5 days. Error bars indicate the standard deviation of Triplicate experiments. One-way ANOVA test for strains at each incubation time (2days, 3days and 5 days), Tukey’s multiple comparisons test, * p < 0.05; ** p<0.0001.
Table 1.
Antibacterial activity of LAB isolated from various matrices against Escherichia coli O157:H7 CECT 4267 and Listeria monocytogenes CECT 4031, as determined by the agar diffusion method. Diameter halos inhibition expressed in mm. Mean values and standard deviations of three replicates are indicated. The six strains selected for further studies are underlined.
Table 1.
Antibacterial activity of LAB isolated from various matrices against Escherichia coli O157:H7 CECT 4267 and Listeria monocytogenes CECT 4031, as determined by the agar diffusion method. Diameter halos inhibition expressed in mm. Mean values and standard deviations of three replicates are indicated. The six strains selected for further studies are underlined.
| Isolated LAB strain |
Source/matrix |
E. coli (mm) |
L. monocytogenes (mm) |
| 3nm |
Intestines of died Locust |
14.17±0.29 |
13.17±0.57 |
| 1nm |
Intestines of died Locust |
15.00±0.00 |
14.57±2.32 |
| S4 |
Fermented green olive brine |
15.00±1.00 |
11.23±0.68 |
| RSOLV |
Fermented olive |
14.17±0.76 |
13.43±0.93 |
| pepp1 |
Fermented pepper brine |
15.00±0.00 |
15.10±0.26 |
| pepp2 |
Fermented green pepper brine |
15.00±0.00 |
14.50±0.50 |
| S5 |
Horse sausage |
10.00±1.00 |
12.07±0.40 |
| S6 |
Horse sausage |
0.00±0.00 |
0.00±0.00 |
| N8 |
Dried anchovy |
11.00±1.00 |
0.00±0.00 |
| N4c |
Dried fermented anchovy |
12.00±1.00 |
8.03±0.25 |
| F1c |
Infant feces |
9.13±0.71 |
0.00±0.00 |
| F5a |
Infant feces |
12.90±0.17 |
9.97±0.15 |
| LM |
Breast milk |
9.83±0.29 |
9.92±0.14 |
| F1 |
Infant feces |
10.17±0.29 |
11.00±0.00 |
| Rg4a |
Artisanal Tunisian Ricotta cheese |
10.17±0.29 |
11.17±1.26 |
| RL4 |
Tunisian fermented milk Leben |
8.83±0.29 |
12.93±0.12 |
| K10 |
Tunisian Artisanal Gueddid |
0.00±0.00 |
10.60±0.36 |
| LC4 |
Goat milk |
0.00±0.00 |
10.50±0.50 |
| S1 |
Fermented olive brine |
10.80±0.36 |
10.47±0.50 |
| AIB |
Intestines of rabbit |
0.00±0.00 |
11.00±0.00 |
| S5 |
Horse sausage |
11.00±1.00 |
11.00±0.87 |
| N8 |
Dried anchovy |
9.90±0.10 |
9.00±0.00 |
| K10 |
Tunisian Gueddid |
0.00±0.00 |
0.00±0.00 |
| O3 |
Brine fermented olive |
0.00±0.00 |
0.00±0.00 |
Table 2.
The closest species/strain to the identified isolates according to the 16S rRNA sequences.
Table 2.
The closest species/strain to the identified isolates according to the 16S rRNA sequences.
| Isolate |
Source |
Closest species/strain |
Percentage of identity |
Accession number* |
| 3nm |
Died locust (intestines) |
Lactiplantibacillus plantarum |
100% |
OR431596 |
| 1nm |
Died locust (intestines) |
Lactiplantibacillus plantarum |
99.86% |
OR431597 |
| S4 |
Fermented Olive |
Lactiplantibacillus plantarum |
99.91% |
OR431698 |
| RSOLV |
Fermented olive |
Lactiplantibacillus plantarum |
99.93% |
OR431599 |
| pepp1 |
Fermented pepper |
Lactiplantibacillus plantarum |
99.82% |
OR431600 |
| pepp2 |
Fermented pepper |
Lactiplantibacillus plantarum |
100% |
OR431601 |
Table 3.
Physiological and Biochemical Characteristics of the selected Lpb. plantarum strains after growth in different conditions: osmotic stress, acidic stress and growth in different sources of carbon e.g glucose (Glc), fructose (Fruc) and Sucrose (Sucr).
Table 3.
Physiological and Biochemical Characteristics of the selected Lpb. plantarum strains after growth in different conditions: osmotic stress, acidic stress and growth in different sources of carbon e.g glucose (Glc), fructose (Fruc) and Sucrose (Sucr).
| Experimental assay |
3nm |
1nm |
S4 |
RSOLV |
pepp1 |
Pepp2 |
| Mobility |
- |
- |
- |
- |
- |
- |
| 2% NaCl growth |
+ |
+ |
+ |
+ |
+ |
+ |
| 4% NaCl growth |
+ |
+ |
+ |
+ |
+ |
+ |
| 8% NaCl growth |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Glc 2% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Glc 4% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Fruc 2% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Fruc 4% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Sucr 2% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS.Sucr 4% |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS pH 3 |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS pH 4 |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS pH 5 |
+ |
+ |
+ |
+ |
+ |
+ |
| MRS pH 9.2 |
+ |
+ |
+ |
+ |
+ |
+ |
Table 4.
(1). Antibacterial activity of whole cultures, CFS and cells from the selected Lpb. plantarum strains; the CFS were used crude, or after neutralization with NaOH, Lpb. plantarum cells were tested before and after heat treatment. The indicator bacteria was Escherichia coli CECT 4267. No(-), mild(+), or strong (++) inhibition, showing zone lower than 7 mm, ranging from 7 to 10 mm, or more than 10 mm, respectively. Assays were performed in duplicate. (2) Antibacterial activity of whole cultures, CFS and cells from the selected Lpb. plantarum strains; the CFS were used crude, or after neutralization with NaOH; Lpb. plantarum cells were tested before and after heat treatment. The indicator bacteria was Listeria monocytogenes CECT 4031. No(-), mild(+), or strong (++) inhibition, showing zone lower than 7 mm, ranging from 7 to 10 mm, or more than 10 mm, respectively. Assays were performed in duplicate.
Table 4.
(1). Antibacterial activity of whole cultures, CFS and cells from the selected Lpb. plantarum strains; the CFS were used crude, or after neutralization with NaOH, Lpb. plantarum cells were tested before and after heat treatment. The indicator bacteria was Escherichia coli CECT 4267. No(-), mild(+), or strong (++) inhibition, showing zone lower than 7 mm, ranging from 7 to 10 mm, or more than 10 mm, respectively. Assays were performed in duplicate. (2) Antibacterial activity of whole cultures, CFS and cells from the selected Lpb. plantarum strains; the CFS were used crude, or after neutralization with NaOH; Lpb. plantarum cells were tested before and after heat treatment. The indicator bacteria was Listeria monocytogenes CECT 4031. No(-), mild(+), or strong (++) inhibition, showing zone lower than 7 mm, ranging from 7 to 10 mm, or more than 10 mm, respectively. Assays were performed in duplicate.
| |
E. coli |
| Strain |
LAB culture |
Crude
CFS
|
CFS pH neutralized |
CFS treated Temperature |
Crude cells |
Cells treated Temperature |
| 3nm |
++ |
++ |
- |
++ |
- |
- |
| 1nm |
++ |
++ |
- |
++ |
- |
- |
| RSOLV |
++ |
++ |
- |
++ |
- |
- |
| S4 |
++ |
++ |
- |
++ |
- |
- |
| pepp1 |
++ |
++ |
- |
++ |
- |
- |
| pepp2 |
++ |
++ |
- |
++ |
- |
- |
| |
L. monocytogenes |
|
| Strain |
LAB culture |
Crude
CFS
|
CFS pH neutralized |
CFS treated Temperature |
Crude cells |
Cells treated Temperature |
| 3nm |
++ |
++ |
- |
++ |
- |
- |
| 1nm |
++ |
++ |
- |
++ |
- |
- |
| RSOLV |
++ |
++ |
- |
++ |
- |
- |
| S4 |
++ |
++ |
- |
++ |
- |
- |
| pepp1 |
++ |
++ |
- |
++ |
- |
- |
| pepp2 |
++ |
++ |
- |
++ |
- |
- |
Table 5.
Concentration of the main organic acids in CFSs from 24 h cultures of the selected Lpb. plantarum strains grown in MRS broth, as determined by HPLC. Results (mg L−1) are presented as the mean and SD of the two measures.
Table 5.
Concentration of the main organic acids in CFSs from 24 h cultures of the selected Lpb. plantarum strains grown in MRS broth, as determined by HPLC. Results (mg L−1) are presented as the mean and SD of the two measures.
| |
Tartaric acid |
Malic acid |
Ascorbic acid |
Lactic acid |
Acetic acid |
| MRS |
503.27±20.85 |
3 786.64±95.81 |
51.80±1.30 |
0.00±0.00 |
17 906.53±162.88 |
| 3nm |
1 202.55±30.22 |
1 326.48±176.26 |
100.09±42.11 |
14 931.50±133.44 |
16 421.03±2 236.89 |
| 1nm |
1 347.92±25.81 |
1 808.24±42.34 |
102.69±5.66 |
14 620±214.70 |
17 033.25±877.62 |
| S4 |
1 218.73±42.21 |
1 294.83±58.28 |
117.81±1.86 |
15 419.91±89.09 |
16 312.59±1 242.27 |
| RSOLV |
677.69±128.32 |
1 735.10±171.48 |
0.00±0.00 |
14 985.98±195.53 |
19 853.59±541.76 |
| pepp1 |
1 201.36±89.76 |
1 309.51±9.18 |
135.44±4.85 |
13 829.46±132.96 |
19 888.39±809.60 |
| pepp2 |
1 092.72±116.94 |
1 335.78±99.67 |
0.00±0.00 |
15 439.67±140.01 |
42 409.27±4 762.60 |
| |
Succinic acid |
Fumaric acid |
Citric acid |
| MRS |
1 066.96±27.55 |
0.00±0.00 |
1 206.78 |
| 3nm |
5 758.93±729.88 |
15.47±6.71 |
2 662.19±403.92 |
| 1nm |
11 693.70±708.58 |
0.00±0.00 |
1 833.64±134.04 |
| S4 |
9 009.26±1 374.46 |
0.00±0.00 |
2 224.48±19.15 |
| RSOLV |
10 193.35±562.11 |
0.00±0.00 |
1 050.29±200.52 |
| pepp1 |
6 897.52±781.84 |
0.00±0.00 |
446.71±45.63 |
| pepp2 |
7 336.16±473.52 |
0.00±0.00 |
894.15±25.05 |
Table 6.
Antifungal activity of Lpb. plantarum strains against P. expansum, A. niger, F. culmorum CECT 2148, S. cerevisiae and B. cinerea CECT 20973, as determined by the overlay method. No (-), mild (+), or strong (++) inhibition, showing zone lower than 1 mm, ranging from 1 to 5 mm, or more than 5 mm, respectively. Assays were performed in duplicate.
Table 6.
Antifungal activity of Lpb. plantarum strains against P. expansum, A. niger, F. culmorum CECT 2148, S. cerevisiae and B. cinerea CECT 20973, as determined by the overlay method. No (-), mild (+), or strong (++) inhibition, showing zone lower than 1 mm, ranging from 1 to 5 mm, or more than 5 mm, respectively. Assays were performed in duplicate.
| Strain |
P. expansum |
A. niger |
B. cinerea CECT 20973 |
F. culmorum |
S. cerevisiae |
| 3nm |
+ |
++ |
+ |
++ |
- |
| 1nm |
+ |
++ |
+ |
+ |
- |
| RSOLV |
++ |
+ |
++ |
++ |
- |
| S4 |
++ |
++ |
+ |
+ |
- |
| pepp1 |
++ |
++ |
+ |
+ |
- |
| pepp2 |
++ |
++ |
++ |
++ |
- |
Table 7.
Auto-aggregation ability of Lpb. plantarum strains after 4 h and 24 h incubation in 37 °C in PBS at pH 7. The assay was performed in duplicate.
Table 7.
Auto-aggregation ability of Lpb. plantarum strains after 4 h and 24 h incubation in 37 °C in PBS at pH 7. The assay was performed in duplicate.
| |
T4 |
T24 |
| 3nm |
24.31%±6.84% |
53.46%±2.01% |
| 1nm |
16.82%±2.33% |
53.59%±12.61% |
| S4 |
18.19%±2.54% |
70.30%±4.73% |
| RSLOV |
28.87%±3.68% |
60.97%±5.67% |
| pepp1 |
21.50%±1.75% |
34.67%±0.99% |
| pepp2 |
16.70%±1.12% |
69.82%±2.20% |
Table 8.
Co-aggregation of Lpb. plantarum index with the pathogen Listeria monocytogenes CECT 4031 and E. coli after 4 h and 24 h incubation at 37°C in PBS pH7. Assays were performed in duplicate.
Table 8.
Co-aggregation of Lpb. plantarum index with the pathogen Listeria monocytogenes CECT 4031 and E. coli after 4 h and 24 h incubation at 37°C in PBS pH7. Assays were performed in duplicate.
| |
L. monocytogenes |
E. coli |
| Strain |
T4 |
T24 |
T4 |
T24 |
| 3nm |
7.94%±0.94% |
7.28%±0.47% |
17.41%±1.54% |
43.42%±6.08% |
| 1nm |
6.38%±1.21% |
5.52%±0.18% |
17.73% ±7.75% |
54.88%±6.98% |
| S4 |
7.47%±0.14% |
7.37%±0.46% |
15.05%±0.89% |
36.96%±5.72% |
| RSOLV |
7.169%±2.25% |
8.76%±1.16% |
18.27%±4.32% |
51.15%±4.69% |
| pepp1 |
5.38%±0.00% |
5.38%±1.34% |
20.72%±2.67% |
43.53%±0.00% |
| pepp2 |
4.99%±0.55% |
4.61%±0.79% |
28.30%±10.70% |
43.16%±1.65% |
Table 9.
Antibiotic resistance assay. The experiment was done in duplicate. The inhibition zone diameters were measured. Susceptibility was expressed in terms of Resistant (R), Susceptible (S) and Intermediate (I) as mentioned.
Table 9.
Antibiotic resistance assay. The experiment was done in duplicate. The inhibition zone diameters were measured. Susceptibility was expressed in terms of Resistant (R), Susceptible (S) and Intermediate (I) as mentioned.
| ATB/strain |
3nm |
1nm |
S4 |
RSOLV |
pepp1 |
pepp2 |
| Ampicillin |
I |
S |
I |
I |
S |
I |
| Vancomycin |
R |
R |
S |
R |
R |
R |
| Gentamycin |
I |
I |
R |
S |
S |
I |
| Kanamycin |
R |
R |
R |
R |
R |
R |
| Streptomycin |
R |
S |
R |
R |
R |
R |
| Tetracyclin |
R |
R |
R |
R |
R |
R |
| Erythromycin |
I |
R |
R |
I |
R |
I |
| Clindamycin |
I |
R |
R |
I |
I |
I |
| Claromycin |
I |
R |
R |
R |
S |
I |
Table 10.
Half-maximal inhibitory concentration (IC50) of different CFSs on the proliferation of LS cell line after 24h and 72h at 37°C.
Table 10.
Half-maximal inhibitory concentration (IC50) of different CFSs on the proliferation of LS cell line after 24h and 72h at 37°C.
| Strain |
IC50 (after 24h) mg/mL |
IC50 ( after 72h) mg/mL |
| 3nm |
12.59 |
4.00 |
| 1nm |
10.21 |
2.40 |
| S4 |
9.91 |
3.21 |
| RSOLV |
11.11 |
3.66 |
| pepp1 |
18.26 |
3.19 |
| pepp2 |
14.88 |
3.69 |
Table 11.
Pathogen antagonism by Lpb. plantarum on tomatoes. CFU counts of pathogens inoculated alone (control) or in presence of each of the indicated Lpb. plantarum strain are reported. Mean ± standard deviation from from three different experiments. Each experiment was done in duplicate. Two-way ANOVA, Dunnett’s multiple comparisons test comparing strains at different time. $ p<0.05 vs 5 days 1nm-Listeria and 5 days pepp1-Listeria; & p<0.05 vs 1 day RSOLV-E.coli and 1 day pepp1-E.coli.
Table 11.
Pathogen antagonism by Lpb. plantarum on tomatoes. CFU counts of pathogens inoculated alone (control) or in presence of each of the indicated Lpb. plantarum strain are reported. Mean ± standard deviation from from three different experiments. Each experiment was done in duplicate. Two-way ANOVA, Dunnett’s multiple comparisons test comparing strains at different time. $ p<0.05 vs 5 days 1nm-Listeria and 5 days pepp1-Listeria; & p<0.05 vs 1 day RSOLV-E.coli and 1 day pepp1-E.coli.
| |
Number of tomato-attached bacteria (Log CFU/g tomato) |
| |
Medium agar |
1 day |
3 days |
5 days |
|
Listeria (control) |
PALCAM |
1.68±1.39 |
1.67±1.10 |
1.74±0.70$
|
| 3nm-Listeria
|
PALCAM |
1.73±0.87 |
1.44±0.74 |
1.52±1.48 |
| 1nm-Listeria
|
PALCAM |
1.70±1.23 |
1.02±0.62 |
0.00 |
| S4-Listeria
|
PALCAM |
1.46±0.22 |
1.00±0.67 |
0.00 |
| RSOLV-Listeria
|
PALCAM |
1.77±0.80 |
1.614±1.38 |
1.18 |
| pepp1-Listeria
|
PALCAM |
1.49±0.997 |
1.54±1.41 |
1.15±0.45 |
| pepp2-Listeria
|
PALCAM |
1.70±1.14 |
1.16±0.84 |
1.63±1.16 |
|
E.coli (control) |
SMAC |
1.92±1.61&
|
1.72±0.99 |
1.63±1.54 |
| 3nm-E.coli
|
SMAC |
1.82±1.13 |
1.55±1.43 |
1.34±0.94 |
| 1nm-E.coli
|
SMAC |
1.87±1.24 |
1.70±0.97 |
1.38±0.31 |
| S4-E.coli
|
SMAC |
1.88±1.05 |
1.73±1.15 |
1.13±0.45 |
| RSOLV-E.coli
|
SMAC |
1.87±1.04 |
1.51±1.33 |
1.56±1.24 |
| pepper1-E.coli
|
SMAC |
1.81±1.22 |
1.45±1.11 |
1.24±0.45 |
| pepper2-E.coli
|
SMAC |
1.84±1.15 |
1.53±0.96 |
1.22±0.41 |