Submitted:
16 October 2023
Posted:
16 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Pigeon Pea Collection and Sample Preparation
2.2. Genomic DNA Extraction
2.2.1. Amplification of the 16s rRNA V1–V9 Regions
2.2.2. Library Preparation and Sequencing
2.2.3. Bioinformatics Analysis
3. Results and Discussion
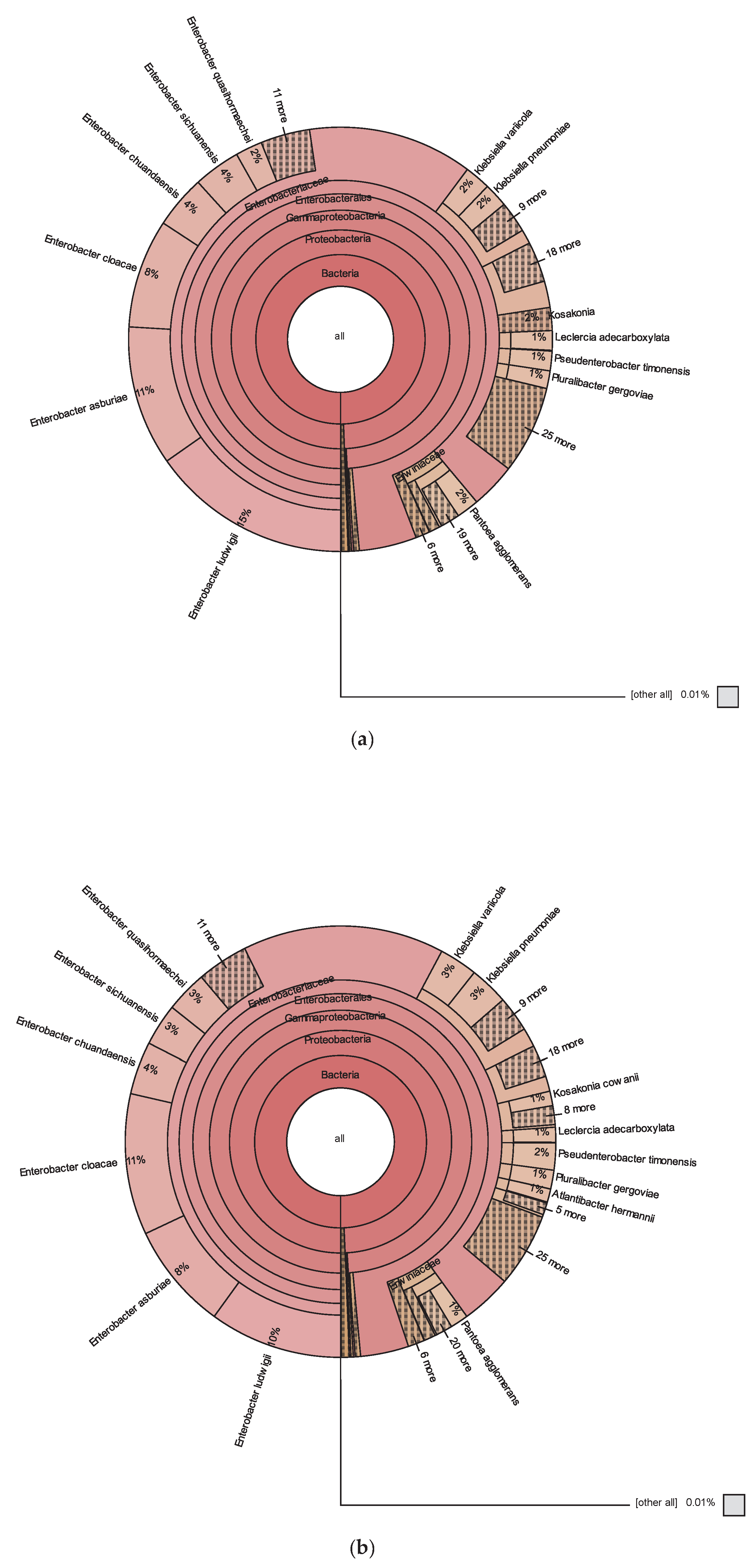
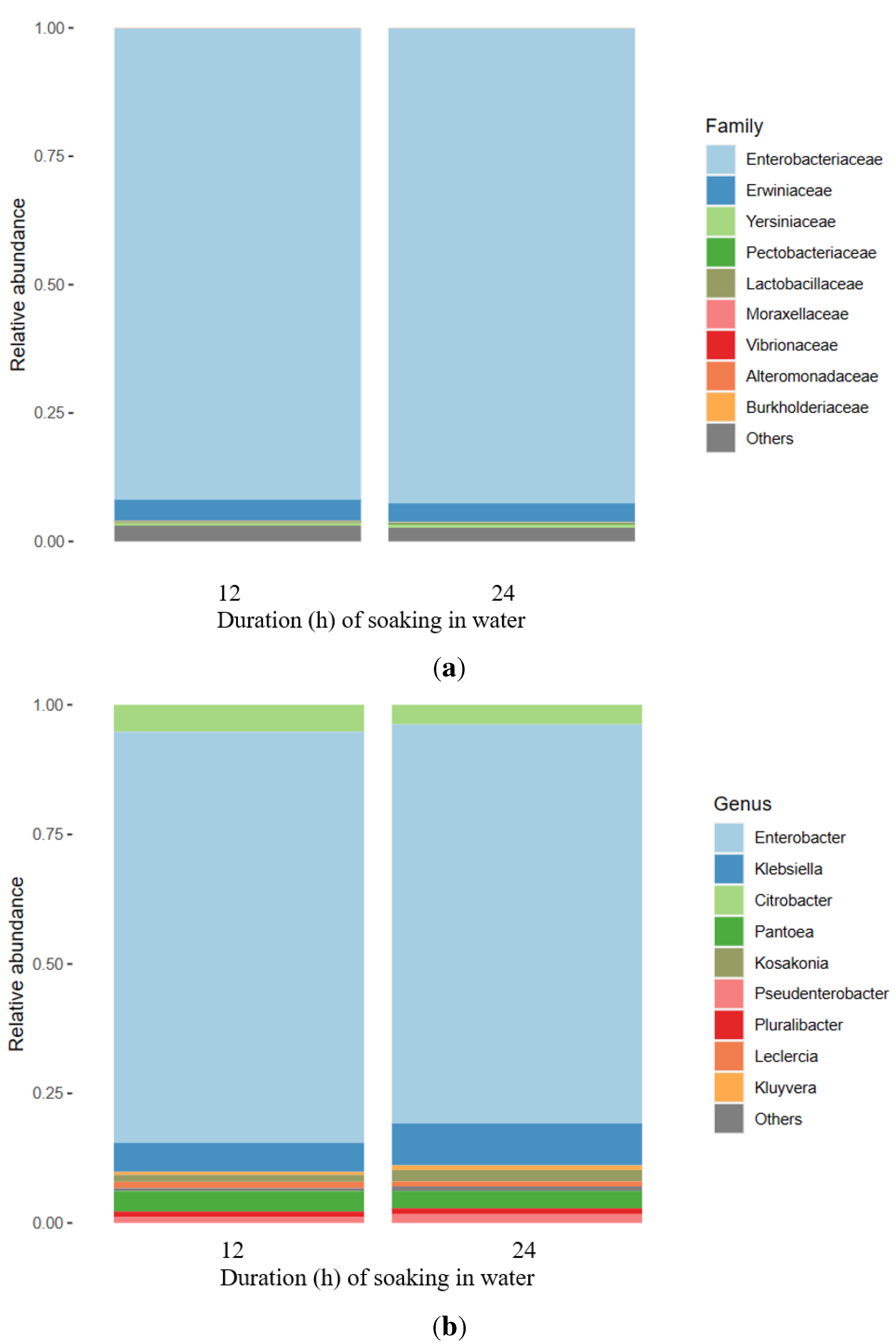
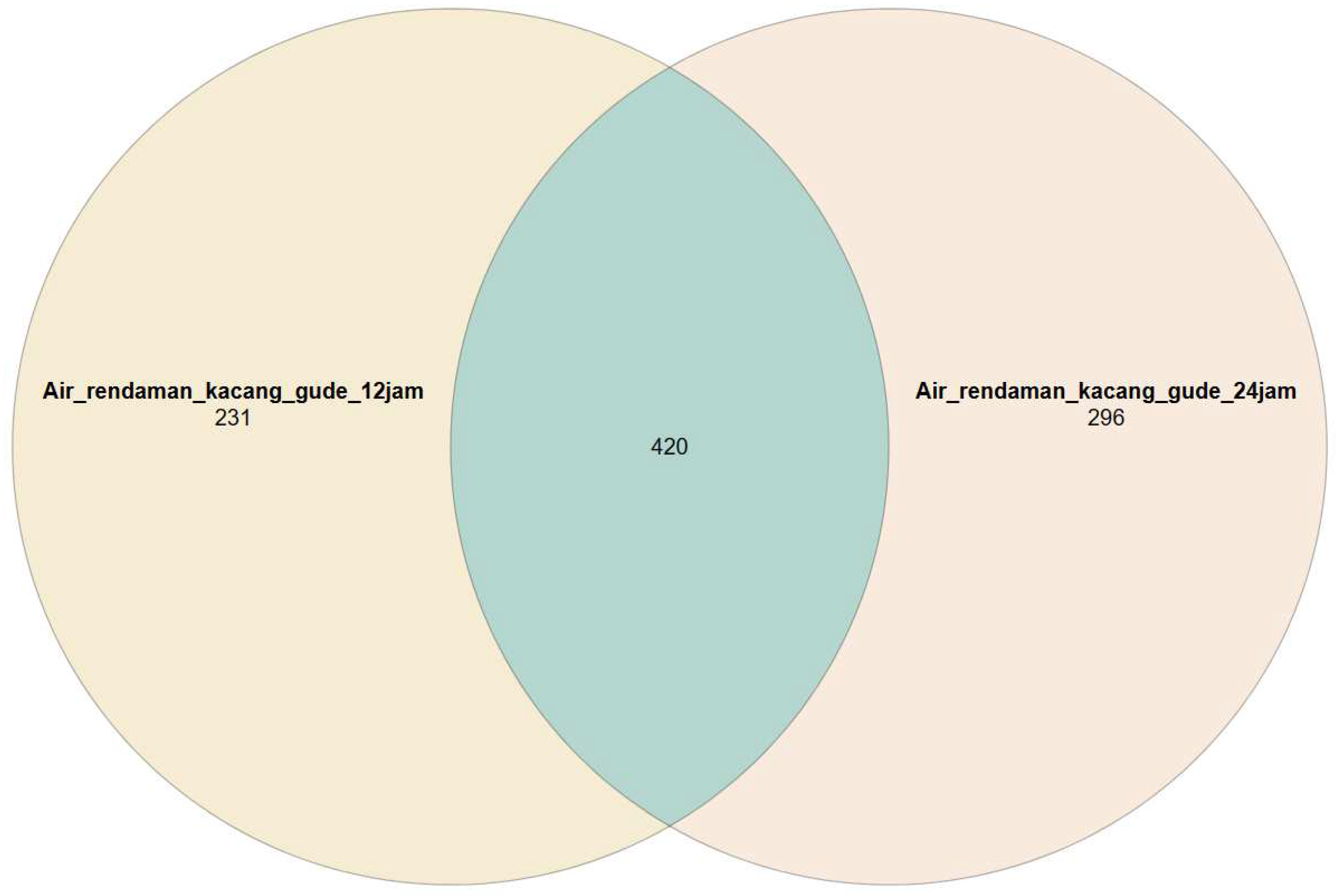
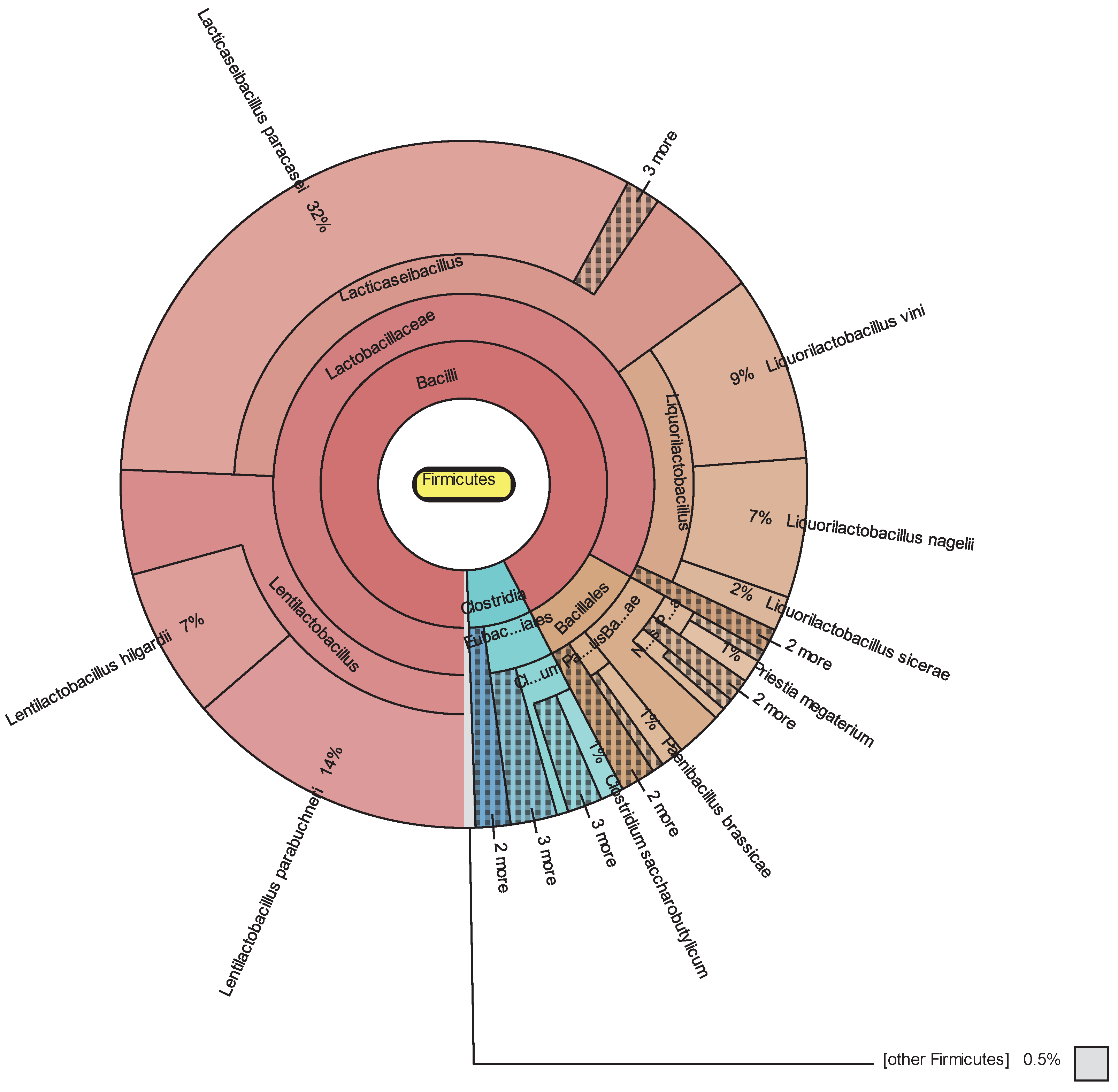
3.1. Metataxonomic Data
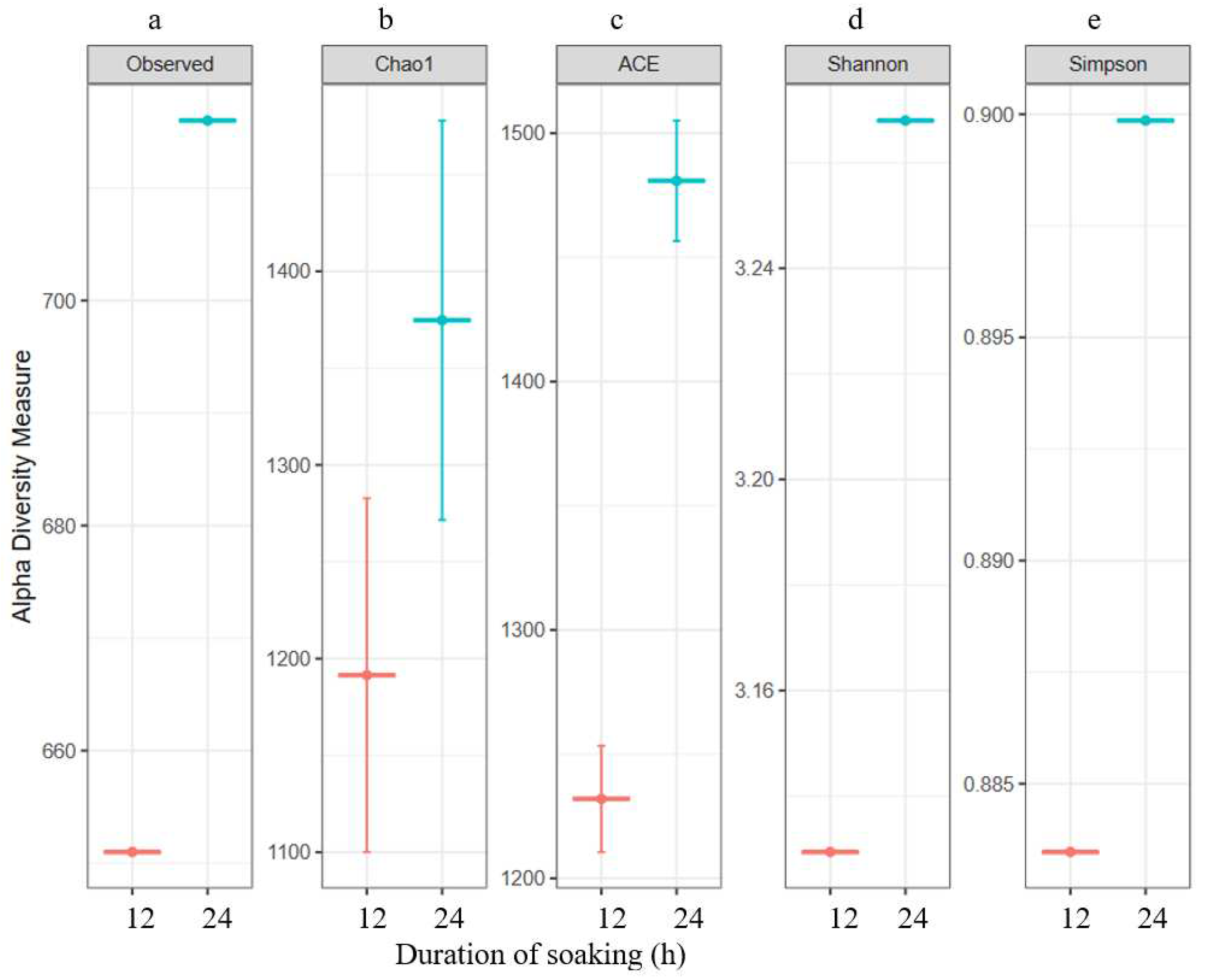
3.2. Bacterial Diversity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiao, Y.; Zhang, K.; Zhang, Z.; Zhang, C.; Sun, Y.; Feng, Z. Fermented soybean foods: A review of their functional components, mechanism of action and factors influencing their health benefits. Food Res. Int. 2022, 158, 111575. [Google Scholar] [CrossRef]
- Nurdini, A.L.; Nuraida, L.; Suwanto, A.; Suliantari. Microbial growth dynamics during tempe fermentation in two different home industries. Int. Food Res. J. 2015, 22, 1668–1674, https://api.semanticscholar.org/CorpusID:27006181. [Google Scholar]
- Valenzuela, H.; Smith, J. Cowpea: Cooperative Extension Service; College of Tropical Agriculture and Human Resources, University of Hawai: Honolulu, HI, USA, 2002; pp. 1–4. [Google Scholar]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.A.; Azam, S.; Fan, G.; Whaley, A.M.; et al. Draft genome sequence of pigeon pea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83–89. [Google Scholar] [CrossRef]
- Odeny, D.A. The potential of pigeon pea (Cajanus cajan (L.) Millsp.) in Africa. Nat. Resour. Forum. 2007, 31, 297–305. [Google Scholar] [CrossRef]
- Sheahan, C.M. Plant Guide for Pigeon pea (Cajanus cajan). USDA-Natural Resources Conservation Service; Cape May Plant Materials Center: Cape May, NJ, USA, 2012; p. 08210. [Google Scholar]
- Van der Maesen, L.J.G. Taxonomy of Cajanus. In Proceedings of the International Workshop on Pigeonpeas, Patancheru, India, 15–19 December 1980; Volume 2, pp. 9–14. [Google Scholar]
- Abebe, B.K. The dietary use of pigeon pea for human and animal diets. Sci. World J. 2022, 4873008. [Google Scholar] [CrossRef]
- Akande, K.E.; Abubakar, M.M.; Adegbola, T.A.; Bogoro, S.E.; Doma, U.D. Chemical evaluation of the nutritive quality of pigeon pea (Cajanus cajan (L). Millsp.). Int. Poult. Sci. 2010, 9, 63–65, https://api.semanticscholar.org/CorpusID:86930724. [Google Scholar] [CrossRef]
- Nwosu, J.N.; Ojukwu, M.; Ogueke, C.C.; Ahaotu, I.; Owuamanam, C.I. The antinutritional properties and ease of dehulling on the proximate composition of pigeon pea (Cajanus cajan) as affected by malting. Int. J. Life Sci. 2013, 2, 60–67, http://www.crdeepjournal.org/wp-content/uploads/2013/05/Vol-2-2-1-IJLS.pdf. [Google Scholar]
- Liu, L.; Chen, X.; Hao, L.; Zhang, G.; Jin, Z.; Li, C.; Yang, Y.; Rao, J.; Chen, B. Traditional fermented soybean products: Processing, flavor formation, nutritional and biological activities. Crit. Rev. Food Sci. Nutr. 2022, 62, 1971–1989. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.Y.; Li, H.B.; Gunaratne, A.; Sui, Z.Q.; Corke, H. Effects of fermented edible seeds and their products on human health: Bioactive components and bioactivities. Compreh. Rev. Food Sci. Food Saf. 2017, 16, 489–531. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.K.; Gowen, A.; McKenna, B. Pulse Foods: Processing, Quality and Nutraceutical Applications; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Hou, J.W.; Yu, R.C.; Chou, C.C. Changes in some components of soymilk during fermentation with bifidobacteria. Food Res. Int. 2000, 33, 393–397. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, L.; Ziao, G.; Feng, J.; Zhou, H.; Huang, F. Changes in some nutritional components of soymilk during fermentation by the culinary and medicinal mushroom Grifola frondosa. LWT 2015, 62, 468–473. [Google Scholar] [CrossRef]
- Gu, Q.; Zhang, C.; Song, D.; Li, P.; Zhu, X. Enhancing vitamin B12 content in soy-yogurt by Lactobacillus reuteri. Int. J. Food Microbiol. 2015, 206, 56–59. [Google Scholar] [CrossRef]
- Desai, A.; Small, D.M.; MCGill, A.E.J.; Shah, N.P. Metabolism of Raffinose and Stachyose in Reconstituted Skim Milk and of n-Hexanal and Pentanal in Soymilk by Bifidobacteria. Biosci. Microflora 2002, 21, 245–250. [Google Scholar] [CrossRef]
- Sandberg, A.S. Developing functional ingredients: A case study of pea protein. In Functional Foods; Woodhead Publishing: Sawston, UK, 2011; pp. 358–382. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, R.; Yang, H.; Chou, C.C. Sugar and acid contents in soymilk fermented with lactic acid bacteria alone or simultaneously with bifidobacteria. Food Microbiol. 2002, 20, 333–338. [Google Scholar] [CrossRef]
- Fredrikson, M.; Andlid, T.; Haikara, A.; Sandberg, A.S. Phytate degradation by micro-organisms in synthetic media and pea flour. J. Appl. Microbiol. 2002, 93, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumari, S.; Wongputtisin, P.; Nout, M.J.; Sarkar, P.K. Optimization of soybean processing into kinema, a Bacillus-fermented alkaline food, with respect to a minimum level of antinutrients. J. Appl. Microbiol. 2015, 119, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Hickisch, A.; Beer, R.; Vogel, R.F.; Toelstede, S. Influence of lupin milk heat treatment and exopolysaccharide-producing lactic acid bacteria on the physical characteristics of lupin-based yogurt alternatives. Food Res. Int. 2016, 84, 180–188. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network base calling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- de Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef]
- Bahram, M.; Anslan, S.; Hildebrand, F.; Bork, P.; Tedersoo, L. Newly designed 16S rRNA metabarcoding primers amplify divers and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 2018, 11, 487–494. [Google Scholar] [CrossRef]
- Fang, R.S.; Chen, Y.C.; Chen, Q.H. Bacterial diversity analysis during the fermentation of traditional Chinese yellow rice wine revealed by 16S rDNA 454 pyrosequencing. J. Food Sci. 2015, 80, M2265–M227. [Google Scholar] [CrossRef]
- Nur, N.; Meryandini, A.; Suhartono, M.T.; Suwanto, A. Lipolytic bacteria and the dynamics of flavor production in Indonesian tempeh. Biodiversitas J. Biol. Divers. 2020, 21, 3818–3825. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The changing face of the family Enterobacteriaceae (Order: “Enterobacterales”): New members, taxonomical issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 2021, 34, 2. [Google Scholar] [CrossRef]
- Sirilun, S.; Sivamaruthi, B.S.; Kesika, P.; Peerajan, S.; Chaiyasut, C. Lactic acid bacteria mediated fermented soybean as a potent nutraceutical candidate. Asian Pac. J. Trop. Biomed. 2017, 7, 930–936. [Google Scholar] [CrossRef]
- Ma, H.; Wang, L.; Yu, H.; Wang, W.; Wu, G.; Qin, G.; Tan, Z.W.Y.; Pang, H. Protease-producing lactic acid bacteria with antibacterial properties and their potential use in soybean meal fermentation. Chem. Biol. Technol. Agric. 2022, 9, 1–17. [Google Scholar] [CrossRef]
- Radita, R.; Suwanto, A.; Wahyudi, A.T.; Rusmana, I. Firmicutes is the predominant bacteria in tempeh. Int. Food Res. J. 2018, 25, 2313–2320, https://api.semanticscholar.org/CorpusID:202544934. [Google Scholar]
- Li, Z.; Rui, J.; Li, X.; Li, J.; Dong, L.; Huang, Q.; Huang, C.; Wang, Z.; Li, L.; Xuan, P.; et al. Bacterial community succession and metabolite changes during doubanjiang-meju fermentation, a Chinese traditional fermented broad bean (Viciafaba L. ) paste. Food Chem. 2017, 218, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Jiang, S.; Chen, J.; Ma, C.; Huo, D.; Shao, Y.; Zhiang, J. Unique microbial diversity and metabolic pathway features of fermented vegetables from Hainan, China. Front. Microbiol. 2018, 9, 309. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Chun, J.B. Bacterial community structure in Kimchi, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 2005, 103, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Demarinis, C.; Verni, M.; Pinto, L.; Rizzello, C.G.; Baruzzi, F. Use of Selected Lactic Acid Bacteria for the Fermentation of Legume-Based Water Extracts. Foods. 2022, 11, 21–3346. [Google Scholar] [CrossRef] [PubMed]
- Balogun, M.A.; Ahmed, R.N.; Akintayo, O.A.; Aruna, T.E.; Omovbude, M.O.; Shittu, T. Microbial, chemical and sensory evaluation of pigeon pea condiment from wild and controlled fermentation. Ceylon J. Sci. 2021, 50, 269–277. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequencing-based analysis of the bacterial and fungal composition of kefir grains and milks from multiple sources. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gulitz, A.; Stadie, J.; Wenning, M.; Ehrmann, M.A.; Vogel, R.F. The microbial diversity of water kefir. Int. J. Food Microbiol. 2011, 151, 284–288. [Google Scholar] [CrossRef]
| Family Name | taxID | taxRank | Number of Uniquely Classified Reads |
|---|---|---|---|
| Enterobacteriaceae | 543 | Family | 2607 |
| Erwiniaceae | 1903409 | Family | 22 |
| Pectobacteriaceae | 1903410 | Family | 10 |
| Calotrichaceae | 2661849 | Family | 4 |
| Halomonadaceae | 28256 | Family | 2 |
| Morganellaceae | 1903414 | Family | 2 |
| Vibrionaceae | 641 | Family | 2 |
| Alteromonadaceae | 72275 | Family | 1 |
| Balneolaceae | 1813606 | Family | 1 |
| Flavobacteriaceae | 49546 | Family | 1 |
| Intrasporangiaceae | 85021 | Family | 1 |
| Kofleriaceae | 224464 | Family | 1 |
| Micrococcaceae | 1268 | Family | 1 |
| Genus Name | taxID | taxRank | Number of Uniquely Classified Reads |
|---|---|---|---|
| Enterobacter | 547 | Genus | 9484 |
| Citrobacter | 544 | Genus | 1516 |
| Klebsiella | 570 | Genus | 819 |
| Pantoea | 53335 | Genus | 138 |
| Kluyvera | 579 | Genus | 100 |
| Kosakonia | 1330547 | Genus | 98 |
| Serratia | 613 | Genus | 85 |
| Tatumella | 82986 | Genus | 54 |
| Erwinia | 551 | Genus | 46 |
| Cronobacter | 413496 | Genus | 30 |
| Vibrio | 662 | Genus | 23 |
| Pectobacterium | 122277 | Genus | 21 |
| Bacillus | 1386 | Genus | 18 |
| Dickeya | 204037 | Genus | 14 |
| Rahnella | 34037 | Genus | 14 |
| Buttiauxella | 82976 | Genus | 13 |
| Mangrovibacter | 451512 | Genus | 12 |
| Planctopirus | 1649480 | Genus | 12 |
| Pseudocitrobacter | 1504576 | Genus | 11 |
| Cedecea | 158483 | Genus | 10 |
| Lentilactobacillus | 2767893 | Genus | 10 |
| Mixta | 2100764 | Genus | 9 |
| Acinetobacter | 469 | Genus | 9 |
| Franconibacter | 1649295 | Genus | 8 |
| Chimaeribacter | 2716544 | Genus | 7 |
| Lelliottia | 1330545 | Genus | 7 |
| Edwardsiella | 635 | Genus | 6 |
| Actinobacillus | 713 | Genus | 5 |
| Escherichia | 561 | Genus | 5 |
| Lacticaseibacillus | 2759736 | Genus | 5 |
| Pseudomonas | 286 | Genus | 4 |
| Yersinia | 629 | Genus | 4 |
| Izhakiella | 1780190 | Genus | 3 |
| Leclercia | 83654 | Genus | 3 |
| Shigella | 620 | Genus | 3 |
| Acetomicrobium | 49894 | Genus | 2 |
| Bradyrhizobium | 374 | Genus | 2 |
| Brenneria | 71655 | Genus | 2 |
| Brevitalea | 2048911 | Genus | 2 |
| Gibbsiella | 929812 | Genus | 2 |
| Haemophilus | 724 | Genus | 2 |
| Lonsdalea | 1082702 | Genus | 2 |
| Paraburkholderia | 1822464 | Genus | 2 |
| Psychromonas | 67572 | Genus | 2 |
| Raoultella | 160674 | Genus | 2 |
| Trabulsiella | 158851 | Genus | 2 |
| Xenorhabdus | 626 | Genus | 2 |
| Actinoplanes | 1865 | Genus | 1 |
| Aeribacillus | 1055323 | Genus | 1 |
| Aeromonas | 642 | Genus | 1 |
| Atlantibacter | 1903434 | Genus | 1 |
| Brucella | 234 | Genus | 1 |
| Burkholderia | 32008 | Genus | 1 |
| Caldimonas | 196013 | Genus | 1 |
| Catenulispora | 414878 | Genus | 1 |
| Crinalium | 241421 | Genus | 1 |
| Croceicoccus | 1295327 | Genus | 1 |
| Flavisolibacter | 398041 | Genus | 1 |
| Gloeobacter | 33071 | Genus | 1 |
| Halomonas | 2745 | Genus | 1 |
| Humidesulfovibrio | 356 | Genus | 1 |
| Hyphomicrobium | 81 | Genus | 1 |
| Iningainema | 1932705 | Genus | 1 |
| Ktedonobacter | 363276 | Genus | 1 |
| Leucothrix | 45247 | Genus | 1 |
| Marichromatium | 85076 | Genus | 1 |
| Massilia | 149698 | Genus | 1 |
| Mathylobacillus | 404 | Genus | 1 |
| Micromonospora | 1873 | Genus | 1 |
| Microvirga | 186650 | Genus | 1 |
| Morganella | 581 | Genus | 1 |
| Motilimonas | 1914248 | Genus | 1 |
| Niastella | 354354 | Genus | 1 |
| Novosphingobium | 165696 | Genus | 1 |
| Oceanisphaera | 225143 | Genus | 1 |
| Paraglaciecola | 1621534 | Genus | 1 |
| Pedomicrobium | 47494 | Genus | 1 |
| Permianibacter | 1649479 | Genus | 1 |
| Phenylobacterium | 20 | Genus | 1 |
| Providencia | 586 | Genus | 1 |
| Pseudoxanthomonas | 83618 | Genus | 1 |
| Rhabdothermincola | 2820403 | Genus | 1 |
| Rhodoplanes | 29407 | Genus | 1 |
| Salmonella | 590 | Genus | 1 |
| Sphingomonas | 13687 | Genus | 1 |
| Thermithiobacillus | 119979 | Genus | 1 |
| Thermosynthropha | 54293 | Genus | 1 |
| Virgisporangium | 65504 | Genus | 1 |
| Family Name | taxID | taxRank | Number of Uniquely Classified Reads |
|---|---|---|---|
| Enterobacteriaceae | 543 | Family | 2836 |
| Yersiniaceae | 1903411 | Family | 34 |
| Erwiniaceae | 1903409 | Family | 23 |
| Vibrionaceae | 641 | Family | 6 |
| Calotrichaceae | 2661849 | Family | 5 |
| Pectobacteriaceae | 1903410 | Family | 5 |
| Bacillaceae | 186817 | Family | 4 |
| Burkholderiaceae | 119060 | Family | 2 |
| Methylococcaceae | 403 | Family | 2 |
| Acidobacteriaceae | 204434 | Family | 1 |
| Aeromonadaceae | 84642 | Family | 1 |
| Anaerolineaceae | 292628 | Family | 1 |
| Bruguierivoracaceae | 2812006 | Family | 1 |
| Flavobacteriaceae | 49546 | Family | 1 |
| Gomontiellaceae | 1892255 | Family | 1 |
| Halanaerobiaceae | 972 | Family | 1 |
| Halomonadaceae | 28256 | Family | 1 |
| Heliobacteriaceae | 31984 | Family | 1 |
| Hyphomicrobiaceae | 45401 | Family | 1 |
| Intrasporangiaceae | 85021 | Family | 1 |
| Microbacteriaceae | 85023 | Family | 1 |
| Morganellaceae | 1903414 | Family | 1 |
| Pasteurellaceae | 712 | Family | 1 |
| Pseudomonadaceae | 135621 | Family | 1 |
| Rhodospirillaceae | 41295 | Family | 1 |
| Succinivibrionaceae | 83763 | Family | 1 |
| Genus Name | taxID | taxRank | Number of Uniquely Classified Reads |
|---|---|---|---|
| Enterobacter | 547 | Genus | 11233 |
| Klebsiella | 570 | Genus | 1061 |
| Citrobacter | 544 | Genus | 842 |
| Kluyvera | 579 | Genus | 150 |
| Kosakonia | 1330547 | Genus | 144 |
| Pantoea | 53335 | Genus | 98 |
| Serratia | 613 | Genus | 68 |
| Tatumella | 82986 | Genus | 58 |
| Erwinia | 551 | Genus | 45 |
| Buttiauxella | 82976 | Genus | 33 |
| Cronobacter | 413496 | Genus | 31 |
| Dickeya | 204037 | Genus | 23 |
| Mangrovibacter | 451512 | Genus | 21 |
| Pectobacterium | 122277 | Genus | 18 |
| Vibrio | 662 | Genus | 17 |
| Planctopirus | 1649480 | Genus | 16 |
| Edwardsiella | 635 | Genus | 13 |
| Rahnella | 34037 | Genus | 12 |
| Shigella | 620 | Genus | 12 |
| Pseudocitrobacter | 1504576 | Genus | 11 |
| Lacticaseibacillus | 2759736 | Genus | 10 |
| Lentilactobacillus | 2767893 | Genus | 9 |
| Acetomicrobium | 49894 | Genus | 8 |
| Lelliottia | 1330545 | Genus | 8 |
| Escherichia | 561 | Genus | 7 |
| Trabulsiella | 158851 | Genus | 6 |
| Chimaeribacter | 2716544 | Genus | 5 |
| Brevitalea | 2048911 | Genus | 4 |
| Rosenbergiella | 1356488 | Genus | 4 |
| Yersinia | 629 | Genus | 4 |
| Gibbsiella | 929812 | Genus | 3 |
| Raoultella | 160674 | Genus | 3 |
| Salmonella | 590 | Genus | 3 |
| Xenorhabdus | 626 | Genus | 3 |
| Bradyrhizobium | 374 | Genus | 2 |
| Burkholderia | 32008 | Genus | 2 |
| Crinalium | 241421 | Genus | 2 |
| Franconibacter | 1649295 | Genus | 2 |
| Gloeobacter | 33071 | Genus | 2 |
| Legionella | 445 | Genus | 2 |
| Microbacterium | 33882 | Genus | 2 |
| Miltoncostaea | 2843200 | Genus | 2 |
| Providencia | 586 | Genus | 2 |
| Pseudaeromonas | 1929090 | Genus | 2 |
| Reyranella | 445219 | Genus | 2 |
| Salinicola | 404432 | Genus | 2 |
| Shewanella | 22 | Genus | 2 |
| Thalassotalea | 1518149 | Genus | 2 |
| Achromobacter | 222 | Genus | 1 |
| Aeromonas | 642 | Genus | 1 |
| Alkalimonas | 265980 | Genus | 1 |
| Ammonifex | 42837 | Genus | 1 |
| Bosea | 85413 | Genus | 1 |
| Caballeronia | 1827195 | Genus | 1 |
| Cephalothrix | 1844514 | Genus | 1 |
| Chloroflexus | 1107 | Genus | 1 |
| Clostridium | 1485 | Genus | 1 |
| Conexibacter | 191494 | Genus | 1 |
| Defluviitalea | 1185408 | Genus | 1 |
| Dongia | 1146845 | Genus | 1 |
| Euryhalinema | 2661529 | Genus | 1 |
| Halomonas | 2745 | Genus | 1 |
| Iningainema | 1932705 | Genus | 1 |
| Laceyella | 292635 | Genus | 1 |
| Leclercia | 83654 | Genus | 1 |
| Lonsdalea | 1082702 | Genus | 1 |
| Lysobacter | 68 | Genus | 1 |
| Marinicella | 863253 | Genus | 1 |
| Massilia | 149698 | Genus | 1 |
| Motilibacter | 1434021 | Genus | 1 |
| Motilimonas | 1914248 | Genus | 1 |
| Neobacillus | 2675232 | Genus | 1 |
| Nitrospira | 1234 | Genus | 1 |
| Oceanisphaera | 225143 | Genus | 1 |
| Paraburkholderia | 1822464 | Genus | 1 |
| Pelagicoccus | 455433 | Genus | 1 |
| Phenylobacterium | 20 | Genus | 1 |
| Photobacterium | 657 | Genus | 1 |
| Pseudoalteromonas | 53246 | Genus | 1 |
| Pseudomonas | 286 | Genus | 1 |
| Psychromonas | 67572 | Genus | 1 |
| Rhizobium | 379 | Genus | 1 |
| Secundilactobacillus | 2767892 | Genus | 1 |
| Shimwellia | 1335483 | Genus | 1 |
| Solirubrobacter | 207599 | Genus | 1 |
| Tepidimonas | 114248 | Genus | 1 |
| Thermoanaerobacter | 1754 | Genus | 1 |
| Thermodesulfovibrio | 28261 | Genus | 1 |
| Trinickia | 2571160 | Genus | 1 |
| Tumebacillus | 432330 | Genus | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





