1. Introduction
Respiratory diseases are common health problems affecting millions of people each year. When individuals come to contact with pathogens like bacteria, viruses, and allergens, their respiratory systems face to a wide range of possibilities causing ill. Certain populations having weak immune systems or those exposed to coronaviruses or rhinoviruses, might be more susceptible to get respiratory problems. Common cold is a convenient term to represent mild upper respiratory diseases, and it has multiple symptoms, like cough, sneezing, and sore throat etc. [
1].
To differentiate these respiratory diseases, physicians use a process called diagnostic reasoning [
2]. They begin by inquiring patient's complaints, symptoms, and past medical histories. Usually in hospital, additional examinations like radiology or hematology tests may be conducted to confirm their diagnostic hypothesis. Based on the lab-test evidence, physicians are able to make better diagnoses. These patients’ information and corresponding diagnoses are recorded in the system of electronic medical records (EMRs), where diagnostic codes follow the format defined by The International Statistical Classification of Diseases and Related Health Problems (ICD) [
3].
Complexities on using recorded data are mostly owing to unstructured or non-normalized recordings. Nowadays, patients’ medical records are recorded electronically in digital form such as HIS (hospital information system). Most medical systems have their unique frameworks and policies on patient’s privacy. As stated in [
4], there is no standard platform utilized among hospitals in using EMRs. In addition, the unstructured information and related medical abbreviation is a challenge for non-medical engineers to utilize and realize these data. This causes difficulty on using the recorded data for research, usually the data has to be purified.
Remote diagnosis on mild respiratory diseases comes to real while in COVID-19 pandemic era in Taiwan. In the pandemic all hospitals in Taiwan are controlled and people having non-urgent illness are encouraged to stay at home or use the remote diagnosis system such as telephone or on-line video consultation. This epidemic prevention measure relieves the pressure on hospitals and medical doctors. Also, it diminishes the contact between patients in hospitals. The motivation of this study is induced. Is it possible to create an AI-based diagnosis system, which can be used for people having mild respiratory complications at home or any place outside hospitals to have a preliminary diagnosis? If yes, this system can be setup in a website or even as an app in any mobile phone.
Compared to text data, quantifiable medical indicators such as biochemical values in blood tests can be analyzed through data mining to identify discriminate thresholds for predicting different diseases [
5]. Similarly, via analyzing relevant features on medical images such as radiomics can serve as image biomarkers extraction for predicting corresponding diseases [
6]. However, non-quantifiable or unstructured text data, it lacks a quantitative analysis approach in the past. Recent rise in natural language processing has led to the era of deep learning on natural language. From word embedding [
7], RNN [8,9] to LSTM [10-12], they deploy a foundation for processing text data without fixed formats and time series. Many algorithms or language models aim to do data mining and extract useful properties in the unconstructed medical texts. For example, The Unified Medical Language System (UMLS) [
13] integrates over 2 million biomedical vocabularies and includes terminologies used for bioinformatics. Similar study in [
14] is to recognize seven categories from the EMRs based on the language model. In [
15] they construct a text generation system called MediExpert to assist differential diagnoses.
Today, with the help of self-attention models, long time series of text data can be effectively processed, allowing the model's performance to get over previous limitations caused by gradient vanish. The drawback of gradient vanish makes it difficult to analyze commonly sequenced sentences. In this feasibility study, we construct the self-attention-based language model, GPT2 [
16], to encode our free-text data in house and predict three respiratory diseases.
2. Materials and Methods
2.1. Dateset
The text data consisting of patient complications and physician diagnoses in the ICD format, in total 30592 patients are collected from Tainan Municipal An-Nan Hospital, China Medical University from 2017 to 2022. Among these data, there are four fields not handled by the system but filled in by physicians from patient interviews to final prescriptions. These fields are diagnosis, past history, symptoms, and treatment. First, physicians record patients' narratives, basic physical conditions, and inquired symptoms in the symptoms field. Then, based on previous medical records or further inquiries, chronic illnesses, family medical history et al. are recorded in the field of past history. Based on the past history and symptoms, physicians make for each outpatient at least one and up to three diagnoses using ICD format for documentation. The field of treatment is filled with the corresponding prescription provided by the physician. Apart from the data in the field of diagnosis, the remaining three fields of the table, namely past history, symptoms, and treatment, are considered to be unstructured text data. This is because for each outpatient different physicians have different writing styles. Besides, Taiwanese physicians are not English native speakers, resulting in records of these three fields containing disorganized descriptions or word fragments. Worse, found in some records, descriptions are Chinese terms specific to traditional Chinese medicine having no corresponding English terminology. It is challenging to translate Chinese descriptions to English using standard medical terminology automatically [
17]. Therefore, our data are purified by excluding those records using Chinese descriptions. In total 16639 records are excluded.
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) of An Nan Hospital with the IRB number TMANH112-REC030.
2.2. Preprocessing
In terms of symptom descriptions, there are many abbreviations and acronyms of specific medical terminology that allow physicians to conveniently record them. However, these abbreviations can pose ambiguity for the word embedding, which may also affect the performance of the model. Therefore, a manual review of medical records from a six-month period is conducted to select 119 medical abbreviations and establish a dictionary for the conversion of all other textual data, by which we want to improve discrimination of word vector produced from the word embedding and decrease probability of misdiagnosis [
18]. During the data purification process, we observe many clinics conducting remote consultations during the pandemic era of COVID-19. These remote consultations in the medical records have no relevant symptom descriptions. This portion of the data (10748 patients) are already excluded because they are all recorded in Chinese. After this data purification, medical records containing the diagnostic codes for nasopharyngitis (460 indicated to ICD), upper respiratory infection (465.9 indicated to ICD), bronchitis and bronchiolitis (466 indicated to ICD) are selected for this research.
2.3. Word embeddding
We adopt the byte pair encoding (BPE) technique used in the GPT-2 paper to embed textual data [
16]. This approach effectively compresses frequently occurring symbol sequences [
19]. First the data is transformed into a UTF-8 encoded format and then processed with BPE, resulting in a vector of encoded tokens. This vector serves as the input to the model.
2.4. Language model
GPT-2, developed by OpenAI in 2018, is a transformer-based language model and an evolution of its predecessor, GPT. [16, 20] The self-attention module, a crucial component of the Transformer architecture, plays a significant role in GPT-2. It generates query (Q), key (K), and value (V) sets for input and applies the following self-Attention in equation 1:
This mechanism enables more effective training and addresses the issue of vanishing gradients when handling long sequences. Notably, GPT models offer numerous advantages over previous language models like RNN or LSTM, as they mitigate several limitations and allow for the training of larger models in an unsupervised manner [
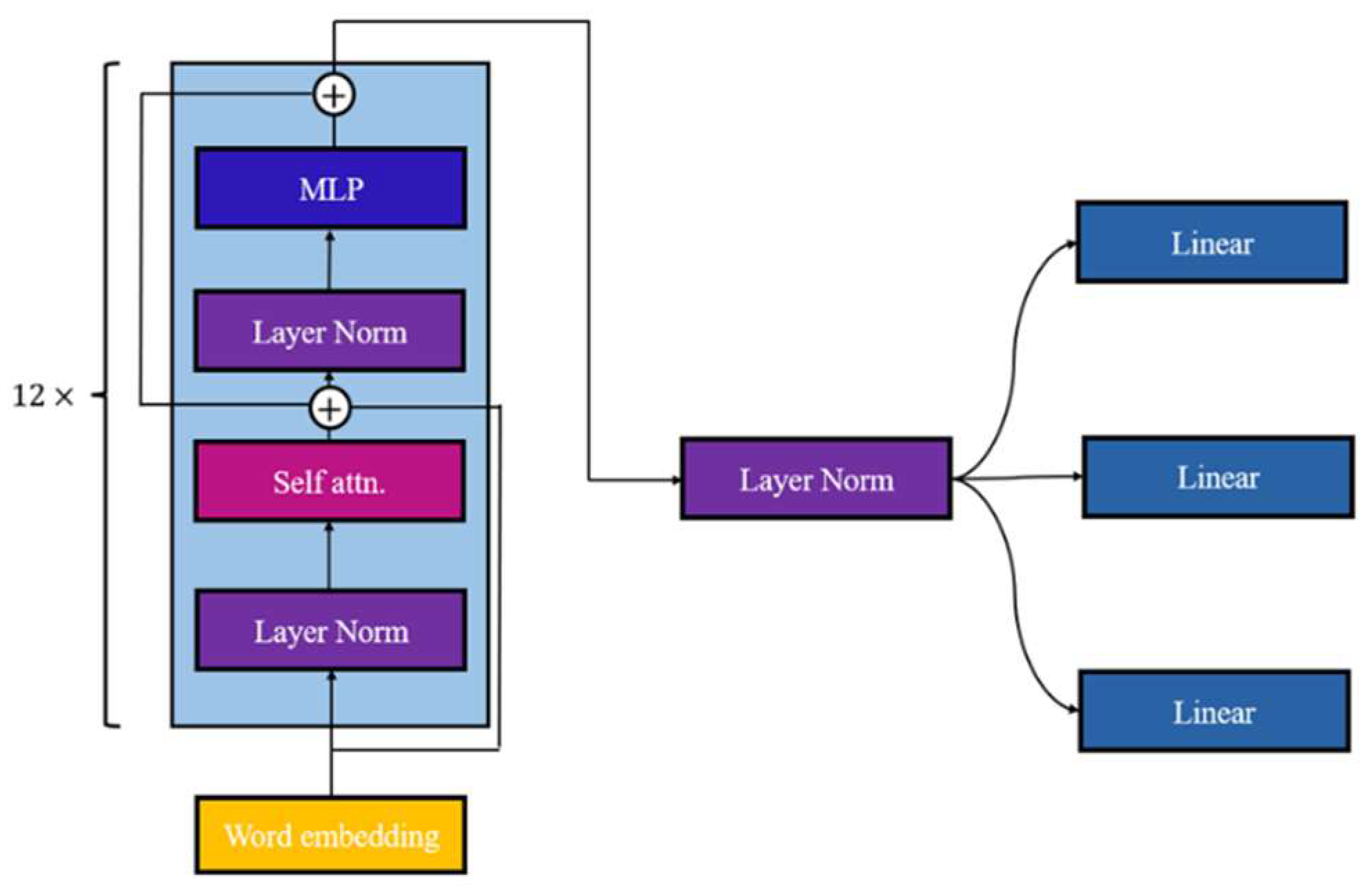
21]. In this study, we employ the GPT-2 model as the backbone and integrate three linear layers, shown in
Figure 1, to extract features from the GPT-2 encoder for the disease classification.
The complete GPT-2 model consists of 12 blocks incorporating self-attention, Multilayer Perception (MLP), and layer normalization [
21]. GPT-2 closely resembles the original GPT structure, with the exception of the placement of layer normalization and positioned before the self-attention and MLP layers [
16]. In our research, each linear layer specifically aims to classify one disease and conducts dimension reduction on the encoder's features, resulting in two channels representing the probability of whether the disease exists. This is our minor modification.
2.5. Training& evaluation
The initial weights of the model consist of 117M parameters pretrained on 40GB of text by OpenAI [
11]. We then conduct fine-tuning of the model using our dataset for 10 epochs. Each linear layer produces a two-channel output that is processed with the SoftMax activation function. We employ cross-entropy as our loss function to measure the disparity between the model's outputs and the ground truths based on the disease. The three cross-entropy values multiplied by 0.2, 0.4, and 0.4 respectively are summed as a joint loss function in equation 2,
The learning rate is set to 1e-5, and we utilize the Adam optimizer [
22].
For evaluation, we employ a confusion matrix to individually analyze the model's performance for each disease, aiming to validate its effectiveness in handling multi-label classification. Additionally, we utilize metrics such as accuracy, sensitivity, specificity, precision and F1-score shown in equations (3)-(7) to assess the model's discriminatory performance across various diseases.
The overall training and evaluation process is performed by 10-fold cross validation. By balancing the differences among the different groups, we can effectively evaluate the model's performance. Through this way, we ensure that the results are not influenced solely by the distribution of the data.
3. Results.
3.1. Screened dataset
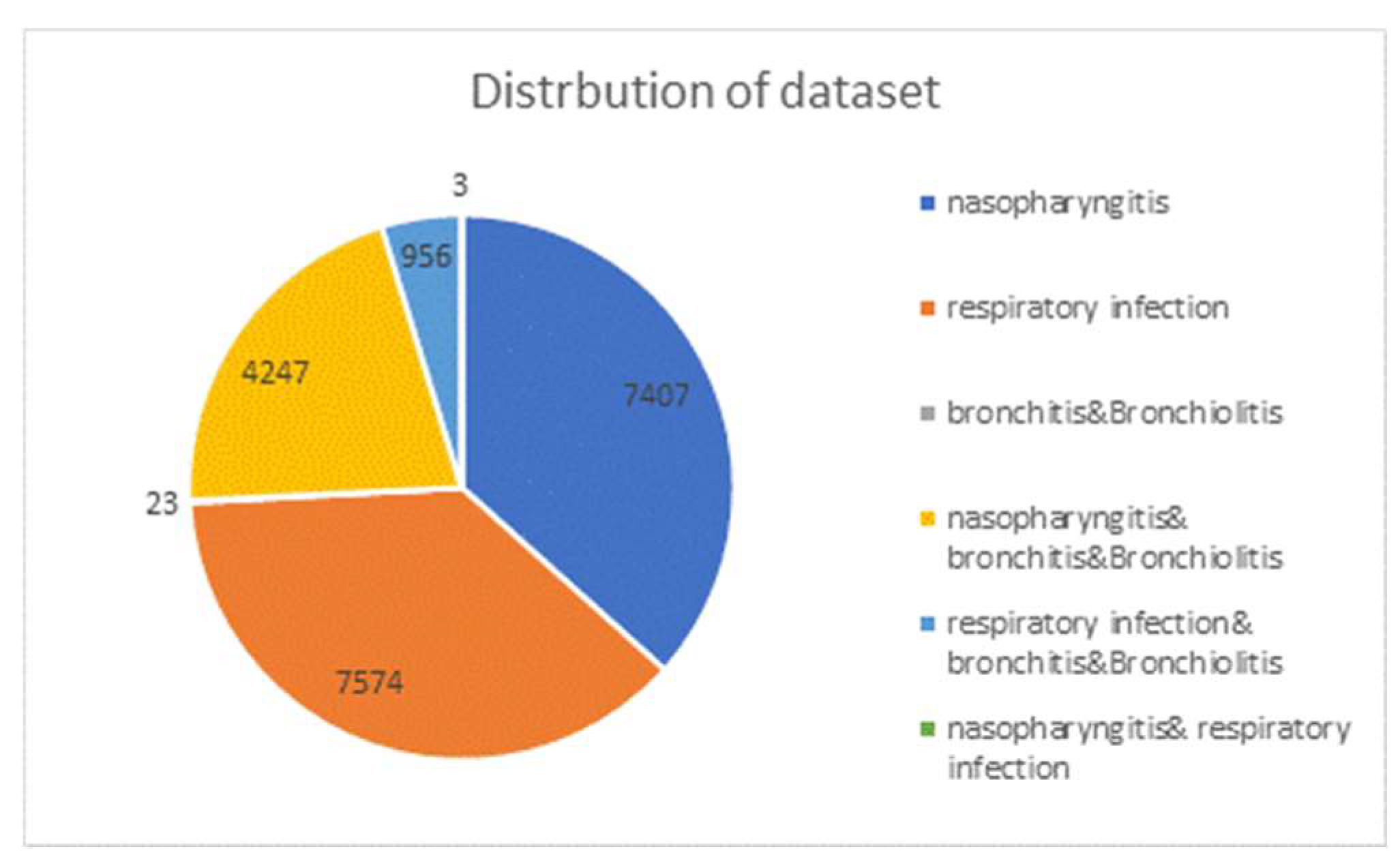
After the data purification described in section 2.1 and 2.2, a total of 20,210 records are collected and its distribution is shown in
Figure 2. There are 7,407 cases of nasal pharyngitis, 7,574 cases of upper respiratory tract infections, and only 23 cases of bronchitis and bronchiolitis in the single-diagnosis group. However, many patients are diagnosed not only with one case but with two cases. In the two-case diagnosis group, the first two majorities are nasal pharyngitis accompanied by bronchitis with 4,247 cases and upper respiratory tract infections accompanied by bronchitis with 956 cases, respectively. Only 3 cases are nasal pharyngeal cancer accompanied by respiratory tract infections. The data ratio for training, validation and test sets is set to be 7:2:1.
3.2. Training and validation
For each patient record, we extract the "symptoms" field from the text data, representing patient's current condition, as input for disease prediction.
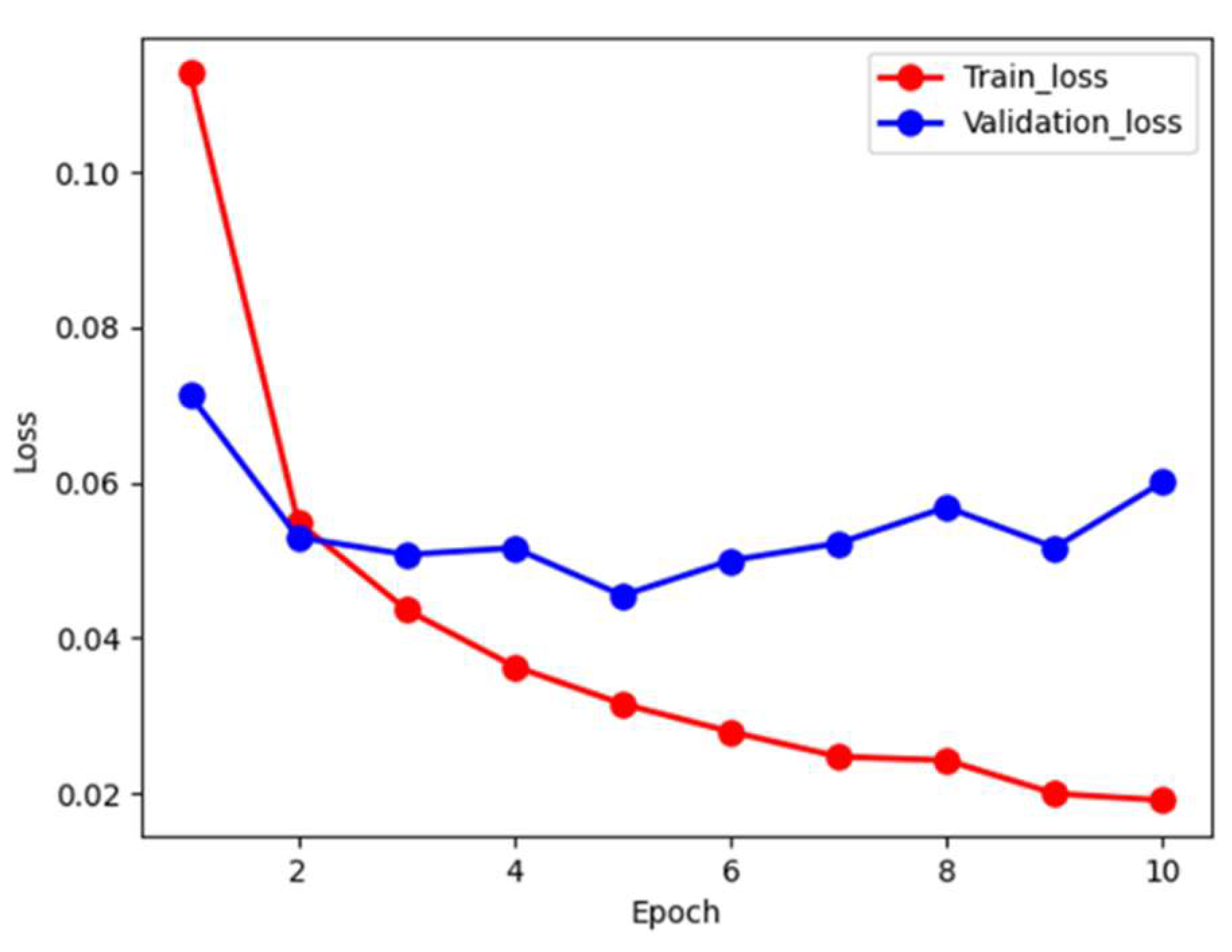
Figure 3 depicts the loss during the training and validation process. The red and blue curves denote the training and validation loss, respectively. We note that the training loss monotonically decreses to approximately 0.02 in 10 epochs. However, the validation loss shows oscillation in a small range. In order to prevent over-fiiting, we stop the training at 10 epochs to allow error tolerance. We futher examine accuracy during training phase, as shown in
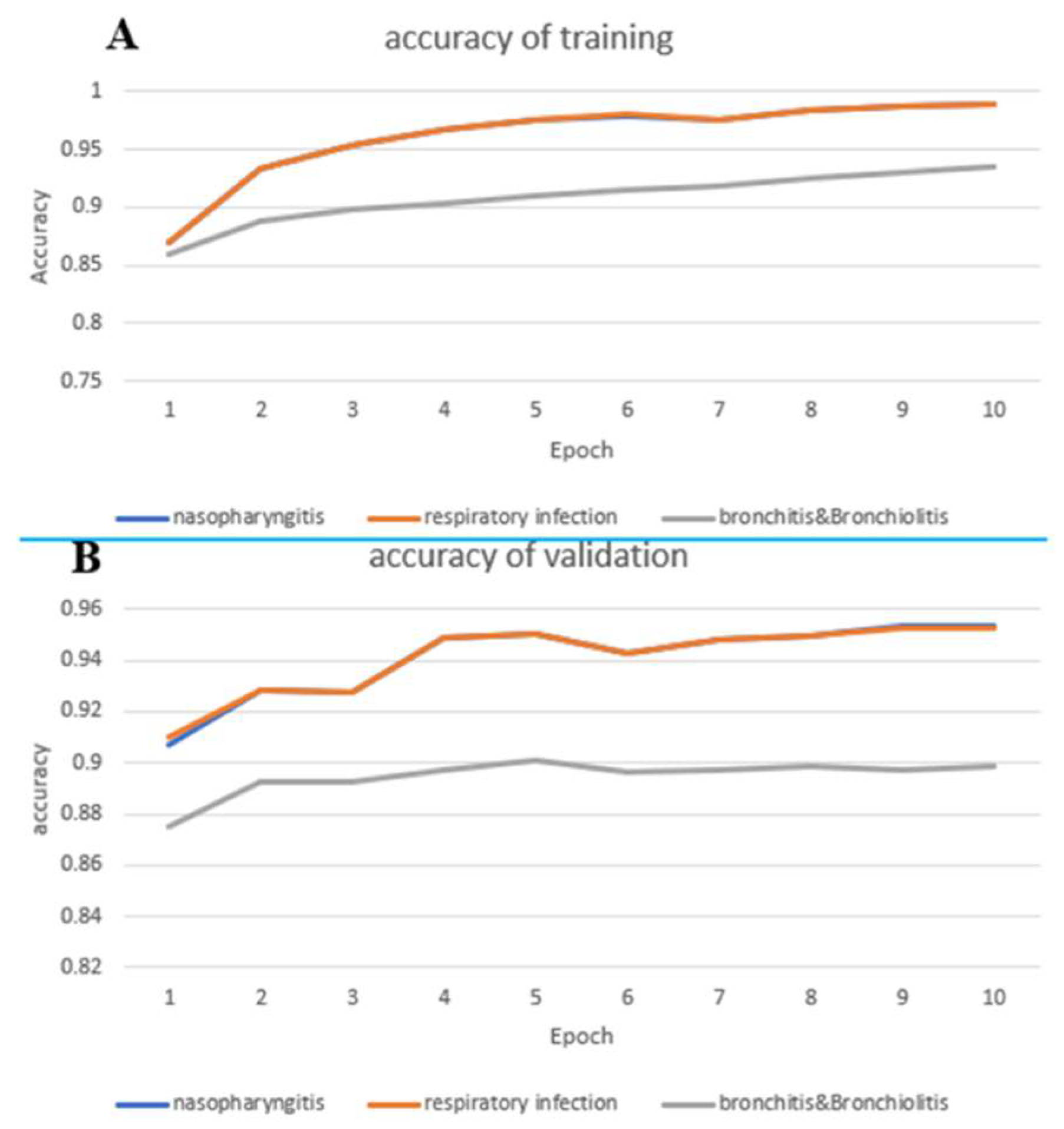
Figure 4. In
Figure 4(A), The prediction accuracy curve for nasal pharyngitis steadily increases from an initial 0.87 to 0.98 with respect to epochs, which is almost overlapped to the prediction accuracy curve for upper respiratory tract infections. However, bronchitis and bronchiolitis showed only 0.93 in accuracy.
Figure 4(B) demonstrates the predication accuracy curves in validation data, which shows lower than the ones in training data. However, the trend keeps as in the training data.
3.3. Evalution
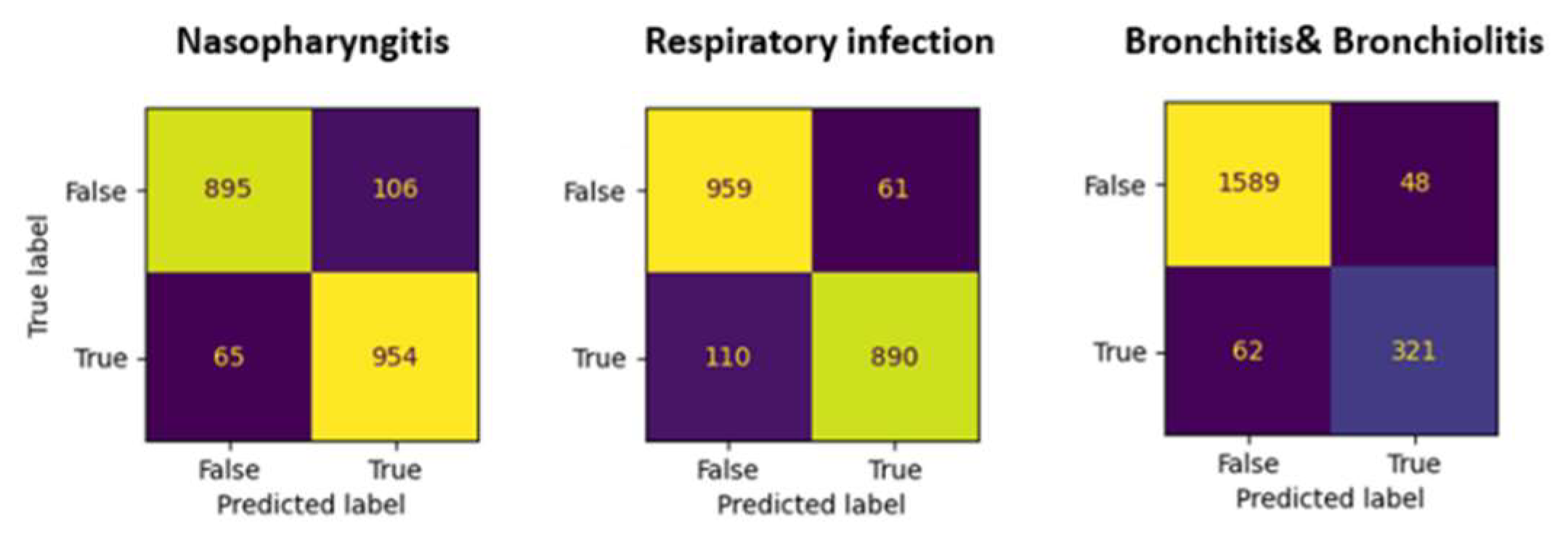
To evaluate model’s performance we use test set, the prediction accuracies are 0.93, 0.93, and 0.89 for nasopharyngitis, respiratory infection, and bronchitis & bronchiolotis, respectively. The sensitivity, specificity, and precision for nasopharyngitis and respiratory infections are above 0.9. For bronchitis and bronchiolitis, the sensitivity is poor at only 0.79. The confusion matrices are shown in
Figure 5. Overall evaluation of performance is listed in
Table 1, they are the averages of ten-fold cross-validation, which are close to the reality.
4. Discussion
This study explores the feasibility using only few symptom words and neural network language model (GPT-2) to predict three upper respiratory tract diseases, which are the most common patients seeking help from family physicians. Computer-aided diagnosis assists outpatients in rapidly identifying common upper respiratory tract diseases, which might offer information for outpatients to seek further medical help. This could be a first step of remote diagnosis on upper respiratory tract problem.
Previous studies[14, 23] have focused on building annotation systems for electronic medical or health records to extract data such as symptoms, treatments, and test results. Those data are used for causal inference or defining standard thresholds for subsequent researches. However, those applications cannot be directly accessible to the general public. Unlike physicians, the general public has less medical knowledge on normal ranges of test results but rather focuses on identifying the specific disease they may be suffering from. In this study, we use symptoms recorded by physicians as input for a language model to infer the disease the outpatient is suffering from. The trained model acts as an artificial diagnostician with extensive experiences, helping outpatients understand their conditions.
Comparing to the handling of unstructured text data, this study takes a different approach from previous research, which extracted various categories of information from the text. Instead, the model directly extracts sequential features from unformatted data to predict diseases. This method reduces the effort required for subsequent data analysis in various categories and eliminates the need for building a guide decision model. Furthermore, the results of this study demonstrate that the model has sufficient capability to make reliable diagnoses from unstructured text data, indirectly suggesting that physicians can maintain their own writing styles since the data can be utilized by the language model without requiring adjustments.
In the dataset used in this study, outpatients diagnosed solely with bronchitis or bronchiolitis accounted for only 0.1% of the total, while the data for outpatients diagnosed with these diseases along with nasopharyngitis or upper respiratory tract infections increased to approximately 25% of the total. This data distribution indicates that only a very small portion of patients are independently diagnosed without nasopharyngeal infections or upper respiratory tract infections. In other words, patients with bronchitis or bronchiolitis are usually diagnosed with multiple diseases. Additionally, we infer that physicians generally do not rely solely on symptoms to diagnose bronchitis or bronchiolitis in the absence of chest X-ray evidence.
Regarding the model's performance, we observe the effectiveness of the GPT-2 model in transfer learning. In terms of diagnostic accuracy shown in the first epoch of the training process, the accuracy rates for various diseases reach a good level as shown in
Figure 4. This result also implies that the model performs well in few-shot learning, greatly increasing the feasibility of expanding the range of diseases to be diagnosed. Especially in the situation when acquiring medical data is not trivial, after fine-tuning with a relatively small dataset, the transformed learning further improves the performance. In this study, particularly for the diagnosis of nasopharyngitis and upper respiratory tract infections, an accuracy rate of 93% is achieved, with sensitivity and specificity are both exceeding 90%. Even for diseases with a smaller amount of data, such as bronchitis or bronchiolitis, an accuracy rate of 0.89 is achieved.
Since 2020, the outbreak of COVID-19 has led to a rapid increase in the number of patients with upper respiratory symptoms, resulting in insufficient medical capacity. To prevent the outbreak of large-scale infections, the Taiwanese government conducted self-health management policies to request positive diagnosed outpatients to quarantining at home if their symptoms were not series [
24]. This study is motivated for the general public who has suffered on respiratory problems anywhere outside hospitals for the preliminary help. In the future, we will extend the language model developed in this study to more respiratory infections classification such as lung infections.
5. Conclusions
We adopt GPT-based language model applied on the unstructured medical text data to classify three common respiratory diseases. This method successfully differentiates different diseases from the symptoms recorded by physicians. The resultant performance suggests that this model has capabilities of dealing with complicated text data through NLP. Currently this is only a feasibility study, which is not mature for clinical usage.
Table 2.
Description of the accuracy of ten folds.
Table 2.
Description of the accuracy of ten folds.
| Fold |
Train |
Validation |
Test |
| 1 |
0.942944 |
0.925672 |
0.875578 |
| 2 |
0.943024 |
0.918951 |
0.925413 |
| 3 |
0.944989 |
0.924897 |
0.931683 |
| 4 |
0.946037 |
0.928888 |
0.918152 |
| 5 |
0.943203 |
0.925656 |
0.939439 |
| 6 |
0.936248 |
0.922093 |
0.930858 |
| 7 |
0.945066 |
0.928888 |
0.927063 |
| 8 |
0.945038 |
0.919932 |
0.915347 |
| 9 |
0.945832 |
0.930496 |
0.914851 |
| 10 |
0.945104 |
0.929614 |
0.925578 |
Author Contributions
Conceptualization, D.-C.C.; methodology, D.-C.C. and K.-H.L.; software, K.-H.L.; validation, K.-H.L.; formal analysis, K.-H.L.; investigation, K.-H.L.; resources, C.-H.T.; data curation, K.-H.L.; writing—original draft preparation, K.-H.L. and D.-C.C.; writing—review and editing, D.-C.C.; visualization, D.-C.C.; supervision, D.-C.C.; project administration, C.-H.T and D.-C.C.; funding acquisition, C.-H.T and D.-C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by TAINAN MUNICIPAL AN-NAN HOSPITAL-CHINA MEDICAL UNIVERSITY, grant number ANHRF111-05.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) of An Nan Hospital with the IRB number TMANH112-REC030.
Informed Consent Statement
Patients’ consent was waived by IRB due to this is a retrospective study, and all electronic medical records about the patients’ identification was anonymized.
Data Availability Statement
Not applicable.
Acknowledgments
We thank National Center for High-performance Computing (NCHC) for providing computational and storage resources in the TAIWAN COMPUTING CLOUD (TWCC).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heikkinen, T.; Järvinen, A. The common cold, (0140-6736 (Print)).
- Kassirer, J.P. Diagnostic reasoning, (0003-4819 (Print)).
- World Health, O. International statistical classification of diseases and related health problems, 10th revision, Fifth edition, 2016 ed.; World Health Organization: Geneva, 2015. [Google Scholar]
- Evans, R.S. Electronic Health Records: Then, Now, and in the Future, (2364-0502 (Electronic)).
- Kourou, K.; et al. Machine learning applications in cancer prognosis and prediction, (2001-0370 (Print)).
- Chan, H.P.; et al. Deep Learning in Medical Image Analysis, (0065-2598 (Print)).
- Mikolov, T.; et al. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems 2013, 26. [Google Scholar]
- Elman, J.L. Finding structure in time. Cognitive science 1990, 14, 179–211. [Google Scholar] [CrossRef]
- Mikolov, T.; et al. Recurrent neural network based language model. In Interspeech; Makuhari, 2010. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural computation 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Sak, H.; Senior, A.W.; Beaufays, F. Long short-term memory recurrent neural network architectures for large scale acoustic modeling. 2014. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. Advances in neural information processing systems 2014, 27. [Google Scholar]
- Bodenreider, O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Kormilitzin, A.; et al. Med7: A transferable clinical natural language processing model for electronic health records. Artif Intell Med 2021, 118, 102086. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, A.; Kondylakis, H.; Marakakis, E. MediExpert: An Expert System based on Differential Diagnosis focusing on Educational Purposes. EAI Endorsed Transactions on e-Learning 2020, 6. [Google Scholar] [CrossRef]
- Radford, A.; et al. Language models are unsupervised multitask learners. OpenAI blog 2019, 1, 9. [Google Scholar]
- Pritzker, S.E.; Hui, K.K. Introducing considerations in the translation of Chinese medicine. Journal of Integrative Medicine 2014, 12, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Grossman Liu, L.; et al. A deep database of medical abbreviations and acronyms for natural language processing. Scientific Data 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Sennrich, R.; Haddow, B.; Birch, A. Neural Machine Translation of Rare Words with Subword Units. arXiv 2016. [Google Scholar]
- Vaswani, A.; et al. Attention is all you need. Advances in neural information processing systems 2017, 30. [Google Scholar]
- Radford, A.; et al. Improving language understanding by generative pre-training. 2018. [Google Scholar]
- Diederik; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017. [Google Scholar]
- Percha, B. Modern Clinical Text Mining: A Guide and Review. Annu Rev Biomed Data Sci 2021, 4, 165–187. [Google Scholar] [CrossRef] [PubMed]
- Control, T.C.f.D. Self-Health management notice (Coronavirus disease 2019, COVID-19). 2020. Available online: https://www.cdc.gov.tw/File/Get/iNSs2KX3g4NbUwirtn80aQ.
- Umakanthan, S.A.-O.; et al. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19), (1469-0756 (Electronic)).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).