Submitted:
22 October 2023
Posted:
24 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Physiological tests of the Gagr gene knockdown mutant
2.1.1. Knockdown of the Gagr gene does not affect embryonic and larval mortality of flies
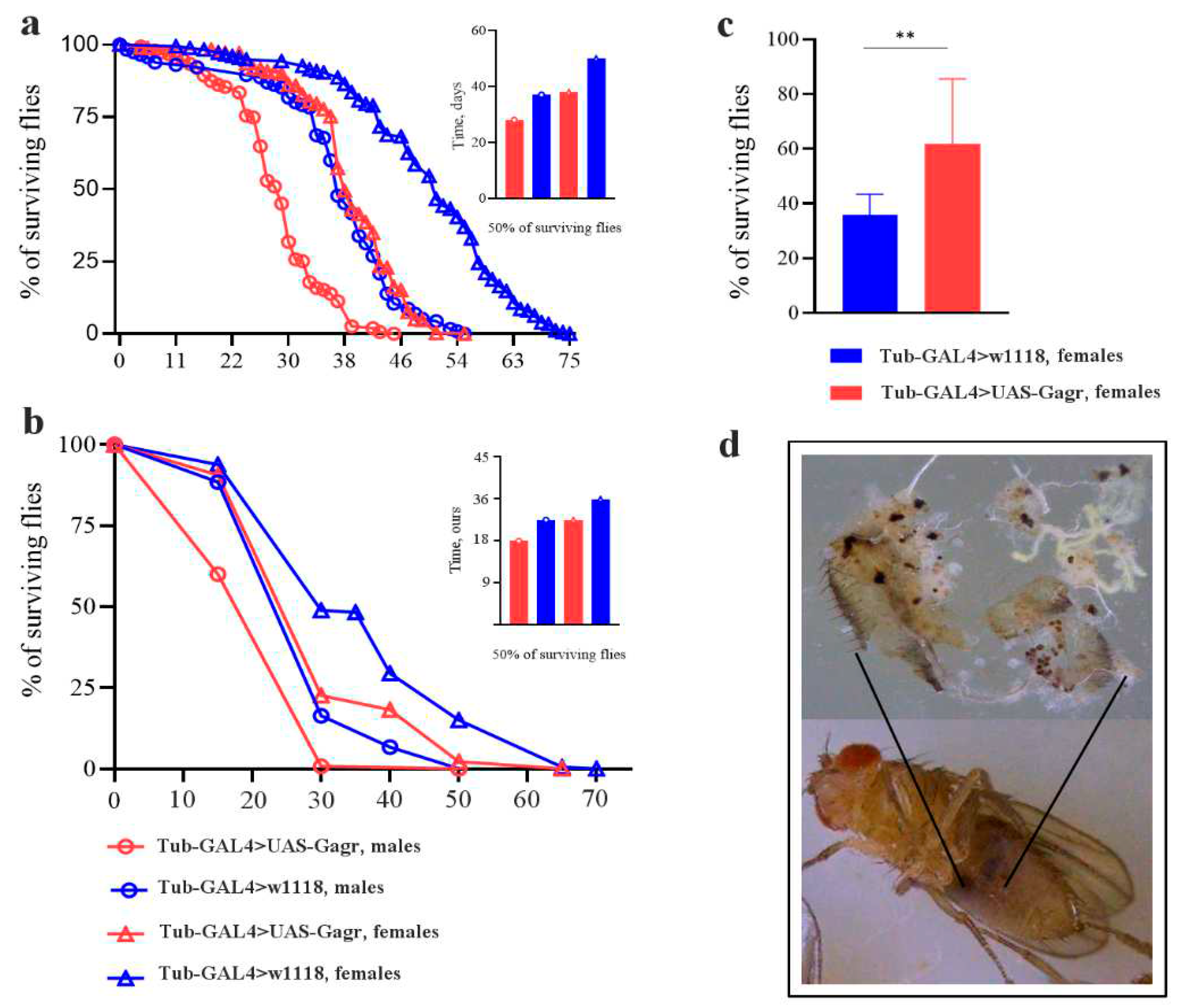
2.1.2. Knockdown of the Gagr gene affects the lifespan of flies under standard and stress conditions
2.1.3. Knockdown of the Gagr gene does not lead to changes in adult motility
2.1.4. Knockdown of the Gagr gene leads to increased resistance to heat stress
2.1.5. Knockdown of the Gagr gene in females promotes the occurrence of melanized masses in the fat body
2.2. Transcriptomic analysis of the Gagr gene knockdown mutant
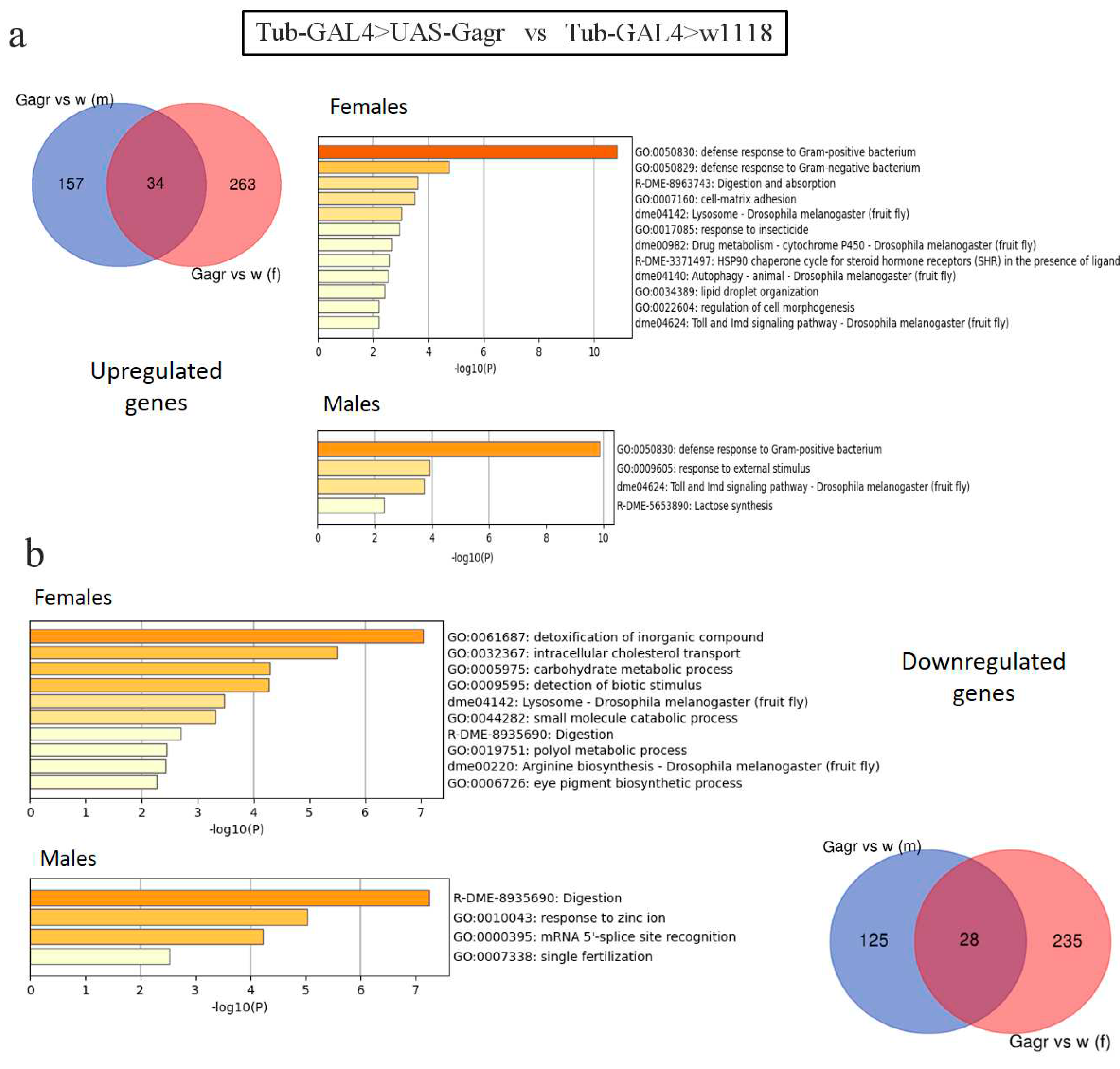
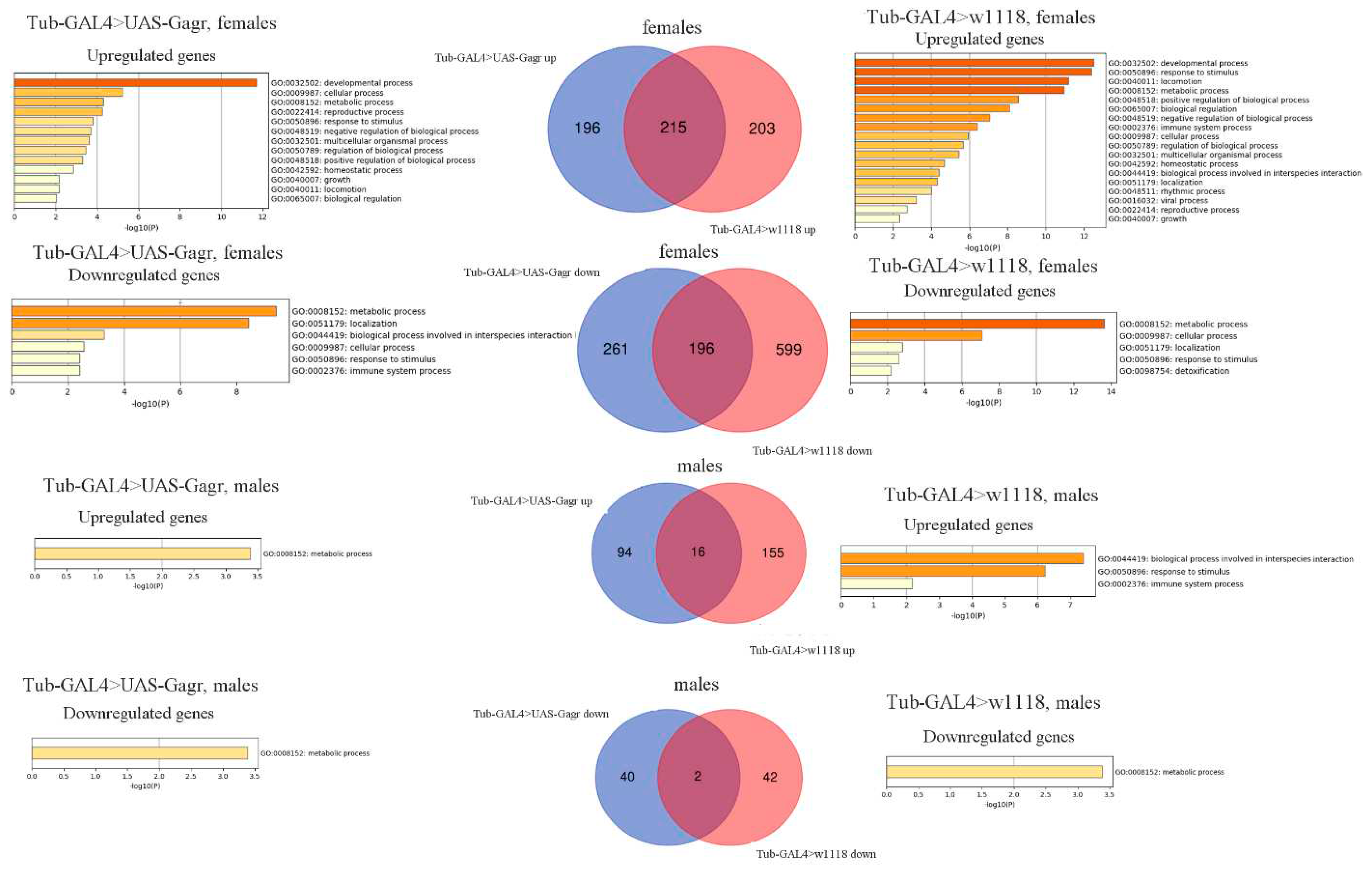
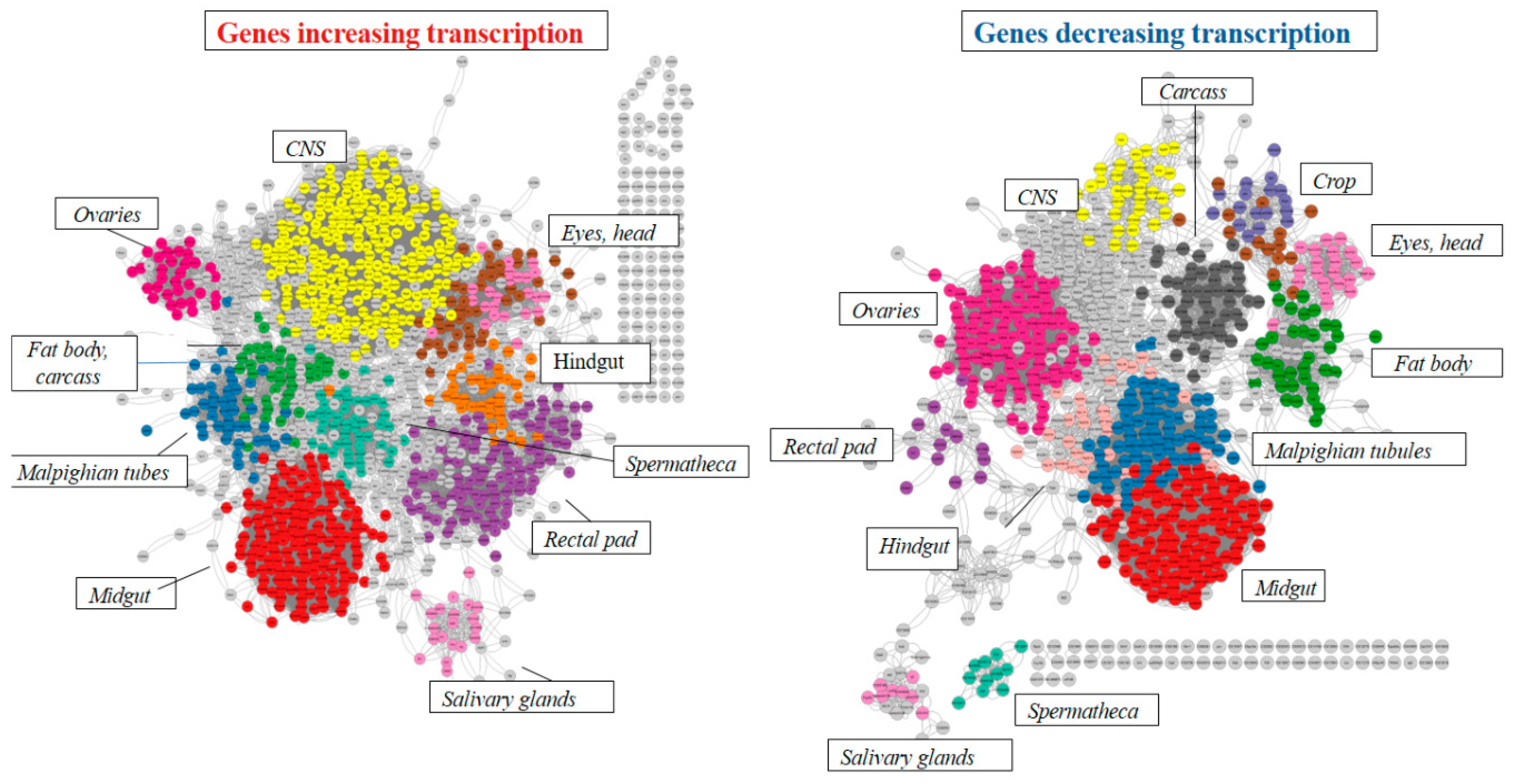
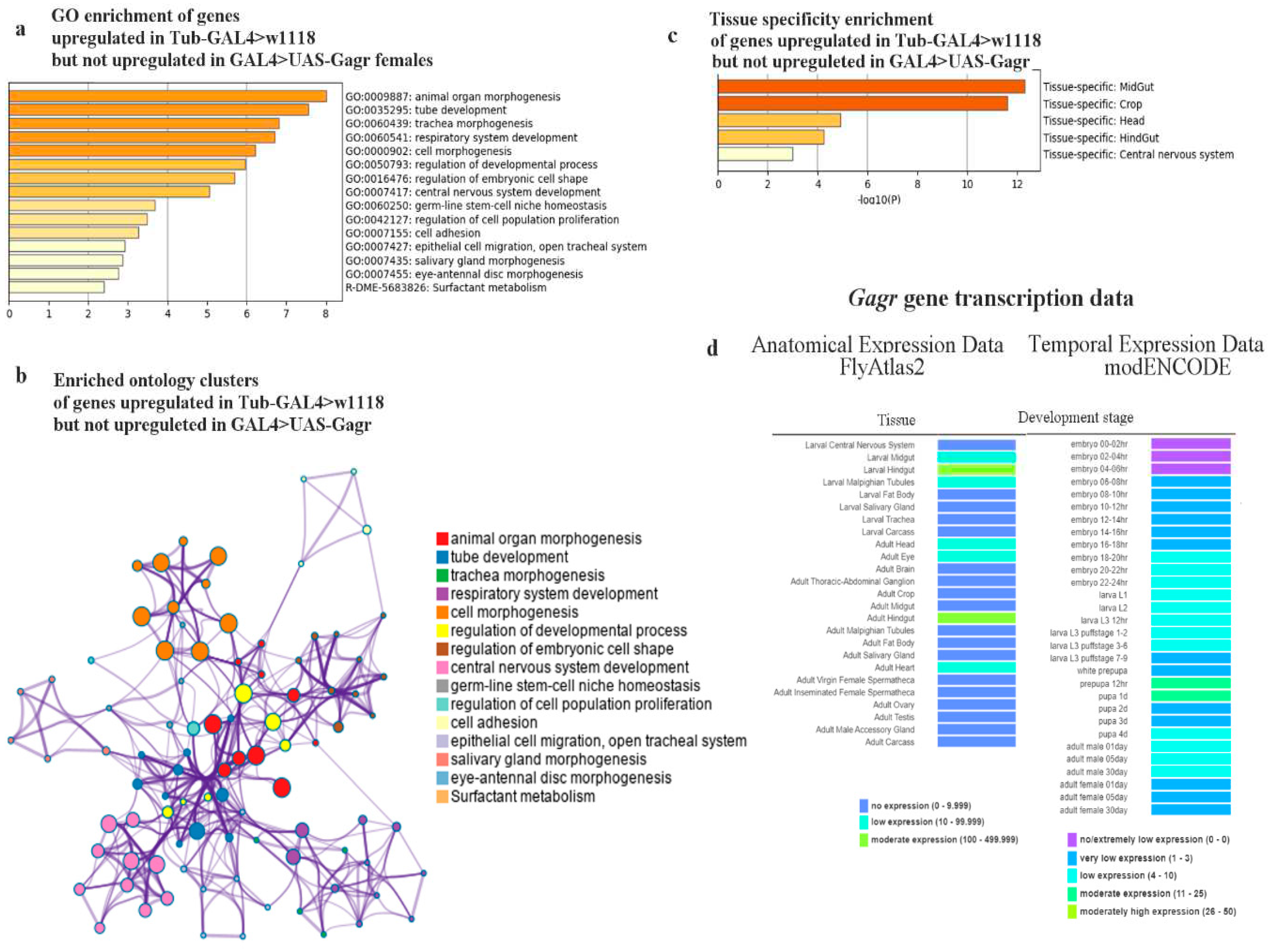
2.2.1. Analysis of differentially expressed genes in the Gagr knockdown strain
2.2.2. Analysis of the transcriptomic response to the action of ammonium persulfate
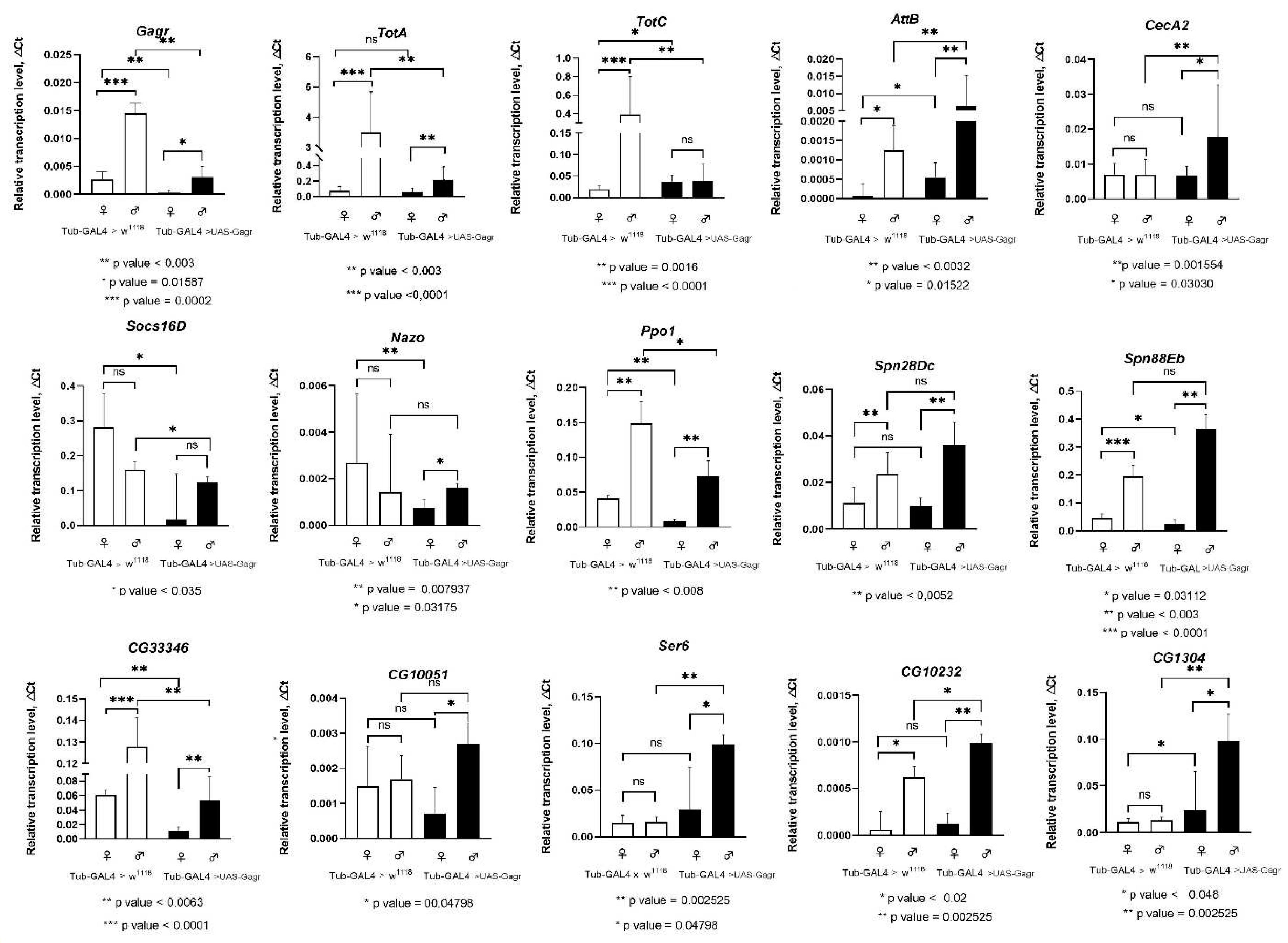
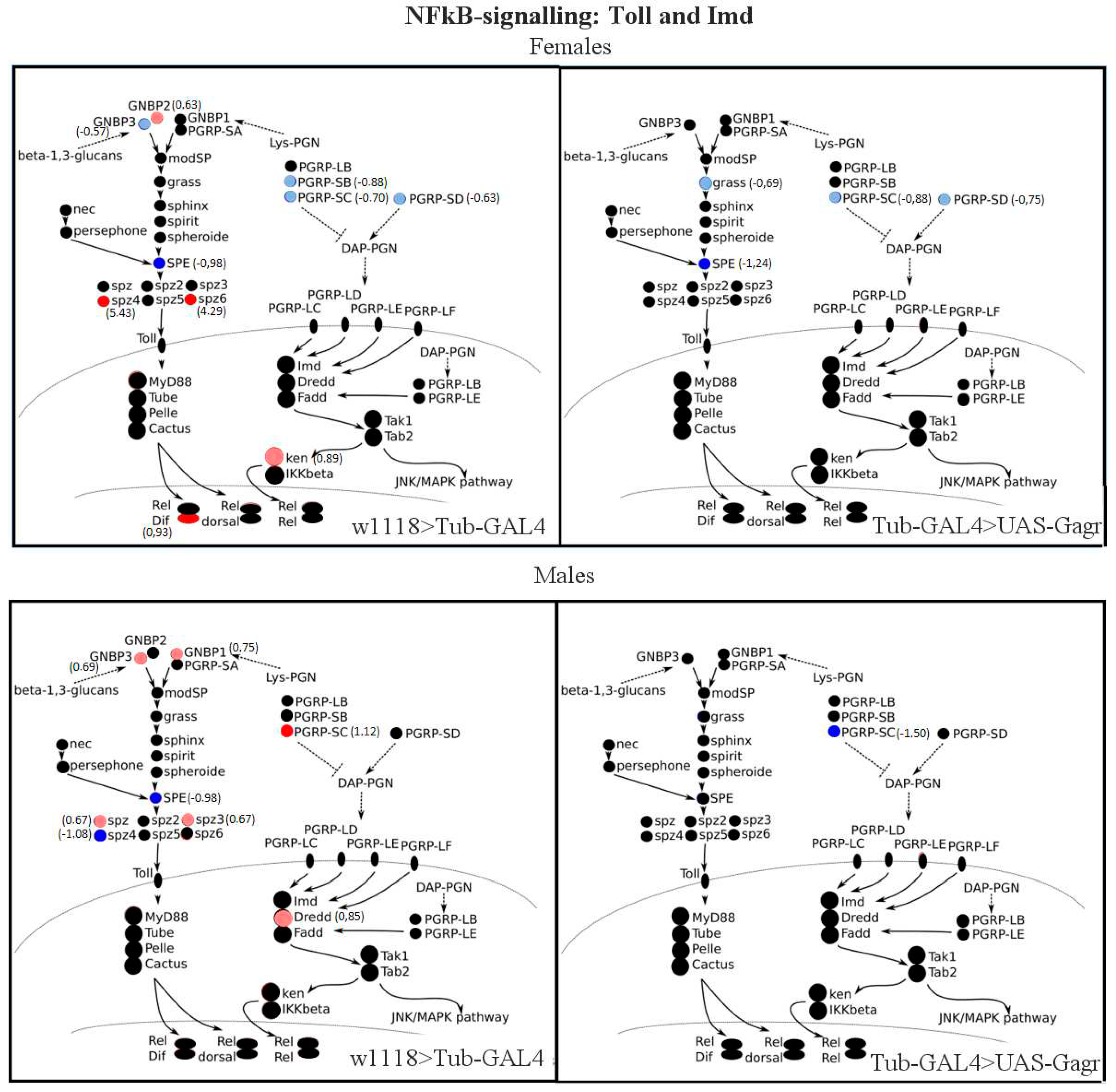
2.2.3. Transcription analysis of genes involved in immune pathways
2.2.4. Genes whose expression is not induced by stress in the Gagr mutant
3. Discussion
4. Materials and Methods
4.1. Drosophila melanogaster strains and conditions
4.2. Physiological tests
4.3. RNA extraction and RT-PCR
4.4. RNA-sequencing and data processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dupressoir, A.; Marceau, G.; Vernochet, C.; Benit, L.; Kanellopoulos, C.; Sapin, V.; Heidmann, T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. PNAS 2005, 102, 725. [Google Scholar] [CrossRef] [PubMed]
- Emera, D.; Wagner, G.P. Transposable element recruitments in the mammalian placenta: Impacts and mechanisms. Briefings in functional genomics 2012, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.W.; Colbeck, E.; Ellis, S.A.; Stoye, J.P. Evolution of the retroviral restriction gene Fv1: Inhibition of non-MLV retroviruses. PLoS pathogens 2014, 10, e1003968. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The impact of transposable elements in adaptive evolution. Molecular ecology. 2018, 28, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Schwalie, P.C.; Pankevich, E.V.; Gubelmann, C.; Raghav, S.K.; Dainese, R.; Cassano, M.; Imbeault, M.; Jang, S.M.; Russeil, J.; Delessa, T.; Duc, J.; Trono, D.; Wolfrum, C.; Deplancke, B. ZFP30 promotes adipogenesis through the KAP1-mediated activation of a retrotransposon-derived Pparg2 enhancer. Nature Communications 2019, 10, 1809. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Xu, W.; Wang, Z.; Liu, C.; Lin, P.; Li, B.; Huang, Q.; Yang, J.; Zhou, H.; Qu, L. An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO reports 2019, 20, e47650. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, L.N.; Kuzmin, I. V, Makhnovskii, P.A.; Kim, A.I. Domesticated retroviral GAG gene in Drosophila: New functions for an old gene. Virology 2014, 450–451, 196. [Google Scholar] [CrossRef]
- Makhnovskii, P.; Balakireva, Y.; Nefedova, L.; Lavrenov, A.; Kuzmin, I.; Kim, A. Domesticated gag gene of drosophila LTR retrotransposons is involved in response to oxidative stress. Genes 2020, 11, 396. [Google Scholar] [CrossRef]
- Silverman, N.; Zhou, R.; Erlich, R.L.; Hunter, M.; Bernstein, E.; Schneider, D.; Maniatis, T. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem, 2003, 278, 48928–48934. [Google Scholar] [CrossRef]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef]
- Ashton-Beaucage, D.; Udell, C.M.; Gendron, P.; Sahmi, M.; Lefrancois, M.; Baril, C.; Guenier, A.S.; Duchaine, J.; Lamarre, D.; Lemieux, S.; et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 2014, 12, e1001809. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.S.; Uehara, T.; Nomura, Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J. Biol. Chem. 2002, 277, 35386–35392. [Google Scholar] [CrossRef] [PubMed]
- Grek, C. Townsend, D.M. Protein disulfide isomerase superfamily in disease and the regulation of apoptosis in endoplasmic reticulum. Stress. Dis. 2014, 1, 4–17. [Google Scholar]
- Majzoub, K.; Hafirassou, M.L.; Meignin, C.; Goto, A.; Marzi, S.; Fedorova, A.; Verdier, Y.; Vinh, J.; Homann, J.A.; Martin, F.; et al. RACK1 controls IRES-mediated translation of viruses. Cell 2014, 159, 1086–1095. [Google Scholar] [CrossRef]
- Schnorrer, F.; Schönbauer, C.; Langer, C.C.; Dietzl, G.; Novatchkova, M.; Schernhuber, K.; Fellner, M.; Azaryan, A.; Radolf, M.; Stark, A.; et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 2010, 464, 287–291. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Song, W.; Zhu, L.; Dong, Z.; Ow, D.W. A Pap1–Oxs1 signaling pathway for disulfide stress in Schizosaccharomyces pombe. Nucleic Acid Res. 2017, 45, 106–114. [Google Scholar] [CrossRef]
- McEwen, D.G.; Peifer, M. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development 2005, 132, 3935–3946. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.E.; Sousa, L.R.; Martins, I.K.; Rodrigues, N.R.; Posser, T.; Franco, J.L. Data on the phosphorylation of p38MAPK and JNK induced by chlorpyrifos in Drosophila melanogaster. Data Brief. 2016, 9, 32–34. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Poidevin, M.; Lemaitre, B. The p38c gene is expressed in the midgut and upregulated upon intestinal infection. PLoS Genet. 2014, 10, e1004659. [Google Scholar]
- Lindsay, S.A.; Lin, S.J.H.; Wasserman, S.A. Short-form bomanins mediate humoral immunity in Drosophila. J. Innate Immun. 2018, 10, 306–314. [Google Scholar] [CrossRef]
- Loch, G.; Zinke, I.; Mori, T.; Carrera, P.; Schroer, J.; Takeyama, H.; Hoch, M. Antimicrobial peptides extend lifespan in Drosophila. PLoS ONE. 2017, 12, e0176689. [Google Scholar] [CrossRef]
- Lemaitre, B, Hoffmann, J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007, 2, 697–743. [Google Scholar]
- Becker, T.; Loch, G.; Beyer, M.; Zinke, I.; Aschenbrenner, A.C.; Carrera, P.; et al. FOXO-dependent regulation of innate immune homeostasis. Nature 2010, 463, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Broderick, N.A.; Poidevin, M.; Pradervand, S.; Lemaitre, B. Drosophila intestinal response to bacterial infection: Activation of host defense and stem cell proliferation. Cell Host Microbe. 2009, 5, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Kim, S.H.; Lee, H.Y.; Bai, J.Y.; Nam, Y.D.; Bae, J.W.; et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Ekengren, S.; Tryselius, Y.; Dushay, M.S.; Liu, G.; Steiner, H.; Hultmark, D. A humoral stress response in Drosophila. Curr. Biol, 2001, 11, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Amstrup, A.B.; Bæk, I.; Loeschcke, V.; Givskov Sørensen, J. A functional study of the role of Turandot genes in Drosophila melanogaster: An emerging candidate mechanism for inducible heat tolerance. J Insect Physiol. 2022, 143, 104456. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.J.; Frey, F. and Carton, Y. Drosophila serpin 27A is a likely target for immune suppression of the blood cell-mediated melanotic encapsulation response. J. Insect Physiol. 2005, 51, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Tingvall, T.O.; Roos, E. and Engstrom, Y. The GATA factor Serpent is required for the onset of the humoral immune response in Drosophila embryos. PNAS 2001, 98, 3884–3888. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M. and Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Lemaitre, B.; Meister, M.; Govind, S.; Georgel, P.; Steward, R.; et al. ; Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 1995, 14, 536–545. [Google Scholar] [CrossRef]
- Lavine, M.D.; and Strand, M.R. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef]
- Takehana, A.; Katsuyama, T.; Yano, T.; Oshima, Y.; Takada, H.; et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. PNAS 2002, 99, 13705–13710. [Google Scholar] [CrossRef] [PubMed]
- Asha, H.; Nagy, I.; Kovacs, G.; Stetson, D.; Ando, I.; et al. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 2003, 163, 203–215. [Google Scholar] [CrossRef]
- Zettervall, C.J.; Anderl, I.; Williams, M.J.; Palmer, R.; Kurucz, E.; et al. A directed screen for genes involved in Drosophila blood cell activation. PNAS 2004, 101, 14192–14197. [Google Scholar] [CrossRef]
- Luo, H.; Hanratty, W.P.; Dearolf, C.R. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995, 14, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.J.; Vass, E.; Malagoli, D.; Carton, Y. The effects of parasite-derived immune-suppressive factors on the cellular innate immune and autoimmune responses of Drosophila melanogaster. J. Parasitol. 2004, 90, 1139–1149. [Google Scholar] [CrossRef]
- Rizki, T.M.; Rizki, R.M. Developmental analysis of a temperature-sensitive melanotic tumor mutant in Drosophila melanogaster. Wilhelm Roux Arch. Dev. Biol. 1980, 189, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, N.T.; Fischer, M.L.; Waring, A.L.; Kr, P.; Kacsoh, B.Z.; Brantley, S.E.; Keebaugh, E.S.; Hill, J.; Lark, C.; Martin, J.; Bains, P.; Lee, J.; Vrailas-Mortimer, A.D.; Schlenke, T.A. Extracellular matrix protein N-glycosylation mediates immune self-tolerance in Drosophila melanogaster. PNAS 2021, 118, e2017460118. [Google Scholar] [CrossRef]
- Kärre, K. Natural killer cell recognition of missing self. Nat. Immunol. 2008, 9, 477–480. [Google Scholar] [CrossRef]
- Belmonte, R.L.; Corbally, M.-K.; Duneau, D.F.; Regan, J.C. Sexual Dimorphisms in Innate Immunity and Responses to Infection in Drosophila melanogaster. Frontiers in Immunology 2020, 10, 3075. [Google Scholar] [CrossRef]
- Pignatti, P.; Frossi, B.; Pala, G.; Negri, S.; Oman, H.; Perfetti, L.; Pucillo, C.; Imbriani, M.; Moscato, G. Oxidative activity of ammonium persulfate salt on mast cells and basophils: Implication in hairdressers’ asthma. Int Arch Allergy Immunol. 2013, 160, 409–419. [Google Scholar] [CrossRef]
- Song, C.; Wang, L.; Ye, G.; Song, X.; He, Y.; Qiu, X. Residual ammonium persulfate in nanoparticles has cytotoxic effects on cells through epithelial-mesenchymal transition. Sci Rep. 2017, 7, 11769. [Google Scholar] [CrossRef]
- Pandey, V.K.; Sharma. R.; Prajapati, G.K.; Mohanta, T.K.; Mishra, A.K. N-glycosylation, a leading role in viral infection and immunity development. Mol Biol Rep. 2022, 49, 8109–8120. [Google Scholar] [CrossRef] [PubMed]
- Vagin, O.; Kraut, J.A.; Sachs, G. Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am J Physiol Renal Physiol. 2009, 296, F459–69. [Google Scholar] [CrossRef] [PubMed]
- Santabarbara-Ruiz, P.; Lopez-Santillan, M.; Martinez-Rodriguez, I.; Binagui-Casas, A.; Perez, L.; Milan, M.; Corominas, M.; Serras, F. ROS-Induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 2015, 11, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.L.; Nakamura, K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim. Biophys. Acta 2007, 1773, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Zeitlinger, J.; Kockel, L.; Peverali, F.A.; Jackson, D.B.; Mlodzik, M.; Bohmann, D. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 1997, 16, 7393–7401. [Google Scholar] [CrossRef] [PubMed]
- Santabarbara-Ruiz, P.; Lopez-Santillan, M.; Martinez-Rodriguez, I.; Binagui-Casas, A.; Perez, L.; Milan, M.; Corominas, M.; Serras, F. ROS-Induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 2015, 11, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Dudzic, J.P.; Li, X.; Collas, E.J.; Boquete, J.P.; Lemaitre, B. Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 2016, 12, e1006089. [Google Scholar] [CrossRef]
- Fox, D.T.; Spradling, A.C. The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 2009, 5, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Chambers, R.P.; Call, G.B.; Meyer, D.; Smith, J.; Techau, J.A.; Pearman, K.; Buhlman, L.M. Nicotine increases lifespan and rescues olfactory and motor deficits in a Drosophila model of Parkinson’s disease. Behav. Brain Res. 2013, 253, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Leader, D. P, Krause, S.A.; Pandit, A.; Davies, S.A.; Dow J.A.T. FlyAtlas 2: A new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucl. Acids Res. 2018, 46, D809–D815. [Google Scholar] [CrossRef]


| Strain | Sum of flies | Flies with a short body | Flies with a long body | |||||
|---|---|---|---|---|---|---|---|---|
| males | females | sum | males | females | sum | |||
| Tub-GAL4>w1118 | Observed | 897 | 224 | 251 | 475 | 193 | 229 | 422 |
| Expected | 448,5 | 448,5 | ||||||
| Tub-GAL4>UAS-Gagr | Observed | 1351 | 315 | 395 | 710 | 255 | 386 | 641 |
| Expected | 675,5 | 675,5 | ||||||
| Gene | Function according to Flybase | Log2FoldChange of transcription level | |||
| Females | Males | ||||
| w1118 1 | Gagr2 | w1118 1 | Gagr2 | ||
| TotA | Antimicrobial peptide expressed in response to stress by the JAK-STAT pathway | 3,69 | 1,18 | 0,24 | 0,09 |
| TotC | 3,94 | 0,08 | 0,23 | 0,02 | |
| AttB | Antimicrobial peptide induced against Gram+ and Gram- bacterium by the Toll pathway | 1,15 | 0,31 | -0,32 | 0,87 |
| CecA2 | 1,92 | 0,80 | 0,66 | 0,55 | |
| Socs16D | Suppressor of Cytokine Signaling positive regulator of JNK/MAPK cascade | 0,32 | 0,36 | -0,07 | 0,34 |
| Nazo | dIKKb-dependent antiviral effector protein of IMD pathway, expressed downstream Sting and Relish signaling | 0,91 | -0,07 | 3,79 | 0,66 |
| Ppo1 | Propheloloxidase 1 involved in the melanization reaction, regulated by the JAK-STAT, Toll and IMD pathways | 0,30 | -0,84 | 0,14 | -0,67 |
| Spn88Eb | Serin endopeptidase inhibitor involved in immune response, regeneration and regulation of stem cells division | 0,86 | 0,43 | 0,35 | -0,26 |
| Spn28D | Serin endopeptidase inhibitor involve induced upon injury, negative regulator of melanization cascade | 0,10 | -0,07 | 0,39 | 0,63 |
| CG33346 | Predicted to enable RNA and single DNA endonuclease activity, involved in apoptotic DNA fragmentation, most active in digestive system | 0,95 | 0,04 | 0,30 | 0,37 |
| CG10051 | Predicted to enable metalloexopeptidase activity, to be involved in proteolysis, most active in digestive system | 0,22 | -1,88 | 1,92 | 0,38 |
| Ser6 | Predicted to enable serine endopeptidase activity, to be involved in proteolysis, most active in digestive system | 0,53 | -0,85 | -0,84 | -0,68 |
| CG10232 | 1,67 | -0,22 | 1,14 | -0,30 | |
| CG1304 | -2,23 | -1,14 | -0,08 | -1,15 | |
| Gene | Biological function of the protein (according to FlyBase) |
|---|---|
| run | Contributes to axon guidance, dendrite morphogenesis and germ-band extension |
| ss | Plays a key role in defining the distal regions of the antenna and the legs |
| ase | Acts together with other proneural genes in nervous system development, which involves N-mediated lateral inhibition |
| sr | Induces the fate of tendon cells in the embryo as well as in the adult fly |
| Antp | Part of a developmental regulatory system that specifies segmental identity in the pro- and mesothorax |
| Sox21a | Involved in the differentiation of stem cells in the midgut |
| esg | Contributes to stem cell maintenance, tracheal morphogenesis and neuroblast differentiation |
| grh | Responsible for the proper expression of many genes primarily involved in epithelial cell fate, barrier formation, wound healing, tube morphogenesis and proliferation of larval neuroblasts |
| ham | Regulates neuron fate selection in the peripheral nervous system and olfactory receptor neurons |
| Dfd | Involved in proper morphological identity of the maxillary segment and the posterior half of the mandibular segment |
| ich | In tracheal terminal cells, regulates the transcription of factors involved in the formation of a mature apical extracellular matrix which is essential for the integrity and shape of seamless tubes |
| nerfin-1 | Regulates early axon guidance at the embryonic stage and is required for the maintenance of larval neuron differentiation |
| dmrt99B | Involved_in sex differentiation |
| grn | Regulates the expression of receptors and adhesion molecules involved in axon guidance |
| Kr-h1 | Involved in axon pathfinding, neurite and axon remodeling as well as pupal photoreceptor maturation |
| acj6 | Acts in odor receptor gene expression and axon targeting of olfactory neurons |
| rib | Required for development of the salivary gland and trachea, as well as for dorsal closure |
| tap | May play a role in the specification of the sugar-sensitive adult gustatory neuron |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




