Submitted:
18 January 2024
Posted:
19 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
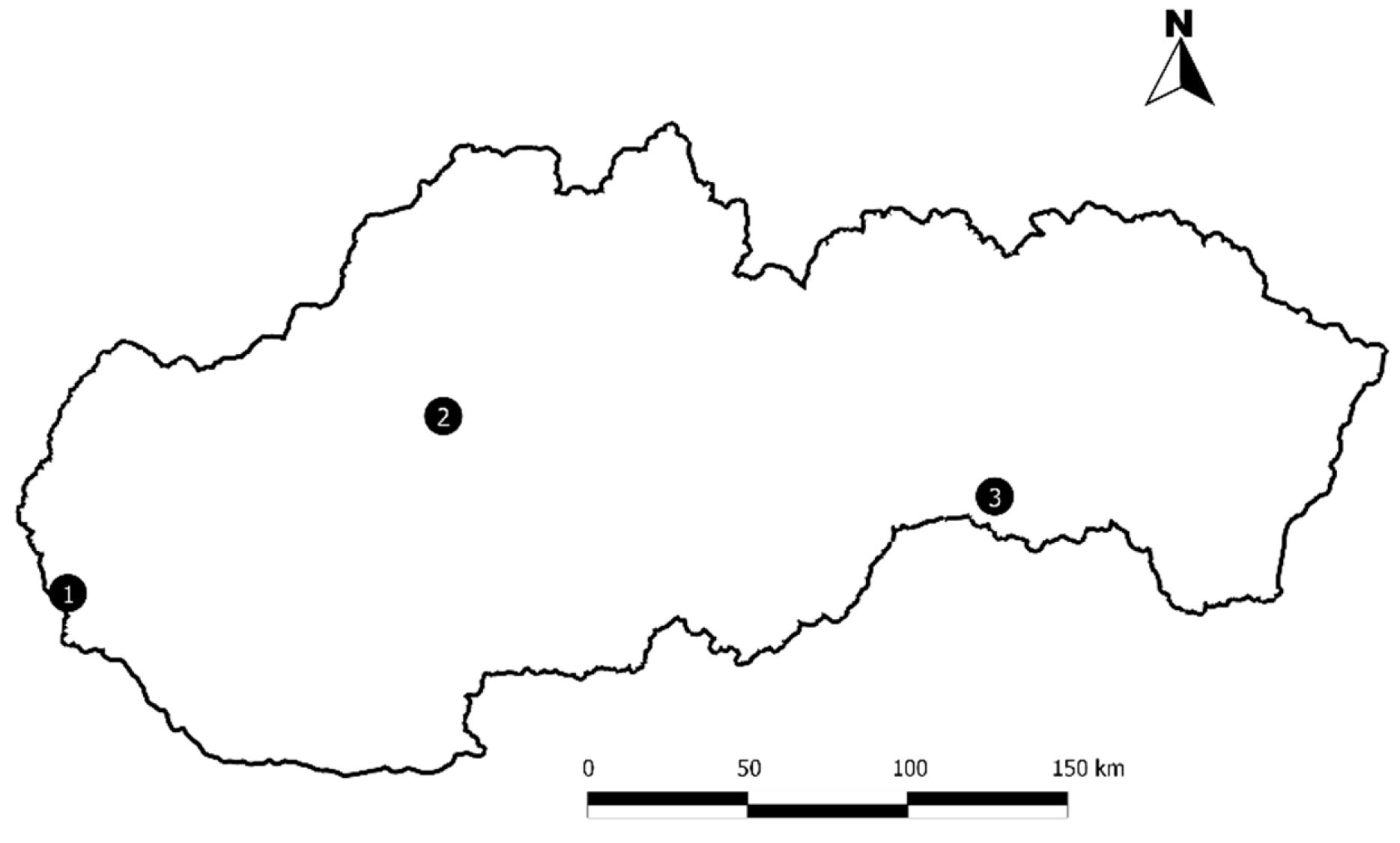
2.1. Study sites
2.2. Mist-netting and sampling birds
2.3. Microscopic analysis of haemosporidian
2.4. Molecular analysis of haemosporidian
2.5. Statistical analysis
3. Results
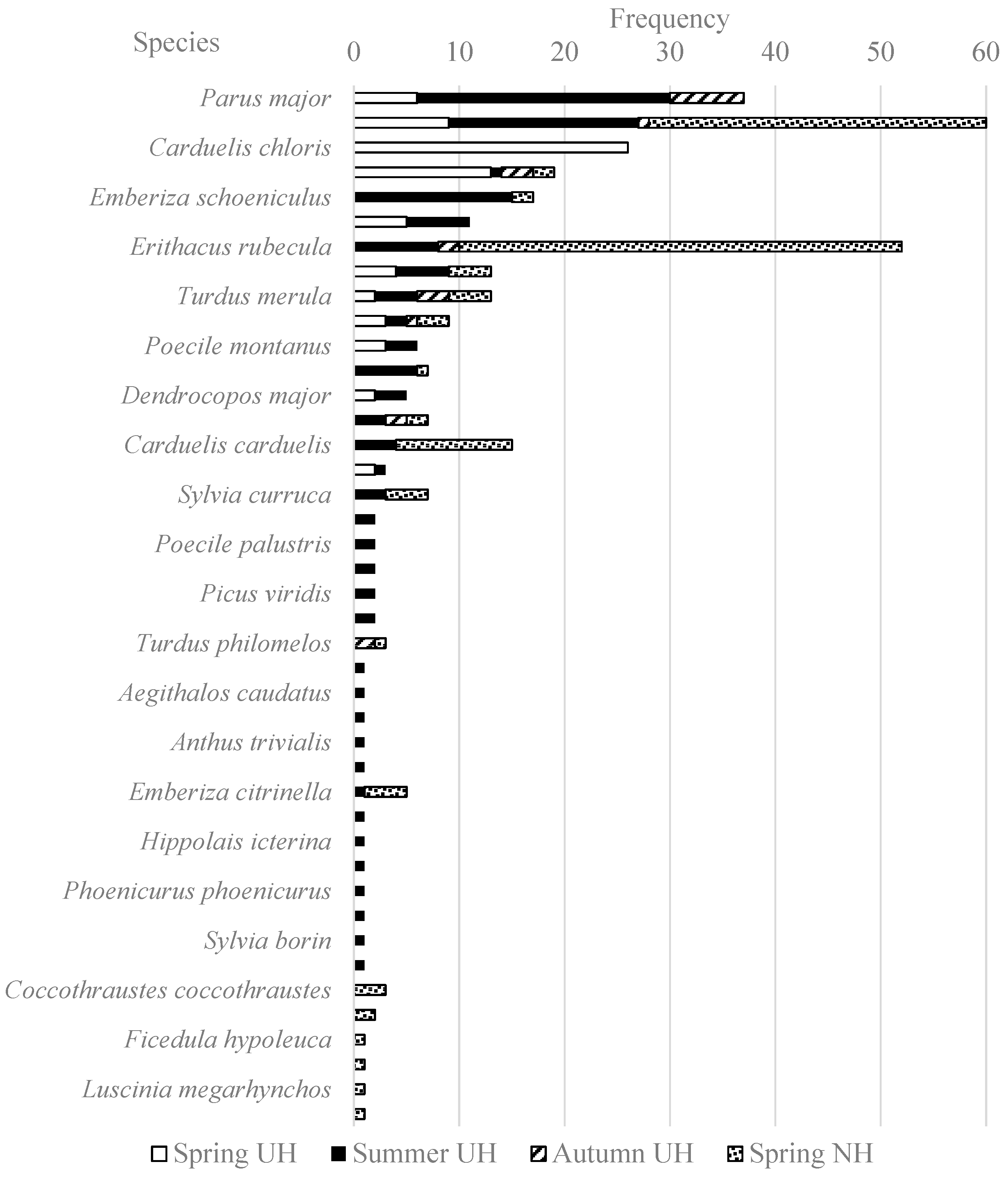
3.1. Haemosporidian infections based on the microscopical and molecular analyses
3.2. Haemosporidian infections in juvenile birds based on the microscopical and molecular analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Compliance with ethical standards
References
- Valkiūnas, G. Avian Malaria Parasites and Other Haemosporidia; CRC press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Valkiūnas, G.; Iezhova, T.A. Keys to the avian Haemoproteus parasites (Haemosporida, Haemoproteidae). Malaria Journal 2022, 21, 269. [CrossRef]
- Kučera, J. Blood parasites of birds in Central Europe. 1.Survey of literature. The incidence in domestic birds and general remark to the incidence in wild birds. Folia Parasitologica 1981a, 28, 13-22.
- Hellgren, O.; Waldenström, J.; Bensch, S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J. Parasitol. 2004, 90, 797–802. [Google Scholar] [CrossRef]
- Bensch, S.; Hellgren, O.; Pérez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef]
- MalAvi database. Available online: http://130.235.244.92/Malavi/index.html (accessed on 28 November 2023).
- Kučera, J. Blood parasites of birds in Central Europe. 2. Leucocytozoon. Folia Parasitologica 1981b, 28, 193-203.
- Kučera, J. Blood parasites of birds in Central Europe. 3. Plasmodium and Haemoproteus. Folia Parasitologica 1981, 28, 303–312. [Google Scholar]
- Hauptmanová, K.; Benedikt, V.; Literák, I. Blood Parasites in Passerine Birds in Slovakian East Carpathians. Acta Protozool. 2006, 45, 105–109. [Google Scholar]
- Haas, M.; Kisková, J. Absence of blood parasites in the Alpine Accentor Prunella collaris. Oecologia Montana 2010, 19, 30–34. [Google Scholar]
- Berthová, L.; Országhová, Z.; Valkiūnas, G. The first report of nine species of haemosporidian parasites (Haemosporida: Haemoproteus, Plasmodium and Leucocytozoon) in wild birds from Slovakia. Biologia 2012; 67, 931-933. [CrossRef]
- Šujanová, A.; Špitalská, E.; Václav, R. Seasonal dynamics and diversity of haemosporidians in a natural woodland bird community in Slovakia. Diversity 2021, 13, 439. [Google Scholar] [CrossRef]
- Kazimírová, M.; Hamšíková, Z.; Kocianová, E.; Marini, G.; Mojšová, M.; Mahríková, L.; Berthová, L.; Slovák, M.; Rosá, R. Relative density of host-seeking ticks in different habitat types of south-western Slovakia. Exp. Appl. Acarol. 2016, 69, 205–224. [Google Scholar] [CrossRef]
- Berthová, L.; Slobodník, V.; Slobodník, R.; Olekšák, M.; Sekeyová, Z.; Svitálková, Z.; Kazimírová, M.; Špitalská, E. The natural infection of birds and ticks feeding on birds with Rickettsia spp. and Coxiella burnetii in Slovakia. Exp. Appl. Acarol. 2016, 68, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L. Identification guide to European passerines, 4th ed.; Lars Svensson, Stockholm. 1992.
- Hromádko, M.; Horáček, J.; Chytil, J.; Pithart, K.; Škopek. J. Guide to the determination of our passerines, Publisher ZUV, Hradec Králové (in Czech). 1992.
- Hromádko, M.; Horáček, J.; Chytil, J.; Pithart, K.; Škopek. J. Guide to the determination of our passerines, Publisher ZUV, Hradec Králové (in Czech). 1993.
- Hromádko, M.; Horáček, J.; Chytil, J.; Pithart, K.; Škopek. J. Guide to the determination of our passerines, Publisher ZUV, Hradec Králové (in Czech). 1998.
- Bensch, S.; Stjernman, M.; Hasselquist, D.; Ostman, O.; Hansson, B.; Westerdahl, H.; Pinheiro, R.T. Host specificity in avian blood parasites: A study of plasmodium and haemoproteus mitochondrial DNA amplified from birds. Proc. Biol. Sci. 2000, 267, 1583–1589. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A., Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725-2729. [CrossRef] [PubMed]
- GenBank database. Available online: http://blast.ncbi.nlm.nih.gov/ (accessed on 22 February 2023).
- Bell, J.A.; Weckstein, J.D.; Fecchio, A.; Tkach, V.V. A new real-time pcr protocol for detection of avian haemosporidians. Parasites Vectors 2015, 8, 383. [Google Scholar] [CrossRef]
- Friedl, T.W.P.; Groscurth, E. A real-time PCR protocol for simple and fast quantification of blood parasite infections in evolutionary and ecological studies and some data on intensities of blood parasite infections in a subtropical weaverbird. J. Ornithol. 2012, 153, 239–247. [Google Scholar] [CrossRef]
- Tiersch, T.R.; Wachtel, S.S. On the evolution of genome size of birds. J. Hered. 1991, 82, 363–368. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeo. Electronica 2001, 4, 4, http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
- EasyCalculation. Available online: https://www.easycalculation.com/statistics (accessed on 22 February 2023).
- Basto, N.; Rodríguez, O.A.; Marinkelle, C.J.; Gutiérrez, R.; Matta, N. Haematozoa in birds from La Macarena National Natural Park (Colombia). Caldasia. 2006, 28, 371–7. [Google Scholar]
- Doussang, D., González-Acuña, D., Torres-Fuentes, L.G. et al. Spatial distribution, prevalence and diversity of haemosporidians in the rufous-collared sparrow, Zonotrichia capensis. Parasites Vectors 2019, 12, 2. [CrossRef]
- Ciloglu, A.; Ergen, A.G.; Inci, A.; Dik, B.; Duzlu, O.; Onder, Z.; Yetismis, G.; Bensch, S.; Valkiūnas, G.; Yildrim, A. Prevalence and genetic diversity of avian haemosporidian parasites at an intersection point of bird migration routes: Sultan Marshes National Park, Turkey. Acta Trop. 2020, 210, 105465. [Google Scholar] [CrossRef] [PubMed]
- Quillfeldt, P.; Arriero, E.; Martínez, J.; Masello, J.F.; Merino, S. Prevalence of blood parasites in seabirds- a review. Front Zool. 2011, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Chakarov, N.; Kampen, H.; Wiegmann, A.; Werner, D.; Bensch, S. Blood parasites in vectors reveal a united blackfly community in the upper canopy. Parasites Vectors 2020, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Schumm, Y.R.; Bakaloudis, D.; Barboutis, C.; Cecere, J.G.; Eraud, C.; Fischer, D.; Hering,J. ; Hillerich, K.; Lormée, H.; Mader, V.; Masello, J.F.; Metzger, B.; Rocha, G.; Spina, F.; Quillfeldt, P. Prevalence and genetic diversity of avian haemosporidian parasites in wild bird species of the order Columbiformes. Parasitol. Res., 2021, 120, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.; Martínez-Gómez, J.E.; Valkiūnas, G.; Loiseau, C.; Bell, D.A.; Sehgal, R.N. Diversity and phylogenetic relationships of hemosporidian parasites in birds of Socorro Island, México and their role in the re-introduction of the Socorro Dove (Zenaida graysoni). J Parasitol 2013, 99, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Heym, E.C.; Kampen, H.; Krone, O.; Schäfer, M.; Werner, D. Molecular detection of vector-borne pathogens from mosquitoes collected in two zoological gardens in Germany. Parasitol Res 2019, 118, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G.; Ilgūnas, M.; Bukauskaitė, D.; Chagas, C.R.F.; Bernotienė, R.; Himmel, T.; Harl, J.; Weissenböck, H.; Iezhova, T.A. Molecular characterization of six widespread avian haemoproteids, with description of three new Haemoproteus species. Acta Tropica 2019, 197, 105051. [Google Scholar] [CrossRef] [PubMed]
- Bennet, G.F.; Peirce, M.A.; Ashford, R.W. Avian haematozoa: mortality and pathogenicity. J. Nat. Hist. 1993, 27, 993–1001. [Google Scholar] [CrossRef]
- Ortiz-Catedral, L.; Brunton, D.; Stidworthy, M.F.; Elsheikha, H.M.; Pennycott, T.; Schulze, C.; Braun, M.; Wink, M.; Gerlach, H.; Pendl, H.; Gruber, A.D.; Ewen, J.; Pérez-Tris, J.; Valkiūnas, G.; Olias, P. Haemoproteus minutus is highly virulent for Australasian and South American parrots. Parasit. Vectors 2019, 12, 40. [Google Scholar] [CrossRef]
- Pérez-Tris, J.; Hellgren, O.; Križanauskienė, A.; Waldenström, J.; Secondi, J.; Bonneaud, C.; Fjeldsȧ, J.; Hasselquist, D.; Bensch, S. Within-Host Speciation of Malaria Parasites. PLoS ONE 2007, 2, e235. [Google Scholar] [CrossRef]
- Hernández-Lara, C.; Duc, M.; Ilgūnas, M.; Valkiūnas, G. Massive Infection of Lungs with Exo-Erythrocytic Meronts in European Robin Erithacus rubecula during Natural Haemoproteus attenuatus Haemoproteosis. Animals 2021, 11, 3273. [Google Scholar] [CrossRef]
- de Jong Y (2016). Fauna Europaea. Fauna Europaea Consortium. Checklist dataset. accessed via GBIF.org on 2024-01-12. [CrossRef]
| Bird species | Infected with Haemoproteus/ Plasmodium/ Analyzed | Infected with Leucocytozoon/ Analyzed | Infected with Haemoproteus/ Plasmodium/ Analyzed | Infected with Leucocytozoon/ Analyzed |
| Urbanized habitats | Natural habitats | |||
| Parus major | 5/37 | 12/37 0/6 3/9 1/28 |
1/4 15/32 |
1/4 0/32 |
| Poecole montanus | 1/6 | |||
| Cyanistes caeruleus | 1/9 | |||
| Sylvia atricapilla | 15/28 | |||
| Sylvia communis | 1/1 | 0/1 | ||
| Sylvia curruca | 1/4 | 0/4 | ||
| Fringilla coelebs | 4/6 |
0/6 0/1 1/9 0/10 0/15 1/2 1/6 |
1/3 | 0/3 |
| Emberiza citrinella | 1/1 | 2/4 | 0/4 | |
| Turdus merula | 8/9 | 4/4 | 0/4 | |
| Erithacus rubecula | 1/10 | 19/42 1/2 |
5/42 0/2 |
|
| Emberiza schoeniculus | 3/15 | |||
| Turdus philomelos | 2/2 | 1/1 | 0/1 | |
| Prunella modularis | 1/6 | |||
| Carduelis carduelis | 3/11 | 0/11 | ||
|
Coccothraustes coccothraustes |
2/3 | 3/3 | ||
| Scolopax rusticola | 1/1 | 1/1 | ||
| Emberiza cia | 1/2 | 0/2 | ||
| Passer montanus | 1/2 | 0/2 | ||
| other Total |
0/66 43/196 |
0/66 19/196 |
0/6 53/121 |
0/6 10/121 |
| Avian species | Haemosporidian species |
| Urbanized habitats | |
| Turdus merula | P. vaughani SYAT05, L. sp. |
| Cyanistes caeruleus | P. relictum SGS1, L. sp. PARUS19 |
| Parus major | P. relictum SGS1, L. sp. PARUS4 |
| Parus major | P. relictum SGS1, L. sp. PARUS18 |
| Parus major | P. relictum SGS1, L. sp. STUR1/TURPEL01 |
| Sylvia atricapilla | P. sp., L. sp. PARUS4 |
| Turdus philomelos | P. circumflexum BT7, L. sp. |
| Natural habitats | |
| Erithacus rubecula | H. sp., L. sp. BT2 |
| Scolopax rusticola | H. sp., L. sp. |
| Erithacus rubecula | H. attenuatus ROBIN1/LULU1, L. sp. SFC8 |
| Coccothraustes coccothraustes | H. tartakovskyi sp. HAWF1, L. sp. |
| Erithacus rubecula | P. circumflexum TURDUS1, L. sp. SYCON06 |
| Coccothraustes coccothraustes | H. concavocentralis HAWF2, L. sp. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





