1. Introduction
Beta coronaviruses represent a significant group of viruses that can potentially cause severe respiratory diseases in humans. They are responsible for several noteworthy outbreaks, including Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and the ongoing Coronavirus Disease 2019 (COVID-19) pandemic [
1].
These viruses have a lipid envelope studded with spike (S) glycoproteins, pivotal for host-cell interaction. The key to their infectivity lies in their ability to interact with specific cellular receptors for entry. An essential characteristic of beta coronaviruses is their distinct propensity to interact with specific cellular receptors, highlighting their unique behaviors rather than uniformity [
2]. Indeed, among them, HCoV-OC43 and HCoV-HKU1 interact with 9-
O-Ac-Sia [
3,
4], whereas SARS and MERS viral strains preferentially target the ACE2 receptor [
5]. ACE2 has been identified as the primary receptor for both SARS-CoV and SARS-CoV-2 [
6]. The receptor-binding domain (RBD) of the spike protein of these viruses specifically recognizes ACE2, facilitating viral entry into host cells. This interaction has significant implications for understanding the pathogenesis and potential therapeutic interventions.
Among these interventions, an emerging strategy is investigating the interaction between SARS-CoV-2 and glycosaminoglycans (GAGs) [
3,
7,
8]. GAGs are an important family of biological molecules like dermatan sulfate, keratan sulfate, heparin (HP), and heparan sulfate (HS) made by an amino sugar and other sugars aggregated in a polymeric chain [
9]. Belonging to this class are Enoxaparin (EX), a molecular fragment extracted from heparin in the porcine intestinal mucosa [
10], and HS, the most abundant GAG in human body with a percentage of 50–90% [
11], are attached on the surface of endothelial cells as a portion of a biological structure called “glycocalyx” an umbrella term for the entirety of free glycans, glycoproteins, proteoglycans and glycolipids present on the cell surface [
12]. Glycocalyx achieves a significant number of functions, such as blastocyst implantation and embryonic development [
13], leukocyte adhesion [
14], and bacterial adhesion [
15], and it is also a key regulator for cellular homeostasis [
16,
17]. HS has recently been known to play an important role in the first step of viral infection, particularly in SARS-CoV-2 infections [
18].
Regarding the interaction among viruses, HS and EX are known to have a different grade of dependence between GAGs and the different virus families. On natural isolates, it has been demonstrated that there was a high dependence between GAGs and viruses like Herpes simplex virus [
19], Dengue virus [
20], Echovirus 5 [
21], Echovirus 6 [
22] and North American eastern equine encephalitis virus [
23]. Other experiments on laboratory strains were conducted and highlighted a correlation also between GAGs and Cytomegalovirus [
24], Pseudorabies virus [
25], Merkel cell polyomavirus [
26], Hepatitis B and Hepatitis Delta virus [
27,
28,
29], and so on.
Considering the persistent threat posed by evolving SARS-CoV-2 variants, the emergence of the Omicron variant and its sublineages, such as BA.2.86, has raised concerns about the efficacy of existing therapeutic strategies. This research aimed to scrutinize a model’s effectiveness in substantiating the antiviral activity of GAGs across different strains of SARS-CoV-2, including the notably diverse variant BA.2.86. The approach involved a synergistic integration of computational simulations with biological assays. Specifically, the in vitro assessment leveraged the cutting-edge BacMam technology, ensuring a comprehensive and reliable evaluation of GAGs’ potential antiviral properties against SARS-CoV-2.
2. Materials and Methods
2.1. Chemicals, Cellular Lines, and Viruses
Heparan sulfate (HS) and enoxaparin sodium (EX) were kindly supplied by Techdow Pharma S.r.l Assago Milanofiori (MI) Italy; A549 CCL-185™ (carcinoma lung epithelial cells) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). A549 cells were cultured in F12K medium supplemented with 2 mM L-glutamine, 100 U/mL penicillin–streptomycin mixture, and 10% fetal bovine serum (FBS), at 37 °C in a 5% CO2 humidified incubator.
Adherent subconfluent cell monolayers were prepared in growth medium (2% FBS) in 96-well plates for cytotoxicity assays and viral inhibition tests.
Pseudo SARS-CoV-2 Reporter was purchased from Montana Molecular and tested on A549 cells.
2.2. Cell Viability Assay by MTT
The evaluation of the cytotoxic effects of HS and EX on A549 cells was performed employing the MTT assay as previously reported [
3]. Briefly, cell viability was measured through the colorimetric reduction of MTT enzymatically catalyzed by the mitochondrial succinate dehydrogenase. The tetrazolium salts that enter the cells are transformed into violet-colored formazan crystals. The level of formazan is used as an indirect index of cell density. The A549 cells were incubated with different concentrations of HS and EX for 24 and 48 h in 5% CO
2 at 37 °C. Then, 100 μL of the tetrazolium salt solution (5 mg/mL) was added for each well and incubated at 37 °C in 5% CO
2 for 3 h. After this incubation, 100 μL of DMSO and the OD was measured at a wavelength of 570 nm with a spectrophotometer (BioTek Synergy HTX Reader). Three assays for each sample were performed, and the results were expressed as mean ±SD.
2.3. Transduction assay
A549 (1×10
4/well) were seeded on 96-well plates in complete F12K medium with 10% FBS and incubated overnight at 37 °C in a humidified 5% CO
2 atmosphere. Then, a transduction mix of ACE2, complete media, and sodium butyrate (
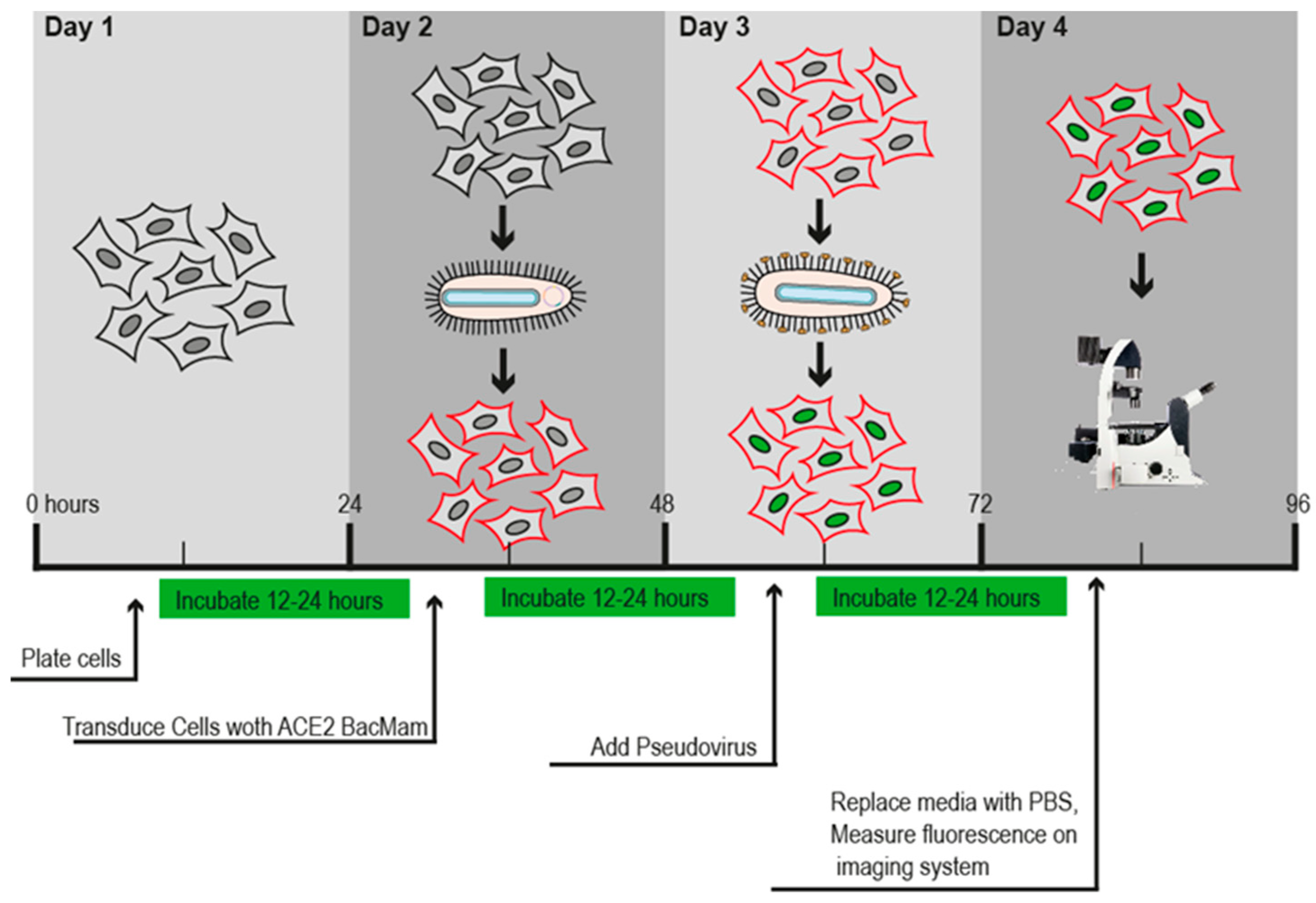
Table 1) was prepared and used as described by the manufacturer (Montana Molecular, Fluorescent Biosensors for Live Cell Discovery). Briefly, 50 μL were added into each well of the plate. The plate was shaken 5–10 times to ensure uniform transduction and incubated at 37 °C overnight. After 24–36 h of incubation, the transduction efficiency was evaluated by Leica DM IL LED (© 2023 Leica Microsystems). Finally, images were analyzed with LAS X Life Science Software.
2.4. Adsorption inhibition assay
A549-ACE2 cells (1×104/well) were seeded on 96-well plates in complete F12K medium with 10% FBS and incubated overnight at 37 °C in a humidified 5% CO2 atmosphere. The medium was then removed from the plate, and a fresh one containing HA and EX in concentrations equal to 5.0 mg/mL and 1.25 mg/mL, respectively, was added.
Cells were treated with the transduction mix described by the manufacturer, including pseudovirus, fresh complete media, and sodium butyrate (
Table 2).
Figure 1 shows the timeline described by the company’s protocol.
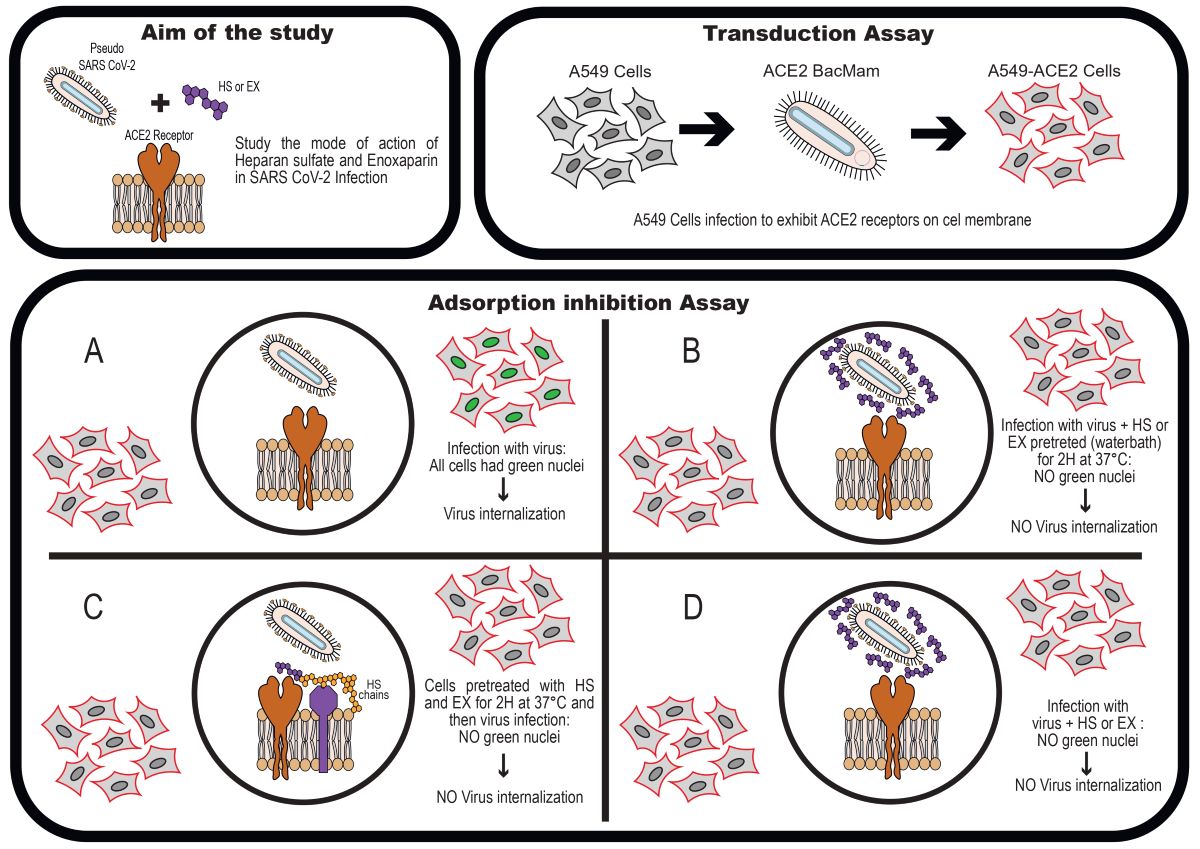
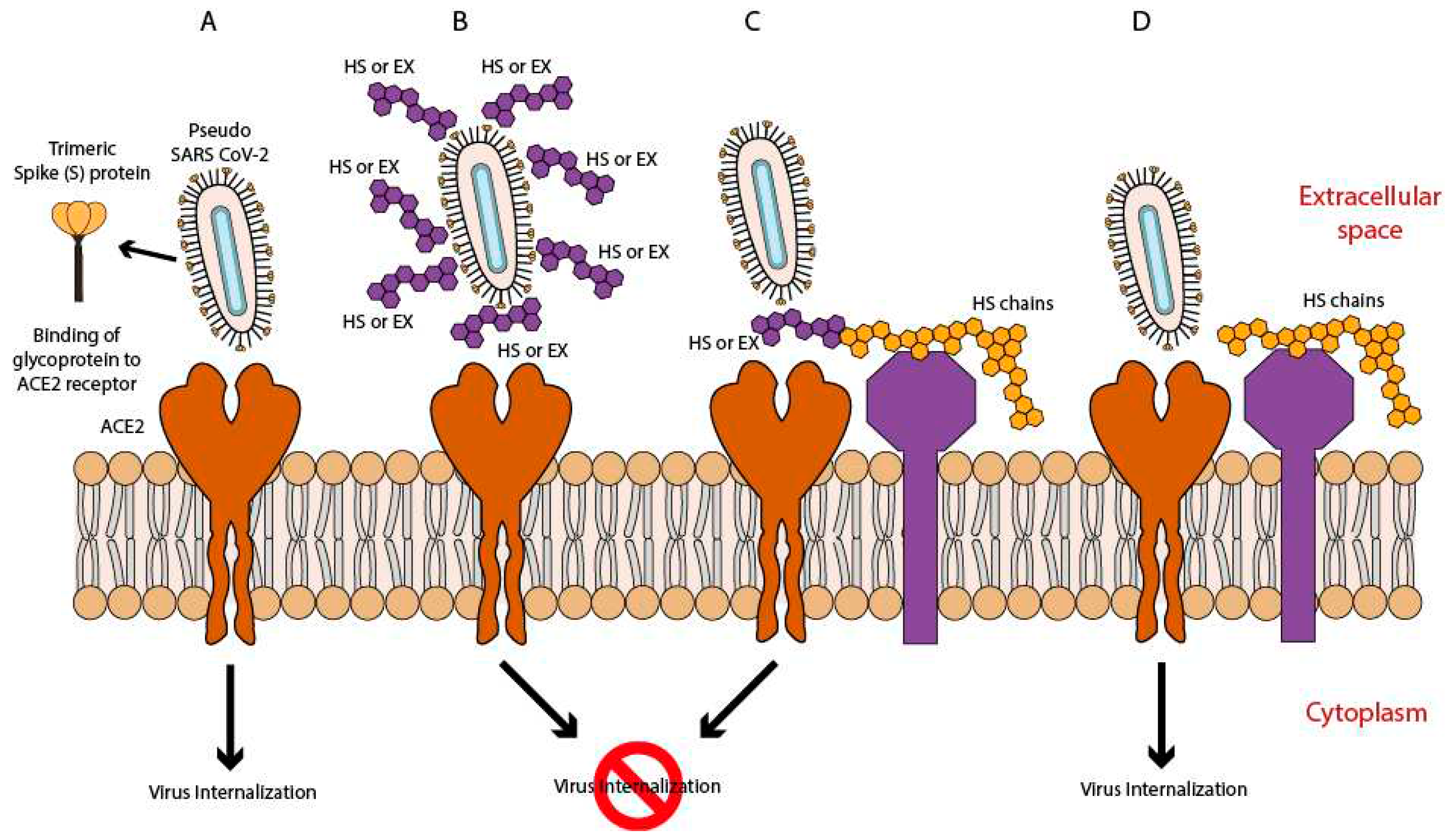
To study the antiviral activity of HS and EX, cells were divided into 3 treatment groups based on the presumptive mode of action of these 2 molecules (as shown in
Figure 2) and on the timing of dosing prior to viral adsorption in co-treatment (Group A), prior to viral adsorption in pre-treatment with the substance, HS or EX respectively (Group B), and pre-treatment of the virus with HS or EX for 2 h (Group C). For all groups, the cell monolayer was washed twice with warm PBS to ensure unbound viral particles were removed. Untreated A549 cells were used as the negative control. All procedures were performed in quadruplicate.
The plate was shaken 5–10 times to ensure uniform transduction across each well. Then, it was incubated at 37 °C in a humidified 5% CO2 atmosphere.
After 12–24 h of incubation, the adsorption inhibition efficiency was evaluated by using a Leica DM IL LED (© 2023 Leica Microsystems). Finally, images were analyzed with LAS X Life Science Software.
2.5. Statistical analyses
All experiments were performed thrice, and data were summarized using the mean (±SD). Where applicable, data were analyzed by one-way ANOVA with correction for multiple comparisons by Bonferroni. Graphs were generated using GraphPad® Prism ver. 8.4.2.0 (GraphPad Software, San Diego, CA, USA) A p-value ≤0.05 was considered significant.
2.6. Molecular modeling
Marvin Sketch was used to create the two molecules’ 2D chemical structures, which were then all subjected to molecular mechanics energy minimization using the MMFF94 force field available in the same software (PubChem CIDs: 70678539 and 60196282) [
30]. The PM3 Hamiltonian was then used to optimize the 3D geometry of all compounds [
31,
32], assuming a pH of 7.0 [
33,
34]. We tested different versions of the heparan sulfate, as reported in the literature [
35]. Working with various variants produced no discernible differences. Since the side chains point outside the binding cavities and the main interactions are accomplished by the same portion of the molecule, they are irrelevant for interactions between proteins and heparan. Molecular docking experiments were achieved with AutoDock 4.2.6 and AutoDock Vina provided in YASARA (v. 22.5.22, YASARA Biosciences GmbH, Vienna, Austria) [
36,
37] using the crystal structures of SARS-CoV-2 spike receptor-binding domain bound with ACE2 (PDB ID: 6M0J) collected from the Protein Data Bank (PDB,
http://www.rcsb.org/pdb, accessed on 05 September 2023) and an already validated protocol [
38,
39,
40,
41,
42,
43]. The structure of the spike of BA.2.86 was created by manual single point mutations of the original spike protein as reported on the European Centre for Disease Prevention and Control website (
https://www.ecdc.europa.eu/en/covid-19/variants-concern, accessed on 06 September 2023). The mutations were the following: I332V, D339H, R403K, V445H, G446S, N450D, L452W, N481K, 483del, E484K, F486P. The proteins have been optimized using YASARA software. The maps were made by AutoGrid (4.2.6) with an architecture of 0.375 Å and an extension encompassing all atoms spanning 5 Å from the exterior of the structure of the ligand. Point charges were originally defined according to the AMBER03 force field and then damped to mimic the less polar Gasteiger charges used to optimize the AutoDock scoring function. All parameters were used at their default settings. In the docking tab, the macromolecule and ligand were selected, and LGA parameters were set as ga_cauchy_beta = 1.0, ga_mutation_rate = 0.02, ga_runs = 100, ga_crossover_mode = two points, ga_cauchy_alpha = 0.0, ga_pop_size = 150, ga_num_generations = 27,000, ga_crossover_rate = 0.8, ga_num_evals = 25,000,000, ga_elitism = 1, number of generations for picking worst individual = 10.
3. Results and Discussion
3.1. Cell Viability
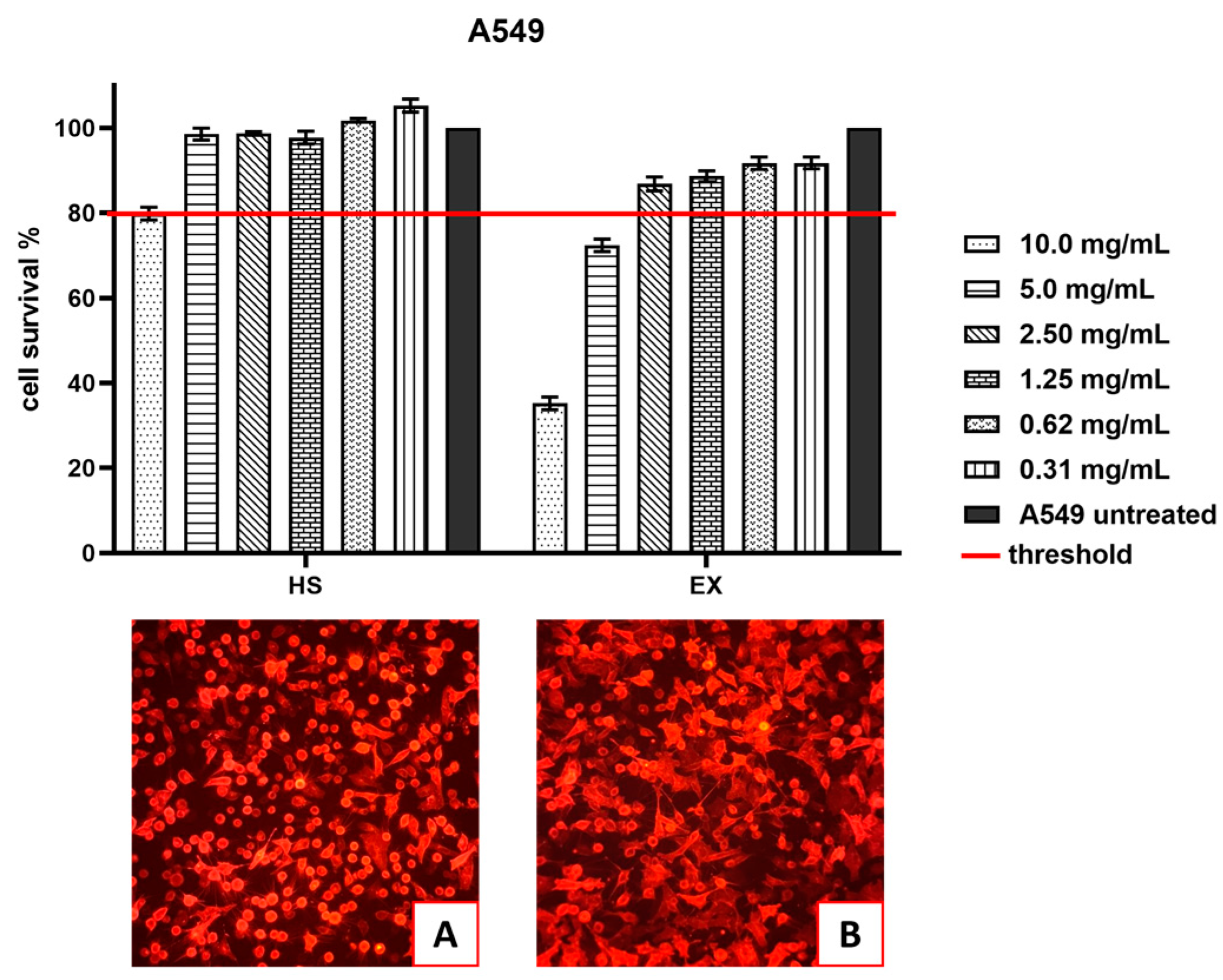
Cytotoxicity evaluation of HS and EX was assessed (tested range 10.0–0.31 mg/mL) by performing an MTT assay on the A549 cell line at 24 h. The results, shown in
Figure 3 demonstrated that HS solution guaranteed cell viability well beyond the chosen reference threshold (80%) at 5.0 mg/mL (98.55%). Instead, EX reduced cell viability at high concentrations, while the 2.5 mg/mL concentration was well tolerated (86.84%).
3.2. Pseudovirus SARS-CoV-2 affinity for ACE2 validation.
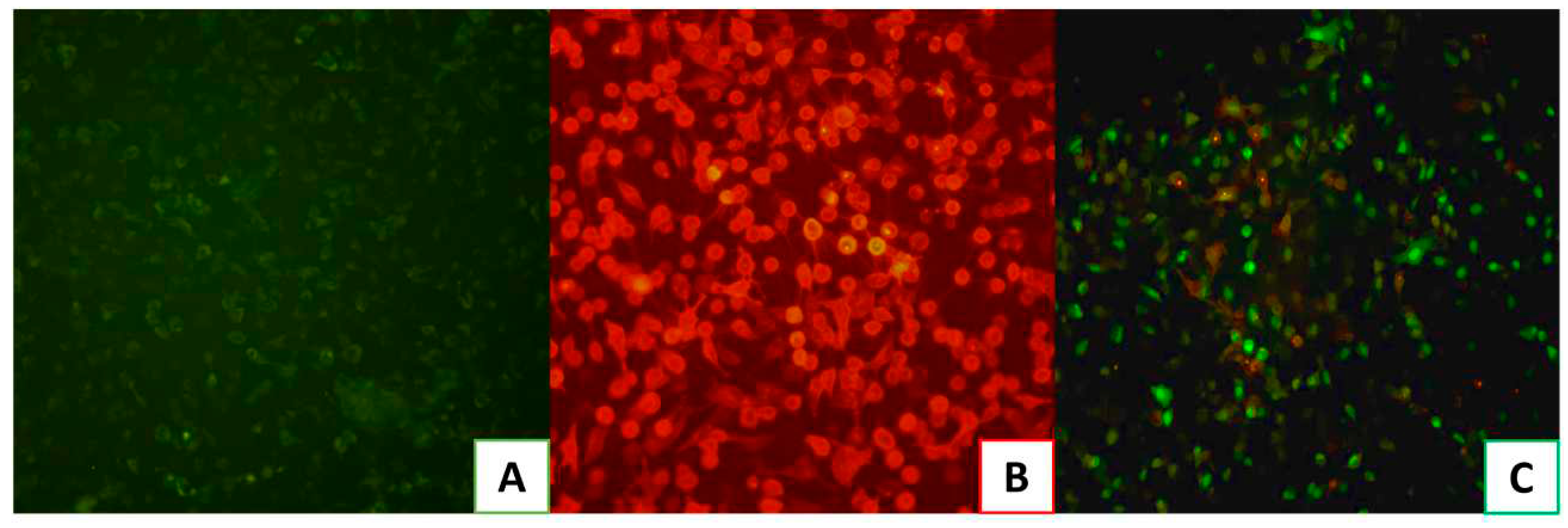
Before testing HS and EX against the pseudovirus, the efficacy of the assay has been confirmed. Indeed, we ensured that we could obtain a specific signal by comparing the transduction efficiency of pseudovirus-exposing SARS-CoV-2 spike in A549 cells with (
Figure 4B) or without ACE2 receptor expression (
Figure 4A). As shown in
Figure 4, the pseudovirus transduction efficiency was significantly higher in A549-ACE2 cells stably expressing ACE2 receptor (
Figure 4C) compared to normal A549 cells. Therefore, these results proved that Pseudo SARS-CoV-2 Reporter entry efficiency in A549 cells without endogenous ACE2 receptor expression was very low, validating its use for testing the molecules HS and EX.
3.3. Antiviral activity of HS and EX
As already said, to delineate whether HS and EX block SARS-CoV-2 viral entry, we used a safe facsimile of the virus that does not replicate in human cells and can be used in pseudo-host cells that express ACE2. Pseudo SARS-CoV-2 Reporter is a baculovirus decorated with the spike protein just like the actual SARS-CoV-2 virus, and it can used in our biosafety Level 2 facility. HS and EX were tested in a SARS-CoV-2 pseudovirus entry assay, dividing A549-ACE2 cells into three groups.
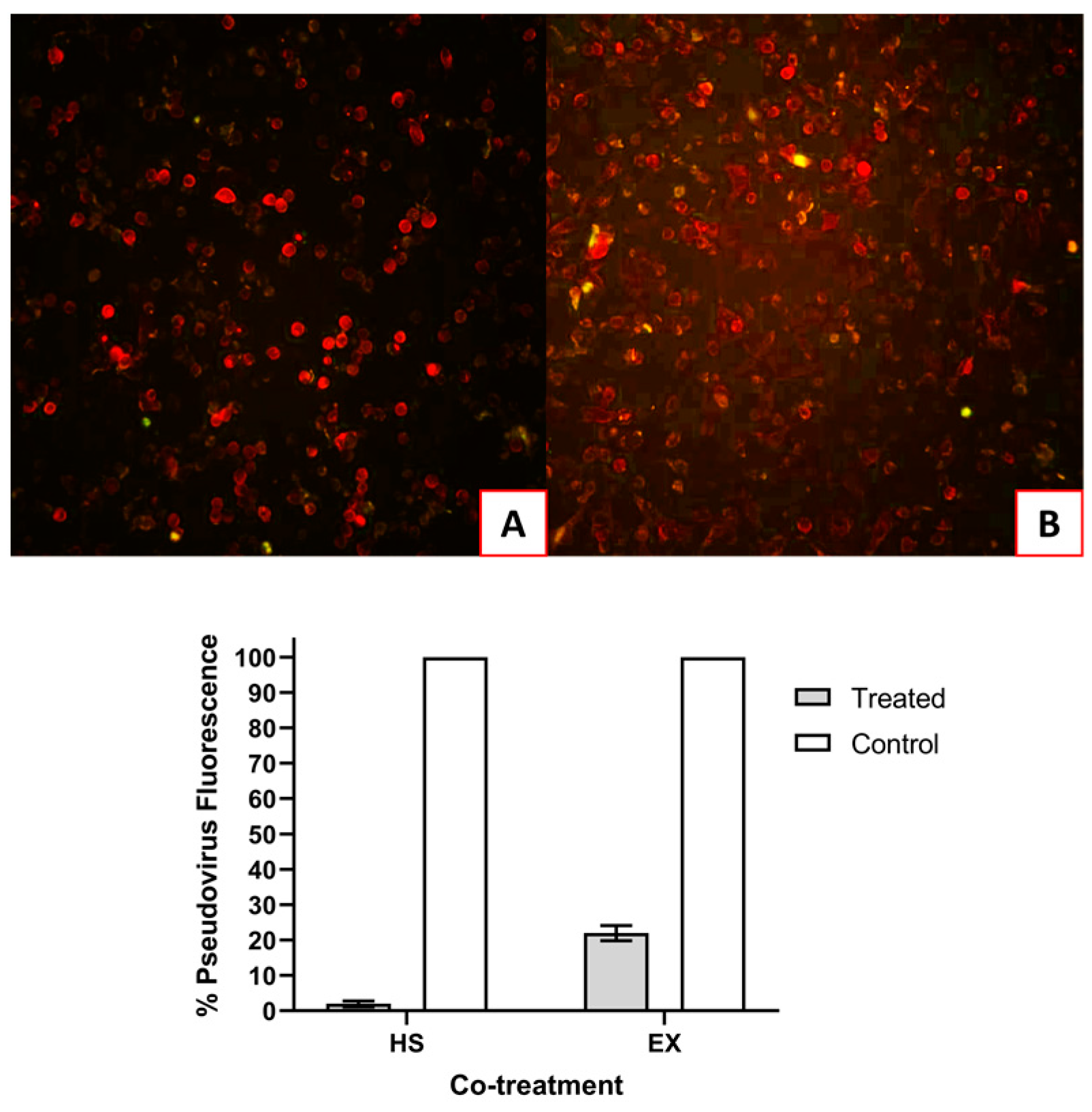
The first group of A549-ACE2 cells were simultaneously infected with SARS-CoV-2 pseudovirus and HS (5.0 mg/mL) or EX (1.25 mg/mL). As shown in
Figure 5, the substances were able to inhibit the absorption of the virus. In
Figure 5B, very few infected cells can be seen in co-treatment with EX, but essentially not significant compared to virus control (22%).
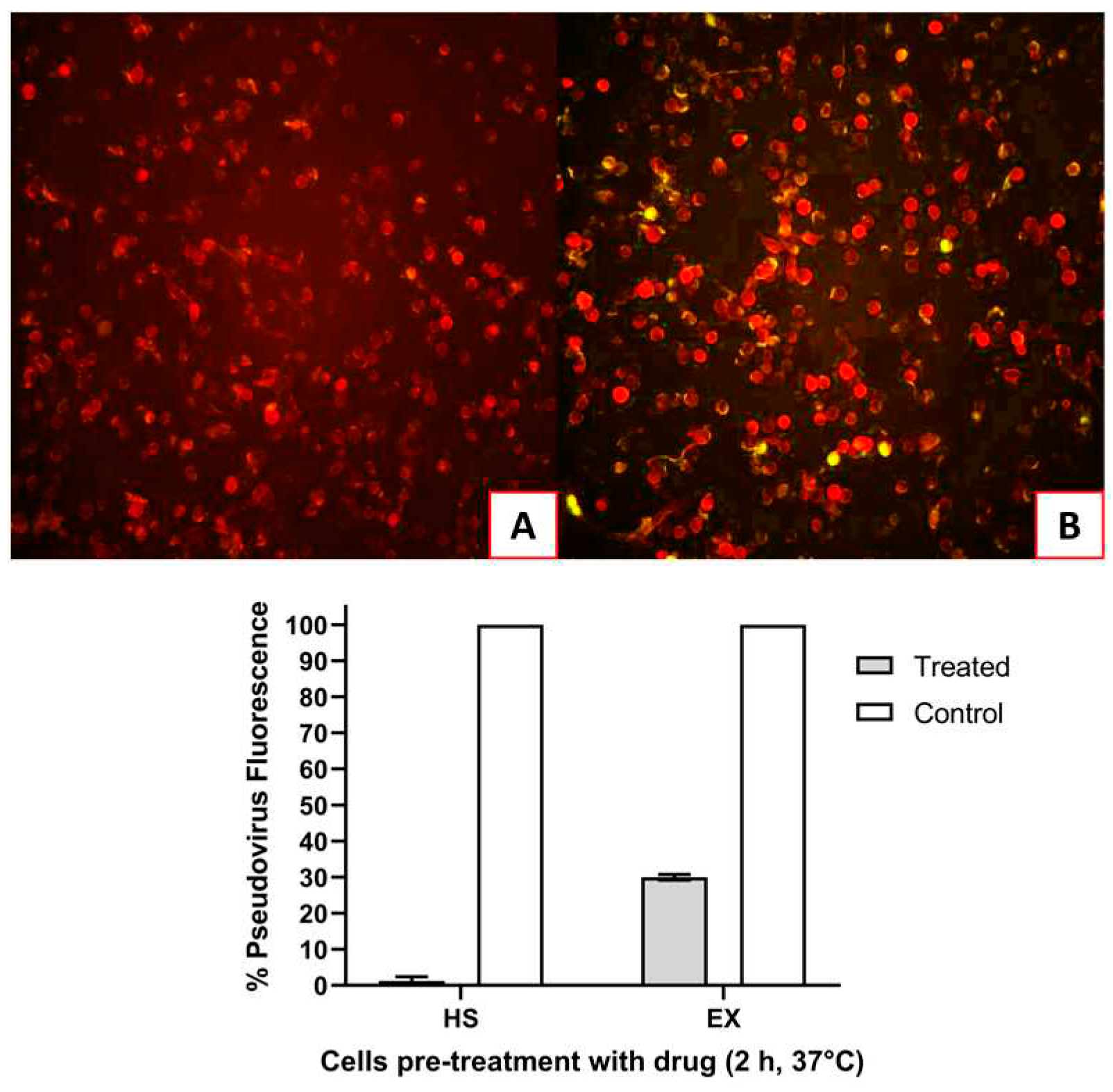
The second group of A549-ACE2 cells were exposed to HS or EX respectively for 2 h and then infected with SARS-CoV-2 Reporter. As shown in
Figure 6, the pre-treatment of the cells with the drugs caused the virus to be unable to enter. Here, as in co-treatment, the antiviral activity was intense: HS caused complete inhibition of absorption (
Figure 6A), while EX decreased it by 70% (
Figure 6B).
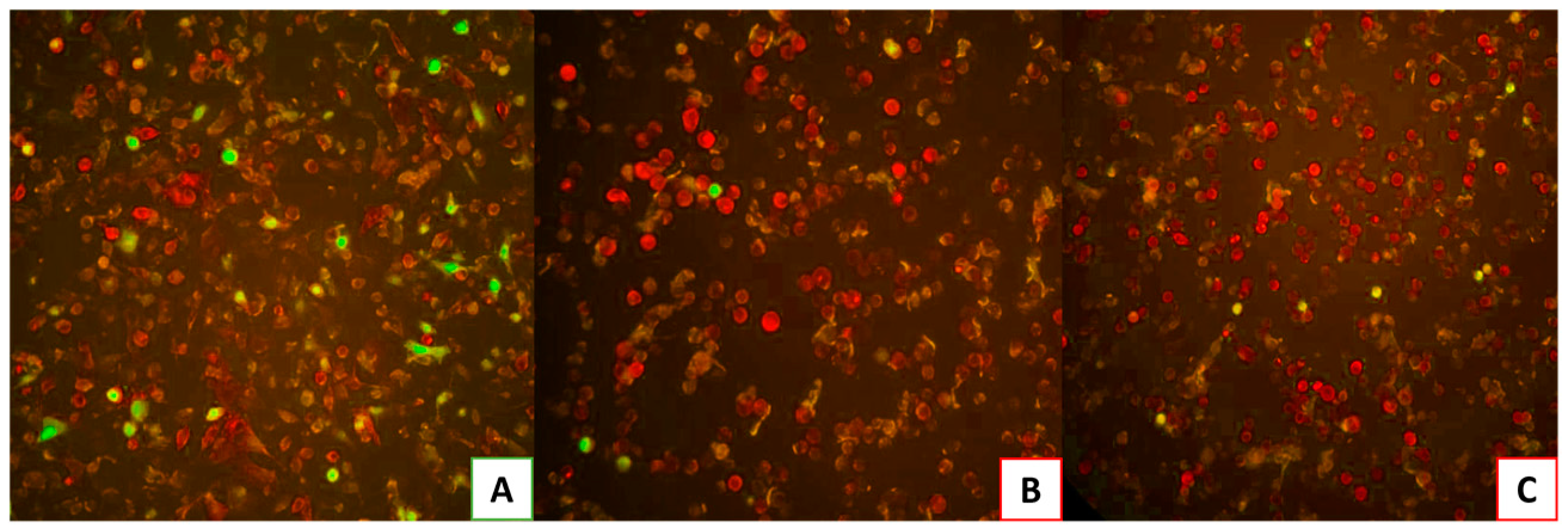
Finally, the virus was added to the last group of cells pre-incubated with HS or EX for 2 hours. As depicted in
Figure 7A, A549-ACE2 cells infected with the virus, which had been pre-treated at a warm temperature of 37 °C for 2 hours, demonstrated the virus’s persistence and survival under these conditions. On the contrary,
Figure 7B,C exhibited no viral infection, indicating that the pre-treatment of substances, respectively HS and EX, with the virus resulted in the virus’s inability to bind to the cellular receptor and enter the cell.
Therefore, the results showed that the examined GAGs exhibited antiviral activity across all performed treatments.
3.4. Molecular modeling
Molecular modeling experiments were performed to confirm the interactions of the two studied molecules at the interface between the spike protein. We initially studied the interaction with the spike protein of the wild type of SARS-CoV-2, and then we performed the same studies with the highly mutated and concerning for of the BA.2.86 variant.
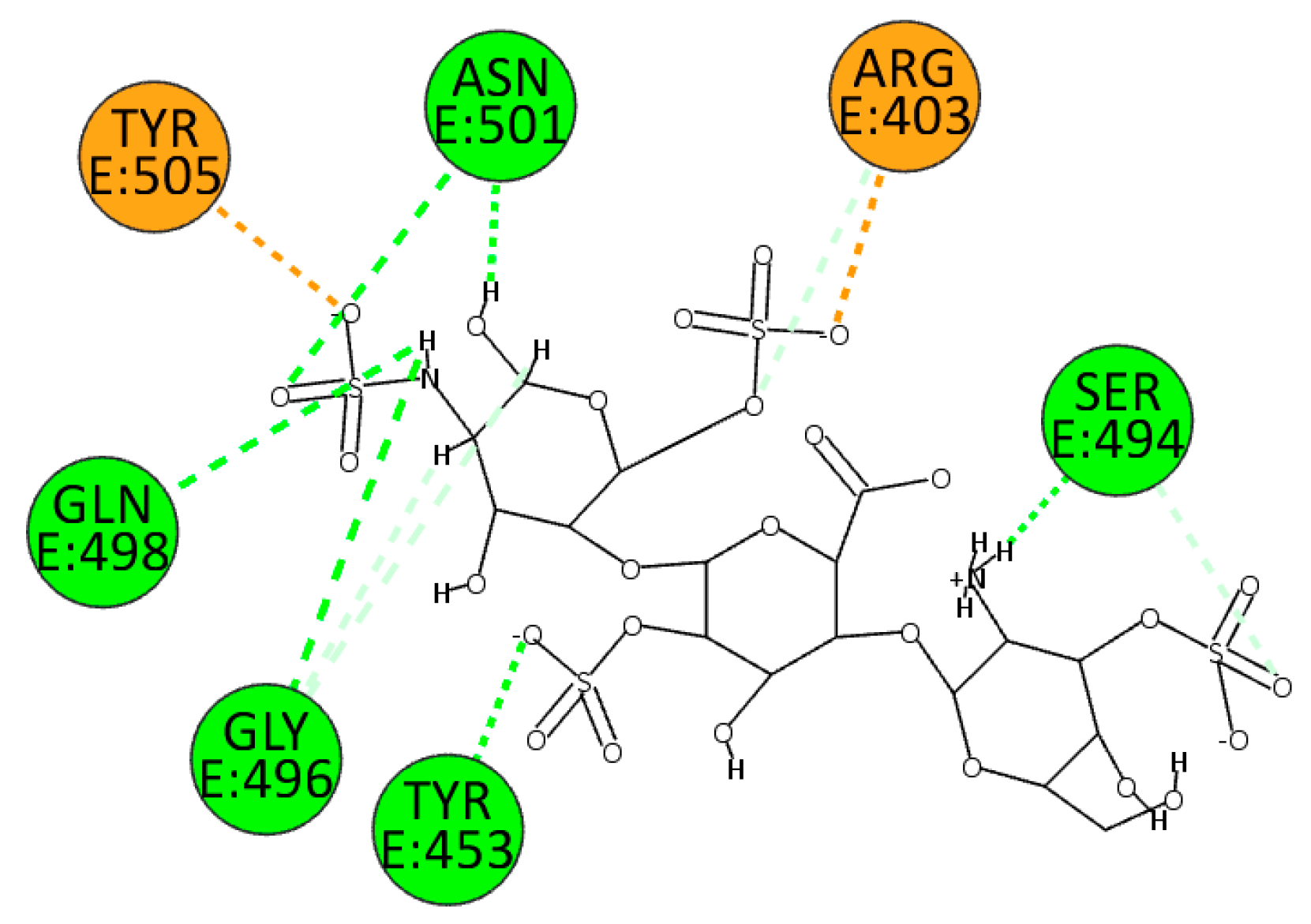
HS establishes many electrostatic interactions with the wild type of SARS-CoV-2 enzyme, in particular interacting with residues Arg403 and Tyr505. Due to the presence of several sulphate groups, it can interact with residues Tyr453, Ser494, Gly496, Gln498, and Asn501 (
Figure 8).
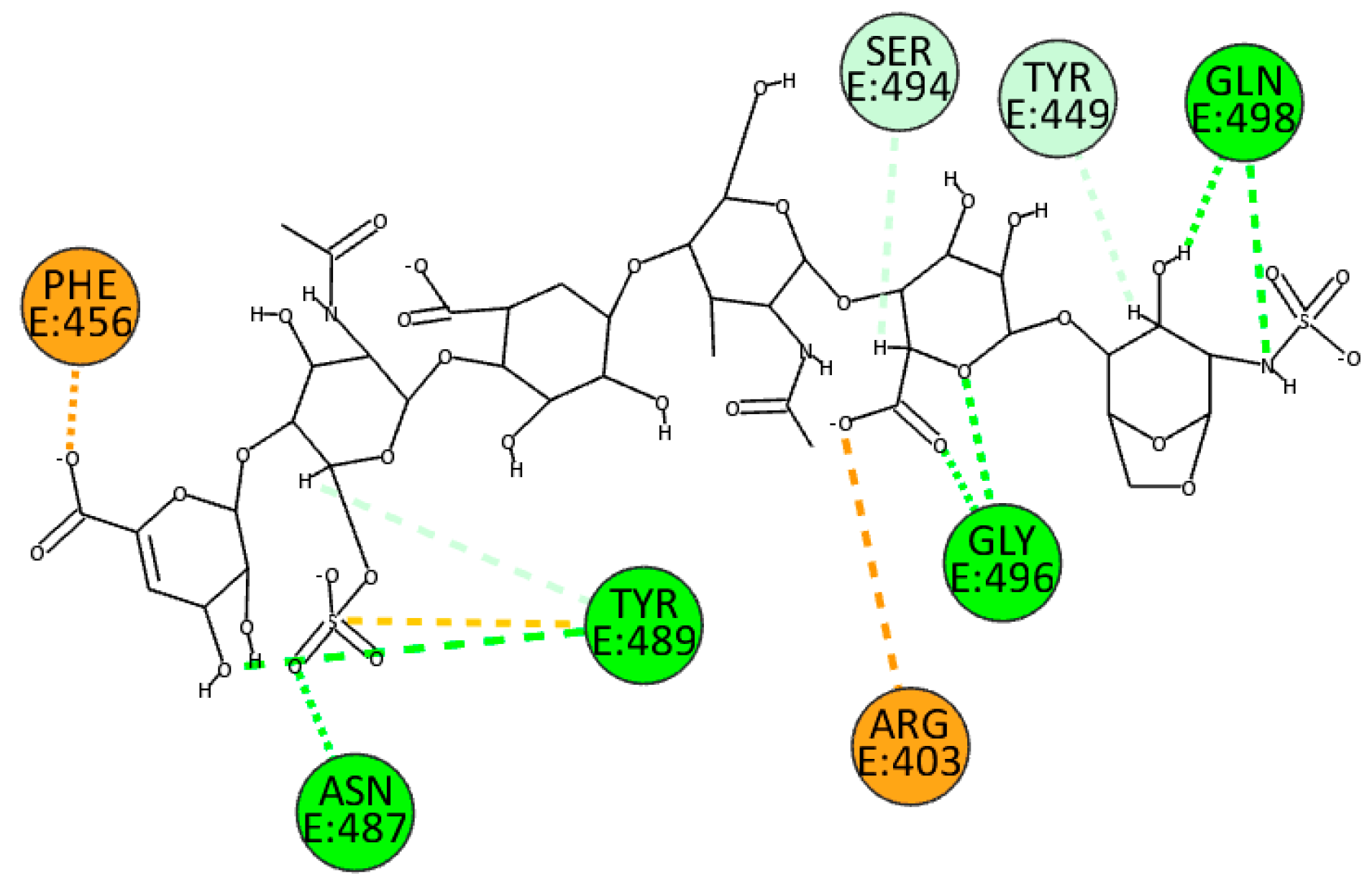
Otherwise, the molecule’s different sulfate and carboxyl groups influence EX’s binding position. In particular, the carboxylate ion allows the formation of a
π-anion and electrostatic interactions with Arg456 and Arg403. In addition, sulfate and hydroxyl groups form several hydrogen bonds, particularly with residues Asn487, Tyr489, Gly496, and Gln498. Finally, it is important to mention that unconventional hydrogen bonds are formed with residues Tyr449 and Ser494 (
Figure 9).
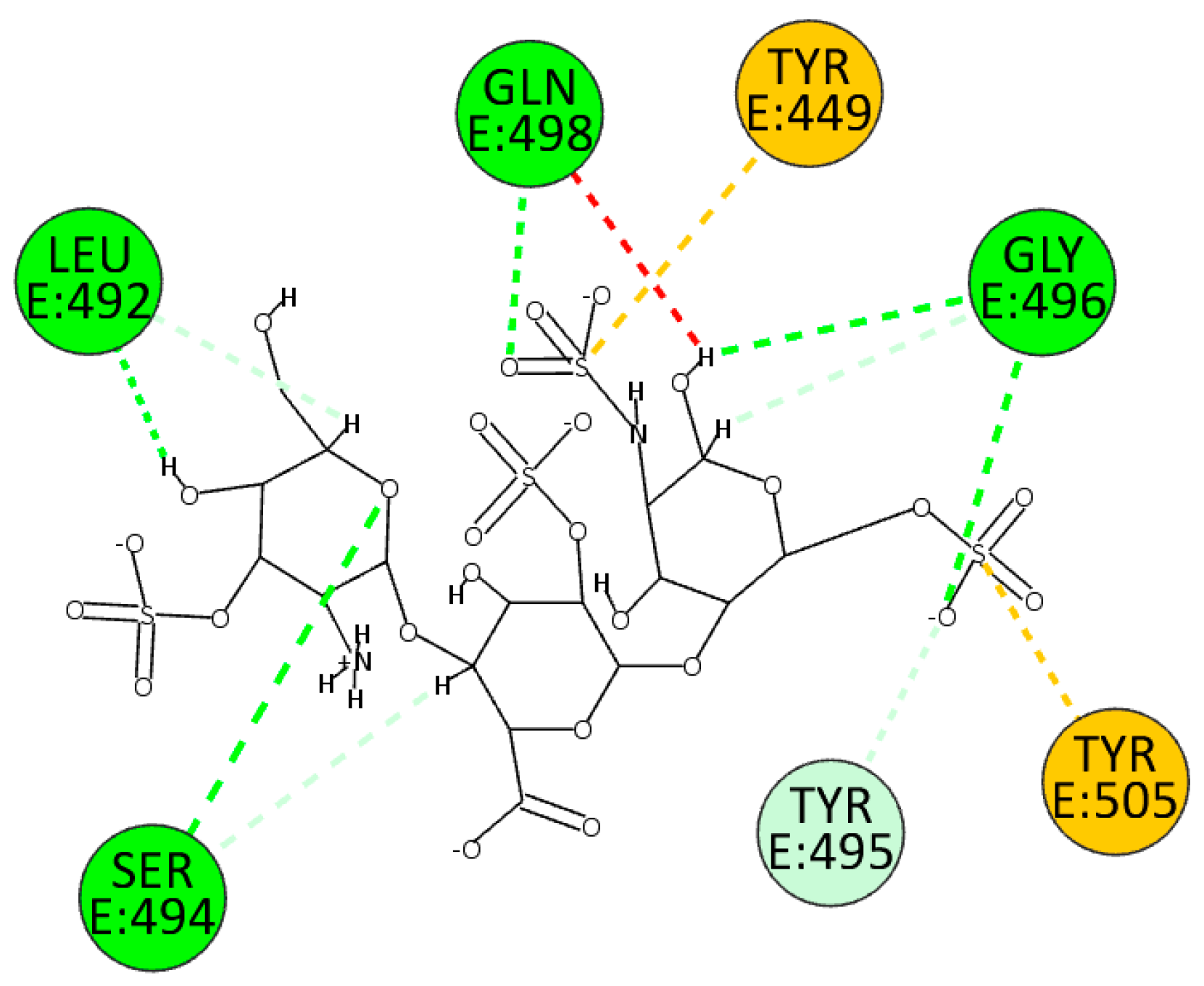
We also performed docking experiments in the enzyme pocket of the BA.2.86 variant. A good interaction between HS and the interface under investigation can also be seen here. HS forms hydrogen bonds with the enzyme due to sulfate and hydroxyl groups with residues Leu492, Ser494, Gly496, and Gln498. In addition, residues Tyr449 and 505 interact through electrostatic interactions, whereas it establishes an unconventional hydrogen bond with residue Tyr495 (
Figure 10).
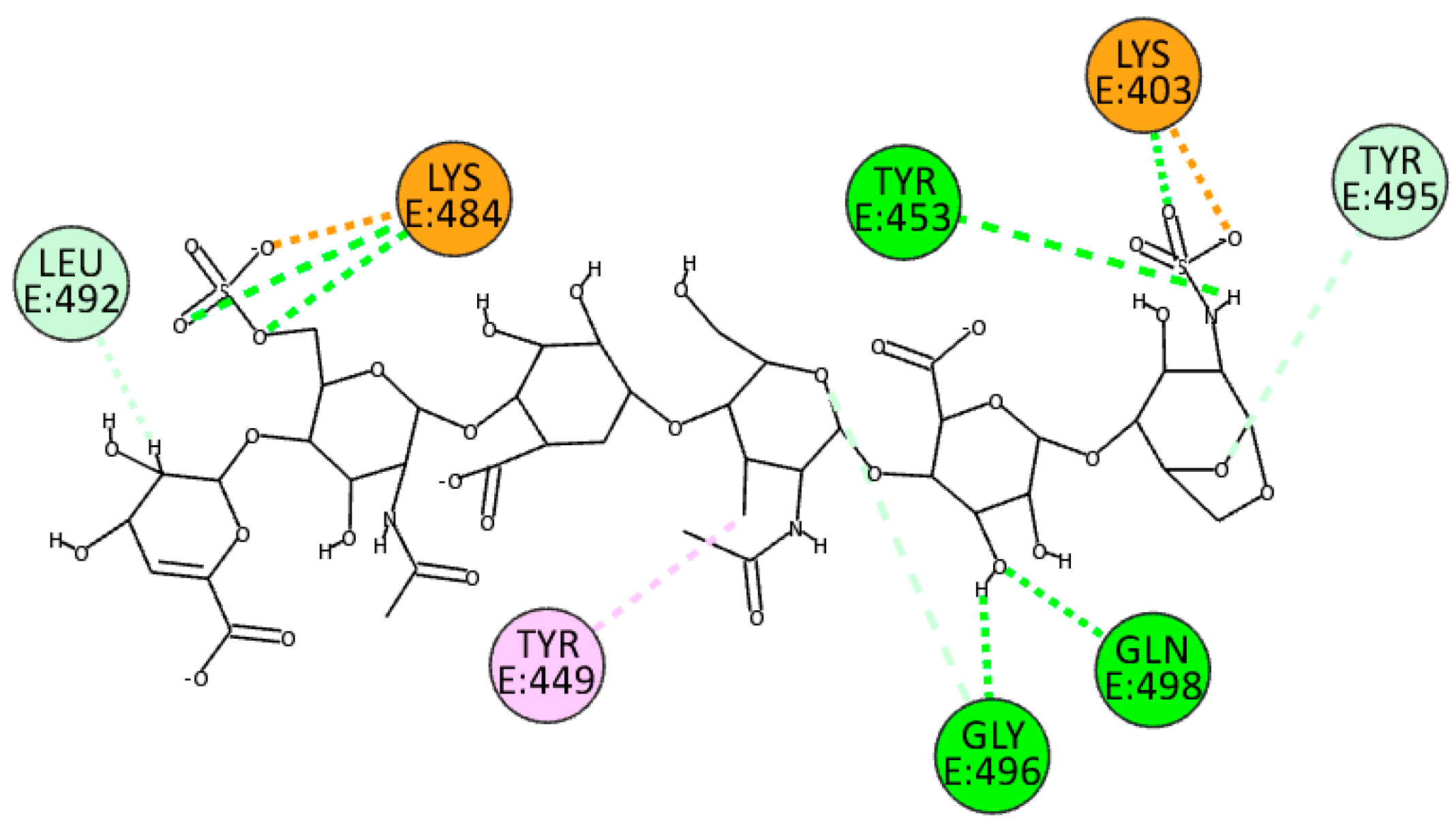
EX exploits sulfate groups to bind at the interface through electrostatic bonds with residues Lys403 and Lys484, whereas exploiting both hydroxyl groups and nitrogen-bonded protons, it establishes hydrogen bonds with residues Tyr453, Gly496, and Gln498. It also forms hydrogen and unconventional bonds with residues Leu492 and Tyr495 while establishing a
π-alkyl interaction with Tyr449 (
Figure 11).
Table 3 summarizes the main interactions between HS and EX with the wild and the BA.2.86 variant interface.
In all four experiments, residues Gly496 and Gln498 consistently establish hydrogen bonds with the ligands, suggesting they are preferred anchorage sites for both ligands.
4. Conclusions
This study has provided valuable insights into potential therapeutic strategies against SARS-CoV-2. It highlights the significance of studying GAGs in the context of viral entry and infection, including for possible future variants of this virus.
Despite the current plateau in SARS-CoV-2 viral infections, this strain has led to significant fatalities and economic losses [
44,
45]. It remains crucial to thoroughly understand this variant to remain vigilant in the event of new mutations, particularly with the approaching winter season, which may bring about a new wave of infections. Moreover, recent studies have presented conflicting results regarding how SARS-CoV-2 enters cells, raising scientific uncertainties and questions [
46,
47].
In this context, our study delved into the roles of ACE2 and GAGs in the virus’s cellular entry. Initially, our work aimed to assess an in vitro model that would allow for precise control over the expression of the ACE2 receptor. To this end, we employed the A549 cell line, which does not naturally express ACE2, and engineered it using BacMam technology. Furthermore, the ability of HS and EX to inhibit virus entry was assessed through three different treatments. Specifically, the virus’s behavior was examined in co-treatment directly on the cells, pre-treatment of the cells with the substances, and pre-treatment of the virus with the substances. This was done to ascertain whether the action of the two GAGs occurred solely on the cell or if there was an interaction with the virus. Some of the interactions shown in
Table 3 were present in both isoforms (6M0J and BA.2.86) despite the mutations such as Tyr505, Ser494, Gly496, and Gln498 for HS. Differently, Gly496 and Gln498 were the residues in common for the interaction of the wild and mutated forms for EX. Therefore, despite the mutation of the spike protein, the calculated interactions and the calculated binding energies suggested that nonrelevant differences can be noted between the two isoforms, so HS and EX should interact very similarly with both proteins. However, in a previous study [
3], HS and EX did not inhibit the infection due to another
β-coronavirus, HCoV-OC43. This was attributed to the relatively lower free binding energies of HS and EX compared to the original ligand, pyranosidonic acid (−6.40 kcal/mol). This suggests that GAGs mode of action is strongly ligand-dependent and appears to be particularly active in the spike-ACE2 interaction as opposed to other interactions that do not involve ACE2.
These results have provided compelling evidence regarding the antiviral potential of HS and EX against SARS-CoV-2. Both substances effectively inhibit viral absorption, with EX demonstrating a slightly less pronounced effect than HS. Although the reduction of infected cells with EX was not as pronounced as that by HS, it still demonstrated a significant impact. Additionally, pre-treating cells with HS or EX before virus exposure resulted in a strong antiviral effect, preventing the virus from entering the cells. HS exhibited complete inhibition of absorption, while EX showed a substantial 70% reduction, further emphasizing their potential as antiviral agents. Finally, the results from the third group are fascinating, as cells exposed to the virus pre-incubated with HS or EX showed no signs of infection, indicating that these molecules may confer a degree of protection even before cellular exposure.
Although these results, consistent with other studies [
7,
48], have demonstrated that HS and EX can interfere with ACE2-mediated virus entry, their effectiveness warrants scrutiny. In fact, the model created aimed to assess whether the findings obtained on the wild type could also be validated for specific virus variants such as the latest BA.2.86
Emphasizing the importance of ongoing vigilance in understanding SARS-CoV-2, particularly in light of potential seasonal variations, is crucial. Recognizing the existing scientific uncertainties surrounding the virus’s entry mechanism is essential. Overall, the obtained results underscore the promising antiviral properties of the examined GAGs in various treatment scenarios.
Author Contributions
Conceptualization, P.M.F., F.D., V.F., G.F. and A.R.; methodology, C.Z., V.F., G.F., V.P., and S.F.; software, G.F., V.P., A.R., and S.F.; data curation, V.F., G.F, and V.P.; writing—original draft preparation, C.Z., V.F., G.F., V.P., P.M.F., and S.F.; writing—review and editing, P.M.F. and A.R.; visualization, V.F., S.F., G.F., and V.P.; supervision, P.M.F. and A.R.; project administration, P.M.F. and A.R.; funding acquisition, P.M.F. and F.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by EU funding within the NextGeneration EU-MUR PNRR Extended Partnership on Emerging Infectious Diseases (Project N. PE00000007, INF-ACT).
Acknowledgments
We gratefully acknowledge ChemAxon Ltd. for the free academic license of their software. We gratefully acknowledge Techdow Pharma Italy S.r.l. for a research grant.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ullah, H.; Ullah, A.; Gul, A.; Mousavi, T.; Khan, M. W. , Novel coronavirus 2019 (COVID-19) pandemic outbreak: A comprehensive review of the current literature. Vacunas 2021, 22, 106–113. [Google Scholar] [CrossRef]
- Hidalgo, P.; Valdés, M.; González, R.A. Molecular biology of coronaviruses: an overview of virus-host interactions and pathogenesis. 78, 58. [CrossRef]
- Fuochi, V.; Floresta, G.; Emma, R.; Patamia, V.; Caruso, M.; Zagni, C.; Ronchi, F.; Ronchi, C.; Drago, F.; Rescifina, A.; et al. Heparan Sulfate and Enoxaparin Interact at the Interface of the Spike Protein of HCoV-229E but Not with HCoV-OC43. Viruses 2023, 15, 663. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.-J.; Bosch, B.-J.; et al. Human coronaviruses OC43 and HKU1 bind to 9- O -acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. 2019, 116, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.; Gautam, R.K.; Chopra, H.; Dubey, A.K.; Singla, R.K.; A Rayan, R.; Kamal, M.A. COMPARATIVE HIGHLIGHTS ON MERS-COV, SARS-COV-1, SARS-COV-2, AND NEO-COV. 21, 1245. [Google Scholar] [CrossRef]
- Rawat, P.; Jemimah, S.; Ponnuswamy, P. K.; Gromiha, M. M. , Why are ACE2 binding coronavirus strains SARS-CoV/SARS-CoV-2 wild and NL63 mild? Proteins 2021, 89, 389–398. [Google Scholar] [CrossRef]
- Tandon, R.; Sharp, J. S.; Zhang, F.; Pomin, V. H.; Ashpole, N. M.; Mitra, D.; McCandless, M. G.; Jin, W.; Liu, H.; Sharma, P.; Linhardt, R. J. , Effective Inhibition of SARS-CoV-2 Entry by Heparin and Enoxaparin Derivatives. J. Virol. 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Patamia, V.; Fuochi, V.; Furneri, P. M.; Rescifina, A. , Natural Substances in the Fight of SARS-CoV-2: A Critical Evaluation Resulting from the Cross-Fertilization of Molecular Modeling Data with the Pharmacological Aspects. Current Medicinal Chemistry 2021, 28, 8333–8383. [Google Scholar] [CrossRef] [PubMed]
- Merry, C. L. R.; Lindahl, U.; Couchman, J.; Esko, J. D. , Proteoglycans and Sulfated Glycosaminoglycans. In Essentials of Glycobiology, Varki, A.; Cummings, R. D.; Esko, J. D.; Stanley, P.; Hart, G. W.; Aebi, M.; Mohnen, D.; Kinoshita, T.; Packer, N. H.; Prestegard, J. H.; Schnaar, R. L.; Seeberger, P. H., Eds. 2022; 217–232. [Google Scholar] [CrossRef]
- Gupta, R.; Ponnusamy, M.P. Analysis of sulfates on low molecular weight heparin using mass spectrometry: structural characterization of enoxaparin. Expert Rev. Proteom. 2018, 15, 503–513. [Google Scholar] [CrossRef]
- Florian, J. A.; Kosky, J. R.; Ainslie, K.; Pang, Z.; Dull, R. O.; Tarbell, J. M. , Heparan sulfate proteoglycan is a mechanosensor on endothelial cells. Circ Res 2003, 93, e136–142. [Google Scholar] [CrossRef]
- Möckl, L. The Emerging Role of the Mammalian Glycocalyx in Functional Membrane Organization and Immune System Regulation. Front. Cell Dev. Biol. 2020, 8, 253. [Google Scholar] [CrossRef]
- Constantinou, P. E.; Morgado, M.; Carson, D. D. , Transmembrane Mucin Expression and Function in Embryo Implantation and Placentation. Adv Anat Embryol Cell Biol 2015, 216, 51–68. [Google Scholar]
- Lipowsky, H.H. The Endothelial Glycocalyx as a Barrier to Leukocyte Adhesion and Its Mediation by Extracellular Proteases. Ann. Biomed. Eng. 2011, 40, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Chandrasekaran, V.; Stamer, I.; Thygesen, M.B.; Terfort, A.; Lindhorst, T.K. Switching of Bacterial Adhesion to a Glycosylated Surface by Reversible Reorientation of the Carbohydrate Ligand. Angew. Chem. Int. Ed. 2014, 53, 14583–14586. [Google Scholar] [CrossRef] [PubMed]
- Wadowski, P.P.; Jilma, B.; Kopp, C.W.; Ertl, S.; Gremmel, T.; Koppensteiner, R. Glycocalyx as Possible Limiting Factor in COVID-19. Front. Immunol. 2021, 12, 607306. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; van Zandvoort, M.A.M.J.; Oude Egbrink, M.G. The endothelial glycocalyx: composition, functions, and visualization. Pflug. Arch. 2007, 454, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Snyder, N.L.; Hartmann, L. Polymers Inspired by Heparin and Heparan Sulfate for Viral Targeting. Macromolecules 2022, 55, 7957–7973. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. A Novel Role for 3-O-Sulfated Heparan Sulfate in Herpes Simplex Virus 1 Entry. Cell 1999, 99, 13–22. [Google Scholar] [CrossRef]
- Artpradit, C.; Robinson, L.N.; Gavrilov, B.K.; Rurak, T.T.; Ruchirawat, M.; Sasisekharan, R. Recognition of heparan sulfate by clinical strains of dengue virus serotype 1 using recombinant subviral particles. Virus Res. 2013, 176, 69–77. [Google Scholar] [CrossRef]
- Israelsson, S.; Gullberg, M.; Jonsson, N.; Roivainen, M.; Edman, K.; Lindberg, A.M. Studies of Echovirus 5 interactions with the cell surface: Heparan sulfate mediates attachment to the host cell. Virus Res. 2010, 151, 170–176. [Google Scholar] [CrossRef]
- Goodfellow, I.G.; Sioofy, A.B.; Powell, R.M.; Evans, D.J. Echoviruses Bind Heparan Sulfate at the Cell Surface. J. Virol. 2001, 75, 4918–4921. [Google Scholar] [CrossRef]
- Gardner, C.L.; Ebel, G.D.; Ryman, K.D.; Klimstra, W.B. Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. 2011, 108, 16026–16031. [Google Scholar] [CrossRef]
- Compton, T.; Nowlin, D.M.; Cooper, N.R. Initiation of Human Cytomegalovirus Infection Requires Initial Interaction with Cell Surface Heparan Sulfate. Virology 1993, 193, 834–841. [Google Scholar] [CrossRef]
- Trybala, E.; Bergström, T.; Spillmann, D.; Svennerholm, B.; Flynn, S. J.; Ryan, P. , Interaction between pseudorabies virus and heparin/heparan sulfate. Pseudorabies virus mutants differ in their interaction with heparin/heparan sulfate when altered for specific glycoprotein C heparin-binding domain. J Biol Chem 1998, 273, 5047–5052. [Google Scholar] [CrossRef]
- Schowalter, R.M.; Pastrana, D.V.; Buck, C.B. Glycosaminoglycans and Sialylated Glycans Sequentially Facilitate Merkel Cell Polyomavirus Infectious Entry. PLOS Pathog. 2011, 7, e1002161. [Google Scholar] [CrossRef] [PubMed]
- Leistner, C.M.; Gruen-Bernhard, S.; Glebe, D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 2007, 10, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Longarela, O.L.; Schmidt, T.T.; Schöneweis, K.; Romeo, R.; Wedemeyer, H.; Urban, S.; Schulze, A. Proteoglycans Act as Cellular Hepatitis Delta Virus Attachment Receptors. PLoS ONE 2013, 8, e58340. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Floresta, G.; Patamia, V.; Chiaramonte, R.; Mauro, G.L.; Rescifina, A.; Vecchio, M. An Integrated Pharmacophore/Docking/3D-QSAR Approach to Screening a Large Library of Products in Search of Future Botulinum Neurotoxin A Inhibitors. Int. J. Mol. Sci. 2020, 21, 9470. [Google Scholar] [CrossRef]
- Cheng, A.; A Best, S.; Merz, K.M.; Reynolds, C.H. GB/SA water model for the Merck molecular force field (MMFF). J. Mol. Graph. Model. 2000, 18, 273–282. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods IV: extension of MNDO, AM1, and PM3 to more main group elements. J. Mol. Model. 2004, 10, 155–164. [Google Scholar] [CrossRef]
- Patamia, V.; Floresta, G.; Zagni, C.; Pistarà, V.; Punzo, F.; Rescifina, A. 1,2-Dibenzoylhydrazine as a Multi-Inhibitor Compound: A Morphological and Docking Study. Int. J. Mol. Sci. 2023, 24, 1425. [Google Scholar] [CrossRef]
- Stewart, J.J.P. MOPAC: A semiempirical molecular orbital program. J. Comput. Mol. Des. 1990, 4, 1–103. [Google Scholar] [CrossRef]
- Ielo, L.; Patamia, V.; Citarella, A.; Efferth, T.; Shahhamzehei, N.; Schirmeister, T.; Stagno, C.; Langer, T.; Rescifina, A.; Micale, N.; et al. Novel Class of Proteasome Inhibitors: In Silico and In Vitro Evaluation of Diverse Chloro(trifluoromethyl)aziridines. Int. J. Mol. Sci. 2022, 23, 12363. [Google Scholar] [CrossRef]
- Wallbrecher, R.; Verdurmen, W.P.R.; Schmidt, S.; Bovee-Geurts, P.H.; Broecker, F.; Reinhardt, A.; van Kuppevelt, T.H.; Seeberger, P.H.; Brock, R. The stoichiometry of peptide-heparan sulfate binding as a determinant of uptake efficiency of cell-penetrating peptides. Cell. Mol. Life Sci. 2013, 71, 2717–2729. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. , YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Cardullo, N.; Catinella, G.; Floresta, G.; Muccilli, V.; Rosselli, S.; Rescifina, A.; Bruno, M.; Tringali, C. Synthesis of Rosmarinic Acid Amides as Antioxidative and Hypoglycemic Agents. J. Nat. Prod. 2019, 82, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Amata, E.; Gentile, D.; Romeo, G.; Marrazzo, A.; Pittalà, V.; Salerno, L.; Rescifina, A. Fourfold Filtered Statistical/Computational Approach for the Identification of Imidazole Compounds as HO-1 Inhibitors from Natural Products. Mar. Drugs 2019, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Ciaffaglione, V.; Intagliata, S.; Pittalà, V.; Marrazzo, A.; Sorrenti, V.; Vanella, L.; Rescifina, A.; Floresta, G.; Sultan, A.; Greish, K.; et al. New Arylethanolimidazole Derivatives as HO-1 Inhibitors with Cytotoxicity against MCF-7 Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 1923. [Google Scholar] [CrossRef] [PubMed]
- Zagni, C.; Pistarà, V.; Oliveira, L.A.; Castilho, R.M.; Romeo, G.; Chiacchio, U.; Rescifina, A. Serendipitous discovery of potent human head and neck squamous cell carcinoma anti-cancer molecules: A fortunate failure of a rational molecular design. Eur. J. Med. Chem. 2017, 141, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Patamia, V.; Zagni, C.; Rescifina, A. Adipocyte fatty acid binding protein 4 (FABP4) inhibitors. An update from 2017 to early 2022. Eur. J. Med. Chem. 2022, 240, 114604. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Patamia, V.; Gentile, D.; Molteni, F.; Santamato, A.; Rescifina, A.; Vecchio, M. , Repurposing of FDA-Approved Drugs for Treating Iatrogenic Botulism: A Paired 3D-QSAR/Docking Approach(dagger). ChemMedChem 2020, 15, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zeng, W.; Kabubei, K. M.; Rasanathan, J. J. K.; Kazungu, J.; Ginindza, S.; Mtshali, S.; Salinas, L. E.; McClelland, A.; Buissonniere, M.; Lee, C. T.; Chuma, J.; Veillard, J.; Matsebula, T.; Chopra, M. , Modelling the economic burden of SARS-CoV-2 infection in health care workers in four countries. Nat Commun 2023, 14, 2791. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.L.; Azizi, A.; Wodarz, D. Network models and the interpretation of prolonged infection plateaus in the COVID19 pandemic. Epidemics 2021, 35, 100463–100463. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; Xiang, Z.; Mu, Z.; Chen, X.; Chen, J.; Hu, K.; Jin, Q.; Wang, J.; Qian, Z. , Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Walls, A. C.; Park, Y. J.; Tortorici, M. A.; Wall, A.; McGuire, A. T.; Veesler, D. , Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735. [Google Scholar] [CrossRef]
- Ennemoser, M.; Rieger, J.; Muttenthaler, E.; Gerlza, T.; Zatloukal, K.; Kungl, A. J. , Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection. Biomedicines 2021, 10. [Google Scholar] [CrossRef]
Figure 1.
Transduction assay timeline.
Figure 1.
Transduction assay timeline.
Figure 2.
A) viral entry ACE2 mediated; B) HS or EX prevented viral entry by the creation of a molecular envelope that inhibits interaction between ACE2 receptor and Spike protein; C) Heparan sulfate or Enoxaparin linked to HS chains, constitutively present in cells membrane, hindered viral entry; D) Viral entry mediated by ACE2 and HS or mediated by an alternative way mediated by HS only.
Figure 2.
A) viral entry ACE2 mediated; B) HS or EX prevented viral entry by the creation of a molecular envelope that inhibits interaction between ACE2 receptor and Spike protein; C) Heparan sulfate or Enoxaparin linked to HS chains, constitutively present in cells membrane, hindered viral entry; D) Viral entry mediated by ACE2 and HS or mediated by an alternative way mediated by HS only.
Figure 3.
A) A549-ACE2 cells treated with HS at 5 mg/mL that, as shown in the graph above, is the first concentration with a percentage of cells availability over the chosen threshold; B) A549-ACE2 cells treated with EX at 2.5 mg/mL that, as shown in the graph above, is the first concentration with a percentage of cells availability over the chosen threshold.
Figure 3.
A) A549-ACE2 cells treated with HS at 5 mg/mL that, as shown in the graph above, is the first concentration with a percentage of cells availability over the chosen threshold; B) A549-ACE2 cells treated with EX at 2.5 mg/mL that, as shown in the graph above, is the first concentration with a percentage of cells availability over the chosen threshold.
Figure 4.
A) A549 cells not exhibiting ACE2 receptors; B) A549 cells expressing ACE2 Receptors (A549-ACE2); C) A549-ACE2 cells infected with Pseudo SARS-CoV-2. The infection can be visualized by green staining of cell nuclei.
Figure 4.
A) A549 cells not exhibiting ACE2 receptors; B) A549 cells expressing ACE2 Receptors (A549-ACE2); C) A549-ACE2 cells infected with Pseudo SARS-CoV-2. The infection can be visualized by green staining of cell nuclei.
Figure 5.
A) A549-ACE2 cells co-treated with HS and Pseudovirus; B) A549-ACE2 cells co-treated with EX and Pseudovirus.
Figure 5.
A) A549-ACE2 cells co-treated with HS and Pseudovirus; B) A549-ACE2 cells co-treated with EX and Pseudovirus.
Figure 6.
A) A549-ACE2 cells pre-treated with HS for 2 h at 37 °C and then infected with Pseudovirus; B) A549-ACE2 cells pre-treated with EX for 2 h at 37 °C and then infected with pseudovirus.
Figure 6.
A) A549-ACE2 cells pre-treated with HS for 2 h at 37 °C and then infected with Pseudovirus; B) A549-ACE2 cells pre-treated with EX for 2 h at 37 °C and then infected with pseudovirus.
Figure 7.
A) A549-ACE2 cells infected with pseudovirus pre-treated for 2 h at 37 °C (water bath) to show the survival of the virus in these conditions; B) A549-ACE2 cells infected with pseudovirus pre-treated with HS for 2 h at 37 °C; C) A549-ACE2 cells infected with pseudovirus pre-treated with EX for 2 h at 37 °C.
Figure 7.
A) A549-ACE2 cells infected with pseudovirus pre-treated for 2 h at 37 °C (water bath) to show the survival of the virus in these conditions; B) A549-ACE2 cells infected with pseudovirus pre-treated with HS for 2 h at 37 °C; C) A549-ACE2 cells infected with pseudovirus pre-treated with EX for 2 h at 37 °C.
Figure 8.
2D pose of HS at the interface between the crystal structures of SARS-CoV-2 spike and ACE2 (PDB ID: 6M0J).
Figure 8.
2D pose of HS at the interface between the crystal structures of SARS-CoV-2 spike and ACE2 (PDB ID: 6M0J).
Figure 9.
2D pose of EX at the interface between the crystal structures of SARS-CoV-2 spike and ACE2 (PDB ID: 6M0J).
Figure 9.
2D pose of EX at the interface between the crystal structures of SARS-CoV-2 spike and ACE2 (PDB ID: 6M0J).
Figure 10.
2D pose of HS at the interface between the crystal structures of SARS-CoV-2 spike (BA.2.86 variant) and ACE2.
Figure 10.
2D pose of HS at the interface between the crystal structures of SARS-CoV-2 spike (BA.2.86 variant) and ACE2.
Figure 11.
2D pose of EX at the interface between the crystal structures of SARS-CoV-2 spike (BA.2.86 variant) and ACE2.
Figure 11.
2D pose of EX at the interface between the crystal structures of SARS-CoV-2 spike (BA.2.86 variant) and ACE2.
Table 1.
ACE2 Red Transduction Mix.
Table 1.
ACE2 Red Transduction Mix.
| |
Amount/ Well |
Final Concentration |
| ACE2 BacMam |
5 μL |
6.6 × 108 VG/mL |
| Sodium butyrate |
0.6 μL |
2 mM |
| Complete media |
44.4 μL |
|
Table 2.
Pseudo SARS-CoV-2 Green Reporter Transduction Mix.
Table 2.
Pseudo SARS-CoV-2 Green Reporter Transduction Mix.
| |
Amount/ Well |
Final Concentration |
| SARS-CoV-2 Pseudovirus |
2.5 μL |
3.3 × 108 VG/mL |
| Sodium butyrate |
0.6 μL |
2 mM |
| Compounds |
|
5.0 mg/mL (HS); 1.25 mg/mL (EX) |
| Complete media |
adjust |
|
Table 3.
Summary of the main residuals between HS and EX with the two interfaces wild and variant.
Table 3.
Summary of the main residuals between HS and EX with the two interfaces wild and variant.
| PDB ID: 6M0J |
| |
Attractive Charge |
Hydrogen Bond |
| HS |
Arg403, Tyr505 |
Tyr453, Ser494, Gly496, Gln498, Asn501 |
| EX |
Arg403, Phe456 |
Asn487, Tyr489, Gly496, Gln498 |
| BA.2.86 variant |
| |
Attractive Charge |
Hydrogen Bond |
| HS |
Tyr449, Tyr505 |
Leu492, Ser494, Gly496, Gln498 |
| EX |
Lys403, Lys484 |
Tyr453, Gly496, Gln498 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).