1. Introduction
Members of the SP24 protein family were first described in chronic bee paralysis virus (CBPV) and anopheline-associated C virus (AACV) (here and below, virus names are given according to the current ICTV rules [
1]), which are RNA viruses related to members of the order
Tolivirales (phylum
Kitrinoviricota) [
2]. This polypeptide contains a poorly conserved positively charged N-terminal region and a conserved domain corresponding to the central region of the protein, including four predicted trans-membrane helical segments [
2,
3]. The SP24 family protein is one of the major structural components of CBPV virions, together with the RNA2-encoded ORF2 putative glycoprotein, which possesses the N-terminal DiSB-ORF2_chro domain in a short conserved region predicted to form disulfide bridges [
4]. CBPV, AACV, and recently described related viruses of the order
Tolivirales, namely
insect tombus bipa-like virus 1, tiger mosquito bi-segmented tombus-like virus, diaphorina citri-associated C virus and
linepithema humile C virus 1 as well as the arachnid-specific virus infecting
Tetragnatha maxillosa (hubei tombus-like virus 28) [
5,
6,
7] are bipartite RNA viruses, in which RdRp and SP24 are encoded by multicistronic RNAs 1 and 2, respectively.
Another group of multipartite RNA viruses encoding SP24 family proteins are members of the family
Kitaviridae (order
Martellivirales, phylum
Kitrinoviricota). Plant viruses of the genera
Cilevirus and
Blunervirus contain the SP24 gene in RNA2 and RNA3 of bipartite and quadripartite genomes, respectively. However, the tripartite genomes of two members of the plant virus genus
Higrevirus (family
Kitaviridae) encode SP24 in either RNA2 or RNA3 [
8,
9].
Several monopartite positive-stranded RNA viruses of insects related to unclassified members of the family
Permutotetraviridae have also been shown to encode SP24. This virus family is characterized by a unique permutated organization of the replicase RdRp domain and is classified as an isolated taxon in the kingdom
Orthornavirae [
10]. Some unclassified viruses of this family, in particular shinobi tetravirus, culex permutotetra-like virus, hubei permutotetra-like virus 3 and sarawak virus, contain genomes with three ORFs, where SP24 and DiSB-chro genes represent ORF2 and ORF3 located downstream of the replicase gene [
10].
The most abundant groups of monopartite positive-stranded RNA viruses encoding SP24 include negeviruses and negev-like (or nege-like) viruses, which have an obvious evolutionary relationship to the multipartite viruses of the family
Kitaviridae [
9,
11]. Initially, viruses of the genus
Negevirus have been considered to be insect-specific viruses that mainly infect species of the order
Diptera [
11]. In particular, negeviruses are among the most abundant virus taxa in many
Culex and
Aedes species, which are the predominant hosts of the eponymous negev virus. Negeviruses are classified into two clades, nelorpiviruses and sandewaviruses, with nelorpiviruses being more closely related to plant kitaviruses [
11,
12].
Negevirus genomes typically encode three ORFs: ORF1 encodes replicase, while ORFs 2 and 3 encode the predicted DiSB-containing glycoprotein and SP24 family polypeptide, respectively. Interestingly, the DiSB-ORF2_chro domain is part of the virion protein of sogatella furcifera hepe-like virus (order
Hepelivirales) (see also below,
Section 3.5.) [
13]. The minority of negeviruses encoding less than three ORFs includes, for example, cordoba virus with a single ORF and buckhurst virus with two ORFs [
11,
12]. Massive metagenomic searches have significantly increased the number of nege- and negev-like viruses and have shown that the genomic structure of negev-like viruses is quite variable [
5,
12,
14,
15]. Moreover, these studies and the in silico studies of public databases have shown that the hosts of negev-like viruses are much more diverse than those of the genus
Negevirus. Negev-like viruses infect hosts belonging to the class
Insecta (phylum
Arthropoda) in addition to the order
Diptera, namely the orders
Hemiptera,
Coleoptera,
Thysanoptera,
Hymenoptera,
Odonata,
Orthoptera, and
Lepidoptera. Several host species belonging to the phylum
Arthropoda (classes
Arachnida, Maxillopoda, Chilopoda and
Malacostraca), as well as to the phyla
Nematoda and
Cnidaria, have also been reported [
5,
12,
14,
15].
Importantly, the vast majority of negeviruses and negev-like viruses encode the SP24 protein, which is predicted to be a virion component [
5,
12,
14,
15]. Indeed, comparative electron microscopy studies of negevirus virions have shown that the particles are pleiomorphic and consist mostly of SP24. These particles have variable sizes, an elliptical shape, and a single projection consisting of glycoprotein with a DiSB signature. Microscopy also reveals stratified layers at the virion surface, probably representing a host-derived lipid bilayer [
16]. Although plant-infecting viruses of the family
Kitaviridae have been shown to encode SP24 and contain lipid membranes, the exact role of SP24 in encapsidation and its function in virus particles remain unclear [
9]. Moreover, cileviruses encode a protein (p29) that is found in virions and viroplasms and is capable of forming spherical virus-like particles when expressed as a single engineered protein
in planta. Strikingly, cileviruses can replicate in insect vector cells in which the accumulation of p29-specific subgenomic RNA is 50 times lower than in plants [
9]. It can be hypothesized that p29 is a plant-specific virus movement factor, whereas SP24 is an animal-specific virus movement factor that may directly interact with the virus genomic RNA [
3,
9], thus facilitating virus transfer in animal (insect) organism.
In this paper, to better understand the taxonomic boundaries of hosts for viruses encoding SP24 and the variability of the negev-like virus genome organizations, we have searched the rapidly increasing amount of new transcriptomic data in the public NCBI database. This search, in our opinion, may open new horizons for planning the future studies on the exact roles of SP24 in virus infection.
3. Results
In the last two years, several papers have been published that further encourage us to perform bioinformatics search of SP24 genes in viruses and virus-like RNA assemblies (VLRAs) of order
Martellivirales infecting all eukaryotes (bikonts and unikonts). First, novel data have shown previously unknown fact that SP24-encoding negev-like viruses also infect flatworms in phylum
Platyhelminthes [
17]. Second, recently obtained transcriptome assemblies from large amount of arthropods, particularly, from 1098 species of spiders (phylum
Arthropoda, class
Arachnidae) [
18] can provide a good starting basis for further integrated analysis of the variability of genome organizations in SP24-encoding negev-like viruses. Third, novel paper published early 2023 [
19] has been reported that a previously undescribed insect virus species—
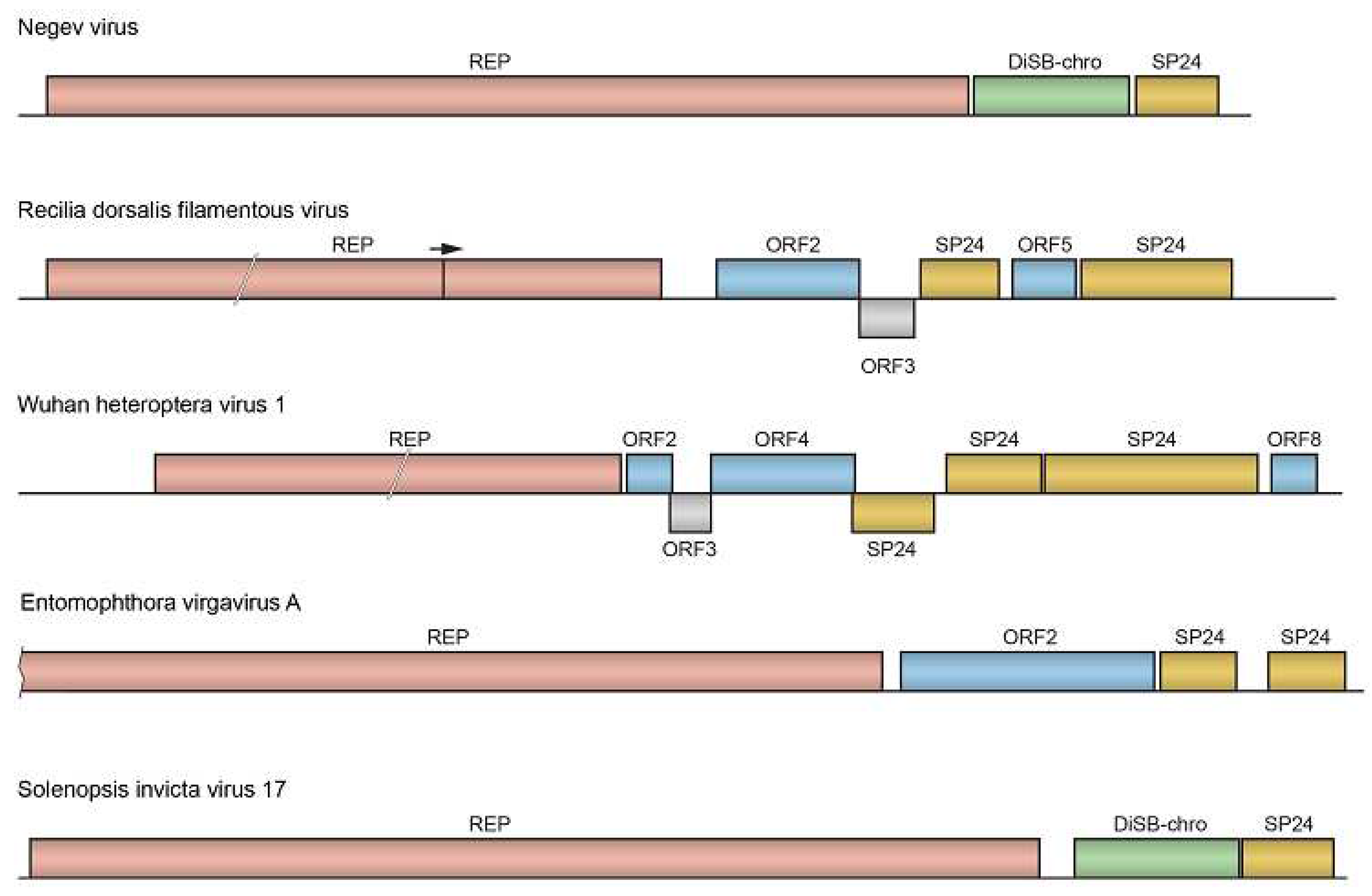
Recilia dorsalis filamentous virus (RdFV) encodes SP24 which is included into filamentous virions unlike elliptical negeviruses and CBPV. Moreover, RdVF monopartite genome includes two copies of distantly related SP24 genes (ORFs 4 and 6) (
Figure 1). Direct interaction of viral capsid SP24 encoded by ORF 6 with the sperm-specific serpin protein of leafhopper
Recilia dorsalis mediates invasion of virus into male reproductive organs and binding virions to insect sperm surfaces [
19]. As a result, these processes are important for subsequent paternal virus transmission, and SP24, thus, represents proteins factor for vertical virus transmission. One can hypothesized that SP24-mediated paternal virus transmission is a novel type of inter-organismal virus movement in insect populations.
Figure 1.
Gene organization of the negevirus and selected negev-like viruses. The replicase protein (REP) is shown in pink (see text for details). SP24 protein is shown in brown. ORFs coding for DiSB-chro domain proteins are in green. Proteins with putative membrane-bound segments are shown in blue. Read-through of leaky terminator codon is shown by arrow above RdFV genome scheme.
Figure 1.
Gene organization of the negevirus and selected negev-like viruses. The replicase protein (REP) is shown in pink (see text for details). SP24 protein is shown in brown. ORFs coding for DiSB-chro domain proteins are in green. Proteins with putative membrane-bound segments are shown in blue. Read-through of leaky terminator codon is shown by arrow above RdFV genome scheme.
3.1. Virus RNA genomes with two or three gene copies encoding SP24
In contrast to many other known negeviruses and negev-like viruses, the genome of a symbiotic virus RdFV infecting rice green zigzag leafhopper
Recilia dorsalis (class
Insecta, order
Hemiptera, family
Cicadellidae) contains two gene copies encoding SP24 [
19] that may correlate with the ability of the virus to form filamentous particles and/or to spread by vertical transmission. In general, this virus encodes seven proteins (
Figure 1). Replicase is encoded by ORF1, which has an internal leaky termination codon and may be translated to form two overlapping polypeptides [
20]. Among non-replicative proteins of RdFV, only the SP24 polypeptides have significant similarity to proteins of annotated viruses. However, a search of the NCBI TSA database revealed that the transcriptome of insect
Xestocephalus desertorum, which belongs to the same family
Cicadellidae as zigzag leafhopper, contains a partial VLRA encoding ORF1 RdRp domain closely related to that of RdFV (e-value 1e-49), a protein homolog of RdFV ORF5 protein (e-value 1e-19) and three copies of SP24 (
Table 1). Three copies of SP24 genes were also revealed in the annotated genomes of pyrrhocoris apterus virus 1 infecting insect
Pyrrhocoris apterus (class
Insecta, order
Hemiptera, family
Pyrrhocoridae) and wuhan heteroptera virus 1 isolated from mixed insects of the suborder
Heteroptera (order
Hemiptera) [
5] (
Figure 1) (
Table 1). It should be noted that the RdRp domain encoded by ORF1 of RdFV, pyrrhocoris apterus virus 1 and wuhan heteroptera virus 1 are clustered together in the phylogenetic tree of Virgaviridae-like RdRp domains (
Supplementary Figure S1).
Another cluster of RdRp domains of negev-like viruses encoding two SP24 gene copies includes, particularly, zeugodacus cucurbitae negev-like virus, broome virga-like virus 1, bactrocera oleae negev-like virus and bactrocera dorsalis negev-like virus isolate Bz infecting mosquitoes and invasive fruit flies from order
Diptera (class
Insecta) [
21,
22] (
Table 1) (
Supplementary Figure S1). Our search also revealed negev-like VLRAs encoding two SP24 gene copies in the transcriptomes of class
Arachnida, specifically
Scotophaeus scutulatus (family
Gnaphosidae) and
Neoscona scylloides (family
Araneidae) (
Table 1).
Table 1.
Examples of the viruses and VLRAs encoding two or three SP24 gene copies.
Table 1.
Examples of the viruses and VLRAs encoding two or three SP24 gene copies.
| Virus and host species |
Host Class/Order |
Accession number |
recilia dorsalis filamentous virus
Recilia dorsalis
|
Insecta/Hemiptera
|
OP326514 |
pyrrhocoris apterus virus 1
Pyrrhocoris apterus
|
Insecta/Hemiptera
|
MK024711 |
| VLRA Xestocephalus desertorum
|
Insecta/Hemiptera
|
GELC01067077 |
| VLRA Neoscona scylloides
|
Arachnida/Araneae |
ICMG01043884 |
| VLRA Scotophaeus scutulatus
|
Arachnida/Araneae |
ICQK01020075 |
atrato virga-like virus 6
Psorophora sp. |
Insecta/Diptera
|
MN661118 |
atrato virga-like virus 7
Psorophora albipes
|
Insecta/Diptera
|
MN661129 |
wuhan heteroptera virus 1
mixed insects |
Insecta/Hemiptera
|
KX883821 |
dougjudy virga-like virus
Calliphora vicina
|
Insecta/Diptera
|
MT129773 |
bactrocera oleae negev-like virus
Bactrocera oleae
|
Insecta/Diptera
|
MW310375 |
broome virga-like virus 1
Culex annulirostris
|
Insecta/Diptera
|
MT498833 |
bactrocera dorsalis negev-like virus
Bactrocera zonata
|
Insecta/Diptera
|
MW310386 |
zeugodacus tau negev-like virus
Zeugodacus tau
|
Insecta/Diptera
|
MW310340 |
zeugodacus cucurbitae negev-like virus
Zeugodacus cucurbitae
|
Insecta/Diptera
|
MW310350 |
entomophthora virgavirus A*
Entomophthora muscae
|
Entomophthoromycetes/ Entomophthorales
|
MK231110 |
3.1.1. Unique fungal virus encoding SP24 gene
Interestingly, the RdRp phylogenetic tree cluster containing Diptera-infecting viruses (in particular, broome virga-like virus 1) also contains the fungal entomophthora virgavirus A (
Supplementary Figure S1) with RNA genome encoding two SP24 gene copies (
Figure 1) (
Table 1). This virus genome has been found in transcriptome of the fungus
Entomophthora muscae, whose hosts are mainly insects of the order
Diptera [
23]. It can be assumed that this virus has evolutionarily originated due to transfer of an insect virus from a dipteran organism to a fungus and its subsequent adaptation to replicate in fungal hyphal bodies. On the other hand, entomophthora virgavirus A can be a true insect virus, and the finding of virus genome in fungal cells is a consequence of frequent passive transfer of virus RNA from host insects.
3.2. Virus RNA genomes encoding SP24 and infecting arthropods of class Arachnida
The transcriptomes of more than 1000 species of spiders (class
Arachnidae) [
18] recently deposited in the NCBI TSA database have the potential to greatly increase the number of novel RNA viruses encoding SP24. First of all, our search of the NCBI TSA database revealed many spiders infected by solenopsis invicta virus 17 (SINV-17) or its closely related strains (
Table 2). This monopartite RNA virus, first described in the red imported fire ant [
24], encodes three genes, namely replicase, ORF2 with DiSB-ORF_chro domain and SP24 gene (
Figure 1). SINV-17 RdRp domain is phylogenetically most closely related to hubei virga-like virus 8 from Myriapoda sub-philum [
5] and VLRA from the insect
Chrysoperla carnea, and more distantly related to sandewaviruses (
Supplementary Figure S1).
Table 2.
Examples of VLRAs with encoded proteins having >90% homology to SINV protein SP24.
Table 2.
Examples of VLRAs with encoded proteins having >90% homology to SINV protein SP24.
| VLRA origin and accession number |
Host Class/Order |
Identity to SINV (%) SP24 * |
|
Acheta domesticus, GHUU01023074 |
Insecta/Orthoptera
|
100 |
|
Orestes mouhotii, GDZN01041390 |
Insecta/Phasmatodea
|
95 |
|
Polistes metricus, GDHQ01029178 |
Insecta/Hymenoptera
|
97 |
|
Karoophasma biedouwense, GINP01229017 |
Insecta/Mantophasmatodea
|
99 |
|
Austrophasmatidae sp., GDYW01015845 |
Insecta/Mantophasmatodea
|
98 |
|
Gryllus bimaculatus, GISW01177432 |
Insecta/Orthoptera
|
99 |
|
Nylanderia pubens, JP785474 |
Insecta/Hymenoptera
|
96 |
|
Bassaniana decorate, ICCJ01037167 |
Arachnida/Araneae |
94 |
|
Weintrauboa contortipes, ICJX01013850 |
Arachnida/Araneae |
94 |
|
Meotipa spiniventris, IBDD01006961 |
Arachnida/Araneae |
94 |
|
Theridion zonulatum, IATV01002901 |
Arachnida/Araneae |
94 |
|
Thiania bhamoensis, IAGH01040226 |
Arachnida/Araneae |
94 |
|
Oxyopes macilentus, IAPY01004984 |
Arachnida/Araneae |
94 |
|
Cyrtophora unicolor, IAYW01028924 |
Arachnida/Araneae |
94 |
|
Argyrodes kumadai, IBBR01021510 |
Arachnida/Araneae |
94 |
|
Mendoza elongate, ICIJ01021408 |
Arachnida/Araneae |
89 |
|
Agelena labyrinthica, IBZQ01034950 |
Arachnida/Araneae |
94 |
|
Tegecoelotes corasides, IASA01013670 |
Arachnida/Araneae |
94 |
|
Octonoba sybotides, IAZZ01004659 |
Arachnida/Araneae |
94 |
|
Leucauge celebesiana, IAWQ01030888 |
Arachnida/Araneae |
94 |
|
Cyclosa mulmeinensis, ICEF01010633 |
Arachnida/Araneae |
94 |
|
Heptathela higoensis, IBEA01018510 |
Arachnida/Araneae |
94 |
|
Oxyopes sp., ICGS01044124 |
Arachnida/Araneae |
94 |
|
Argiope minuta, ICGZ01029236 |
Arachnida/Araneae |
94 |
|
Uloborus sp., IBJL01027593 |
Arachnida/Araneae |
94 |
|
Chikunia albipes, IAUZ01010102 |
Arachnida/Araneae |
94 |
|
Ebrechtella tricuspidata, IBYS01040384 |
Arachnida/Araneae |
94 |
|
Rhomphaea hyrcana, IAOI01013840 |
Arachnida/Araneae |
94 |
|
Latouchia swinhoei, IARH01001083 |
Arachnida/Araneae |
94 |
|
Lycosa ishikariana, IBVL01017570 |
Arachnida/Araneae |
94 |
|
Dolomedes pegasus, ICEZ01008221 |
Arachnida/Araneae |
94 |
|
Araneus seminiger, IAIN01035359 |
Arachnida/Araneae |
94 |
|
Thanatus bungei, IALY01037623 |
Arachnida/Araneae |
94 |
|
Iwogumoa interuna, IBKG01028989 |
Arachnida/Araneae |
94 |
|
Sinopoda okinawana, ICJY01003033 |
Arachnida/Araneae |
94 |
|
Thelcticopis severa, ICJF01009625 |
Arachnida/Araneae |
94 |
|
Heptathela higoensis, IBNB01017401 |
Arachnida/Araneae |
94 |
|
Heptathela yanbaruensis, ICKJ01022680 |
Arachnida/Araneae |
94 |
|
Mendoza elongata, ICIJ01011610 |
Arachnida/Araneae |
94 |
|
Iwogumoa interuna, IBKG01014765 |
Arachnida/Araneae |
94 |
|
Dolomedes triton, GGRN01030862 |
Arachnida/Araneae |
94 |
|
Anolis carolinensis, GAFD01024809 |
Lepidosauria/Iguania |
98 |
|
Sceloporus jarrovii, GIWZ010003422 |
Lepidosauria/Iguania |
99 |
|
Protopterus annectens, GGXP01055242 |
Euteleostomi/Ceratodontiformes |
97 |
Spiders represent only a part of the arthropod hosts for SINV-17. Our transcriptome search revealed seven insect species that can be infected by SINV-17 (
Table 2). Moreover, it is quite unexpected that sequences highly similar to SINV-17 were found in a search of TSA databases of vertebrate species, namely the West African the lungfish (
Protopterus annectens), the Yarrow’s spiny lizard (
Sceloporus jarrovii) and the lizard
Anolis carolinensis (
Table 2).
Additionally, we revealed many novel spider viruses encoding SP24 that are distinct from SINV-17 (
Supplementary Figure S1). Previously, several viruses of this type have only been revealed in the golden orb-weaver spider (
Nephila clavipes) and the spider mite (
Tetranychus urticae) [
25].
3.3. Negev-like virus RNA genomes infecting species of metazoan taxa outside insects
The finding of VLRAs highly similar to the negev-like SINV-17 genome in vertebrates (see above) prompted us to perform another comprehensive search of the transcriptomes of non-arthropod metazoan taxa to find new negev-like virus RNA genomes. Our search revealed that viruses only distantly related to SINV-17 infect at least three fish species (
Table 3). These species are included in the infraclass
Teleostei (teleost fishes), clade
Acanthomorphata.
Table 3.
Examples of negev-like virus RNA genomes infecting species of metazoan taxa outside arthropods *.
Table 3.
Examples of negev-like virus RNA genomes infecting species of metazoan taxa outside arthropods *.
| Host taxonomic position |
Name (Virus and VLRA) |
Accession number |
|
Teleostei; Anabantiformes
|
Parargyrops edita VLRA |
GICI01264178 |
|
Teleostei; Cyprinodontiformes
|
Aphanius dispar VLRA DN-106 |
GJEY01002386 |
|
Teleostei; Anabantiformes
|
Channa argus VLRA |
GEML01178446 |
|
Teleostei; Cyprinodontiformes
|
Aphanius dispar VLRA DN-113 |
GJEY01012897 |
|
Porifera; Demospongiae
|
Halisarca dujardinii VLRA |
GIFI01065598 |
|
Cnidaria; Actiniaria
|
beihai anemone virus 1 |
KX883744 |
|
Spiralia; Entoprocta
|
Loxosomella nordgaardi VLRA |
GIMJ01025245 |
|
Spiralia; Mollusca
|
Octopus vulgaris VLRA |
GKAX01007523 |
|
Spiralia; Mollusca
|
Charonia lampas VLRA |
GIQZ01066080 |
|
Spiralia; Mollusca
|
Potamilus streckersoni VLRA |
GJAA01000017 |
|
Spiralia; Platyhelminthes
|
dicrocoelium nege-like virius |
OP548619 |
|
Spiralia; Platyhelminthes
|
Girardia sp. VLRA |
GHOU01019505 |
|
Spiralia; Platyhelminthes
|
provittati virus |
BK059742 |
|
Spiralia; Platyhelminthes
|
fasciogiga virus |
BK059714 |
|
Spiralia; Platyhelminthes
|
fasciohepa virus 1 |
BK059715 |
|
Spiralia; Platyhelminthes
|
meterori virus |
BK059677 |
|
Spiralia; Platyhelminthes
|
clonorsi virus 1 |
BK059702 |
|
Spiralia; Platyhelminthes
|
fasciohepa virus 3 |
BK059717 |
|
Spiralia; Platyhelminthes
|
clonorsi virus 2 |
BK059703 |
|
Spiralia; Platyhelminthes
|
fasciohepa virus 2 |
BK059716 |
|
Ecdysozoa; Onychophora
|
Principapillatus hitoyensis VLRA |
GJGV01069003 |
|
Ecdysozoa; Nematoda
|
Nippostrongylus brasiliensis VLRA |
VDL74358 |
|
Ecdysozoa; Nematoda
|
Anguina tritici VLRA |
GKDZ01058772 |
|
Ecdysozoa; Nematoda
|
xingshan nematode virus 1 |
KX883837 |
|
Ecdysozoa; Nematoda
|
xingshan nematode virus 2 |
KX883836 |
| Ecdysozoa; Nematoda |
xinzhou nematode virus 1 |
KX883838 |
|
Ecdysozoa; Nematoda
|
Anisakis pegreffii |
HBXC01106013 |
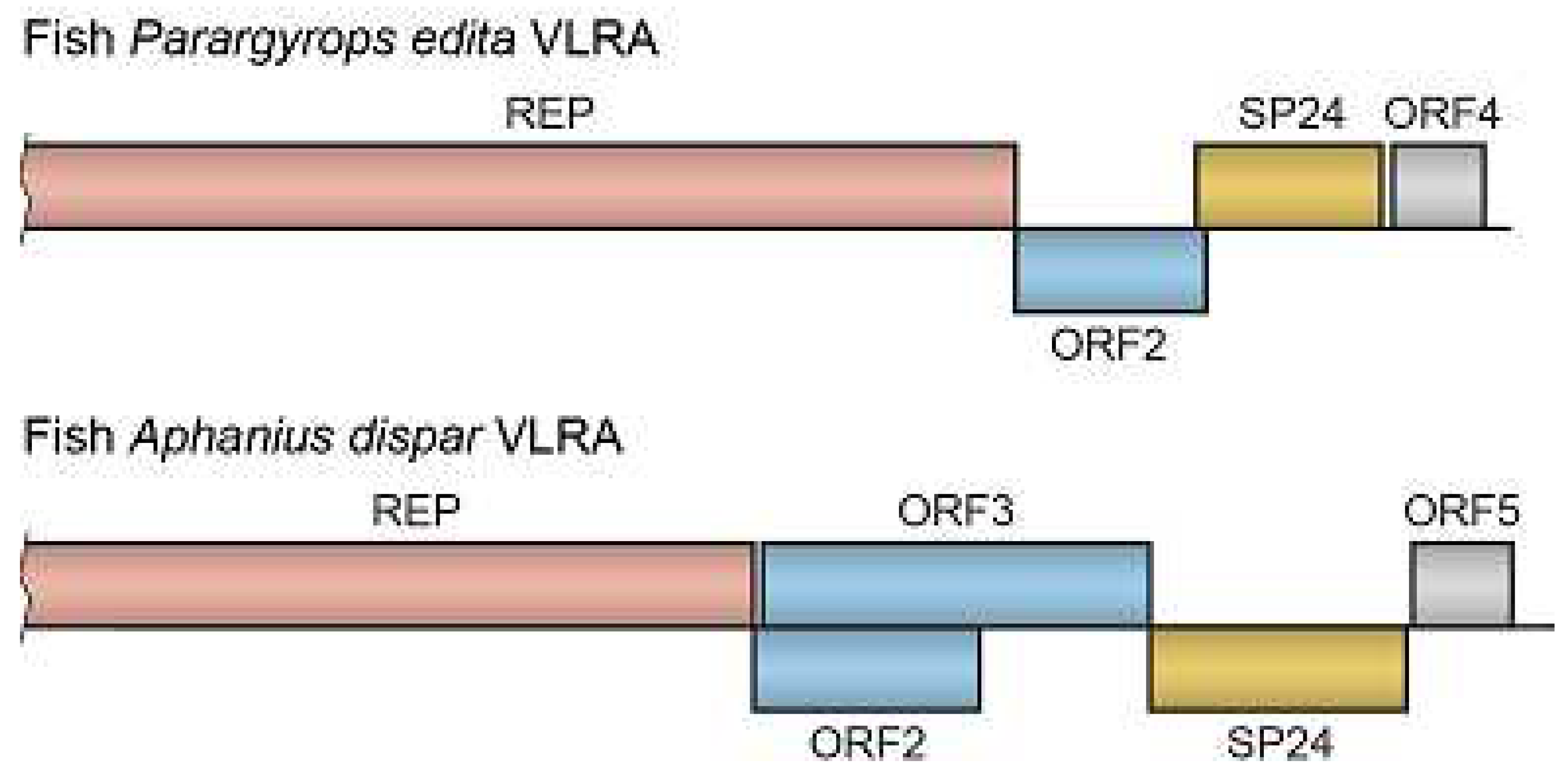
A VLRA of Crimson seabream (
Parargyrops edita, order
Anabantiformes) contains four ORFs (
Figure 2). The incomplete ORF1 encodes the C-terminal part of the replicase with RNA helicase and RdRp domains. ORF2 codes for a protein of 224 amino acids with two long hydrophobic segments and no significant similarity to known viral proteins. ORF3 (221 codons) represents a SP24 gene, while ORF4 encodes a protein of 109 amino acids with distant similarity to the VP-2 capsid protein of jingmen tick virus [
26]. Another fish virus encoding SP24 is represented by a VLRA from euryhaline fish (
Aphanius dispar, order
Cyprinodontiformes) (
Table 3). This virus genome encodes five proteins (
Figure 2). A replicase ORF is followed by overlapping ORF2 and ORF3, which encode hydrophobic proteins of 467 and 268 amino acids, respectively. ORF4 encodes an SP24 protein of 306 amino acid residues, while ORF5 encodes a small protein of 119 residues (
Figure 2). This virus is moderately similar to another negev-like virus (
Table 3), which also infects the fish
A. dispar and encodes five proteins, namely replicase, ORF2 protein (492 residues long), ORF3 protein (382 amino acids), ORF4 protein (209 amino acids) representing SP24, and ORF5 (141 residues). It should be noted that RdRp domains encoded by ORF1 of VLRAs from
Parargyrops edita and
Aphanius dispar are clustered together with negev virus and nelorpiviruses in the phylogenetic tree of
Virgaviridae-like RdRp domains (
Supplementary Figure S1).
Figure 2.
Gene organization of the selected fish VLRAs. See
Figure 1 for details of ORF naming and shading.
Figure 2.
Gene organization of the selected fish VLRAs. See
Figure 1 for details of ORF naming and shading.
Among metazoan species in the clade
Spiralia, negev-like viruses encoding SP24 have only been identified in flatworms (phylum
Platyhelminthes) [
17]. Our searches of the NCBI TSA databases revealed additional flatworm viruses that encode SP244-like proteins (
Table 3). One more SP24-encoding negev-like virus was found by our search in the goblet worm
(Loxosomella nordgaardi), which belongs to
Entoprocta, another phylum of
Spiralia [
27,
28]
(Table 3). In addition, novel viruses encoding SP24 were found in species of phylum
Mollusca (classes
Bivalvia,
Gastropoda and
Cephalopoda). Specifically, these animals include
Octopus vulgaris (class
Cephalopoda, order
Octopoda),
Charonia lampas (triton shell, class
Gastropoda, order
Littorinimorpha) and
Potamilus streckersoni (class
Bivalvia, order
Unionida) [
28] (
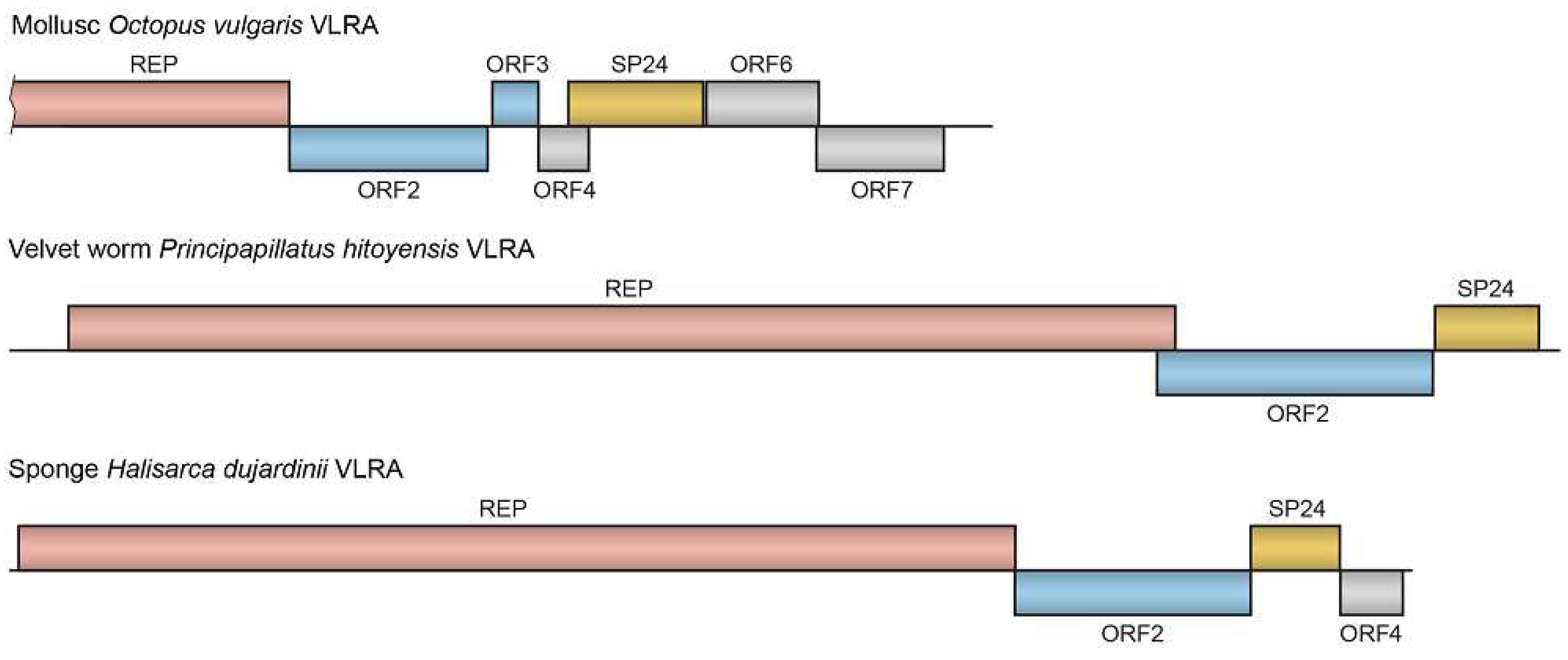
Table 3). The octopus SP24-encoding VLRA contains seven ORFs (
Figure 3). A partial replicase ORF is followed by ORF2, which encodes a hydrophobic protein of 442 residues with a DiSB-like chro domain. ORF3 encodes a protein of 103 residues with a long predicted transmembrane region. ORF4 encodes a small protein (110 residues) and overlaps with ORF5 of the SP24 protein (304 residues), while ORF6 encodes a highly hydrophobic protein of 254 residues and overlaps ORF7 (285 residues) (
Figure 3). In the RdRp phylogenetic tree, RdRp domains of ORF1 in VLRAs from
Octopus vulgaris and
Charonia lampas are positioned in a large cluster that is quite distant from a subtree containing RdRps from flatworm negev-like viruses (
Supplementary Figure S1).
In the animal clade
Ecdysozoa, species infected by negev-like viruses encoding SP24 were found not only in the phylum
Arthropoda, but also in the phylum
Nematoda (
Table 3). Furthermore, we found that the velvet worm
Principapillatus hitoyensis, which belongs to the phylum
Onychophora, which is phylogenetically closest to
Arthropoda [
28], can be infected by a negev-like virus (
Table 3). This velvet worm-specific virus encodes three proteins, the ORF1 replicase with viral methyltransferase domain, macro domain representing an ADP-ribose binding module, OTU domain family (cysteine proteases), superfamily 1 viral RNA helicase domain and the C-terminal RdRp domain; the ORF2 protein with DiSB-like chro domain; the ORF3 protein with SP24 domain (
Figure 3).
Figure 3.
Gene organization of the selected VLRAs. See
Figure 1 for details of ORF naming and shading.
Figure 3.
Gene organization of the selected VLRAs. See
Figure 1 for details of ORF naming and shading.
The phylum
Porifera (sponges) and the phylum
Cnidaria contain species that are among the most primitive metazoan organisms [
28]. In this study, we report for the first time a sponge VLRA that corresponds to a distant relative of negeviruses (
Table 3). This sponge-specific virus infects
Halisarca dujardinii (phylum
Porifera, class
Demospongiae, order
Dendroceratida) and encodes four proteins (
Figure 3). The ORF1-encoded replicase has three domains: a replicase with viral methyltransferase domain, a superfamily 1 viral RNA helicase domain, and the C-terminal RdRp domain. ORF1 is followed by ORF2, which encodes a hydrophobic protein of 524 amino acid residues. ORF3 encodes an SP24-like protein (200 residues), while ORF4 encodes a small protein of 136 residues (
Figure 3). Previously, a transcriptome search of sea anemones (phylum
Cnidaria, class
Hexacorallia, order
Actiniaria) has revealed a negev-like virus encoding SP24 [
5]. This virus, named beihai anemone virus 1 (
Table 3), encodes six proteins. ORF1 encodes a replicase with viral methyltransferase domain, macro domain representing an ADP-ribose binding module, superfamily 1 viral RNA helicase domain and the C-terminal RdRp domain. ORF2 encodes a protein (166 residues) that has no counterparts among known viral proteins and overlaps with ORF3, which encodes a highly hydrophobic protein (163 amino acids). ORF4 protein (438 amino acids) contains DiSB-like chro domain, while ORF5 (203 residues) encodes SP24 and ORF6 encodes a small protein (110 amino acids) with no similarity to known viral proteins. Phylogenetic analysis of the RdRp domain of beihai anemone virus 1 showed that the most similar virus is sanxia atyid shrimp virus 1 (SASV1), which infects shrimps of the family
Atyidae (phylum
Arthropoda, class
Malacostraca) (
Supplementary Figure S1). This shrimp-specific virus encodes 5 proteins. ORF1 encodes a replicase (2629 amino acids) with viral methyltransferase domain, macro domain, superfamily 1 viral RNA helicase domain and RdRp domain. ORF2 encodes a protein (128 residues) that has no counterparts among known viral proteins. ORF3 encodes a highly hydrophobic protein (164 amino acids). ORF4 protein (570 amino acids) contains the DiSB-like chro domain, while ORF5 (208 residues) encodes SP24. Our data show that several novel species of the class
Malacostraca can be infected by negev-like viruses encoding SP24. Some of these viruses appear to be related to SASV1 (
Supplementary Figure S1). In particular, eriocheir sinensis kita-like virus, which infects Chinese hairy crab (phylum
Arthropoda, class
Malacostraca, family
Varunidae), encodes 3 proteins (
Supplementary Figure S1): ORF1 protein is a replicase; ORF2 protein (665 amino acids) is a fusion of SASV1 ORF3 and ORF4 (DiSB) proteins; ORF3 (231 redidues) encodes SP24. However, some other negev-like viruses that infect the class
Malacostraca are only distantly related to SASV1. For example, the RdRp domain of the SP24-encoding VLRA of the amphipod
Gammarus fossarum (class
Malacostraca, family
Gammaridae) is phylogenetically more closely related to a protein encoded by a brown algae VLRA than to other negev-like viruses infecting class
Malacostraca (
Supplementary Figure S1).
3.4. SP24-encoding negev-like viruses infecting species of green plants and brown algae
The discovery of the SP24-encoding kitaviruses in plants [
9] suggests that the SP24 gene may also be a genomic component of other plant viruses. Indeed, a novel negev-like RNA virus, fragaria vesca associated virus 1 (FVaV-1), has been found to contain the SP24 gene, and an SP-24-encoding gene has been found in the genome of recently annotated chrysanthemum kita-like virus (CKLV) [
29]. Our search of plant transcriptomes revealed an additional plant VLRA encoding SP24 (
Table 4). The phylogeny of the RdRp domains in FVaV-1,
Triticum polonicum VLRA and
Glycine dolichocarpa VLRA (
Supplementary Figure S1) shows their rather close relationship to arthropod viruses. We hypothesize that these viruses may have emerged in virus evolution by a host change of an insect virus that was accompanied by an acquired ability to replicate or passively persist in plant cells (see also
Section 3.1.1.).
Table 4.
Negev-like virus RNA genomes infecting species of green plants and brown algae.
Table 4.
Negev-like virus RNA genomes infecting species of green plants and brown algae.
| Host taxonomic position |
Name (Virus and VLRA) |
Accession number |
|
Magnoliopsida, Rosales
|
fragaria vesca associated virus 1 |
MN895062 |
|
Magnoliopsida, Asterales
|
chrysanthemum kita like virus |
OP807956 |
|
Magnoliopsida, Caryophyllales
|
Amaranthus tuberculatus VLRA |
GGGT01091955 |
| Magnoliopsida, Lamiales |
Linaria vulgaris
VLRA* |
HBXK01234259 |
| Magnoliopsida, Caryophyllales |
Silene latifolia VLRA |
JO777742 |
| Magnoliopsida, Poales |
Hordeum vulgare
VLRA |
GGCQ01066052 |
| Magnoliopsida, Poales |
Alloteropsis semialata
VLRA |
GFYF01029264 |
| Magnoliopsida, Ranunculales |
Papaver nudicaule
VLRA |
GJOR01065662 |
| Magnoliopsida, Ranunculales |
Papaver armeniacum
VLRA |
GJOO01068528 |
| Magnoliopsida, Ranunculales |
Papaver pavoninum
VLRA |
GJOU01016337 |
|
Magnoliopsida, Fabales
|
Glycine dolichocarpa VLRA |
GGIW01009300 |
|
Magnoliopsida, Oxalidales
|
Elaeocarpus photiniifolius VLRA |
FX134396 |
| Pinopsida, Pinales |
Pseudotsuga menziesii VLRA |
GFFY01083799 |
|
Magnoliopsida, Apiales
|
Panax ginseng VLRA |
GDQW01045297 |
|
Phaeophyceae, Fucales
|
Sargassum vulgare VLRA |
GEHA01041094 |
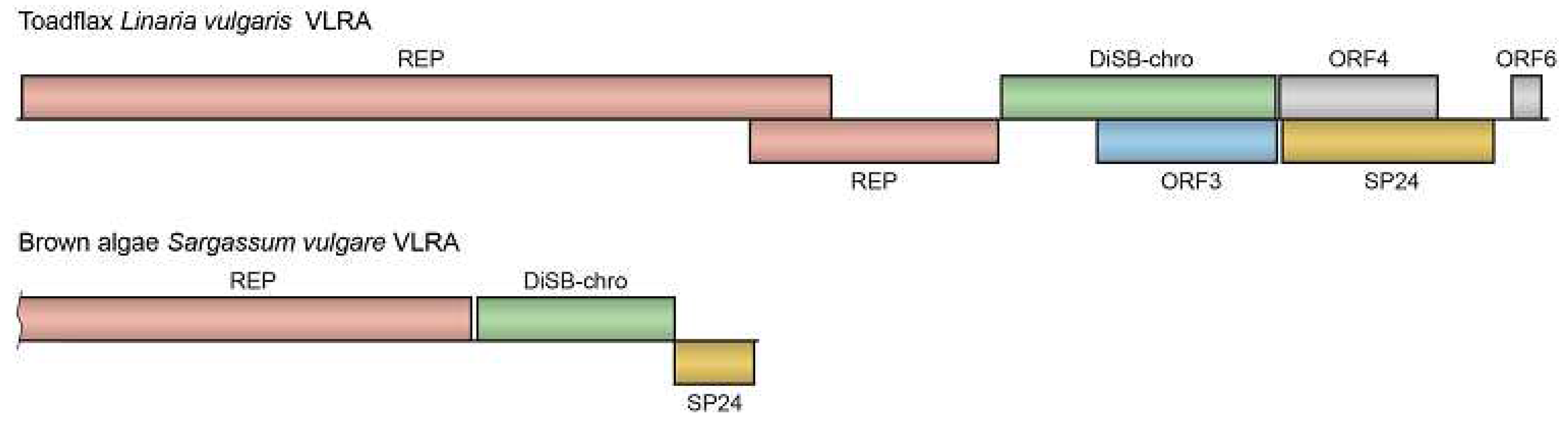
VLRA from the common toadflax
Linaria vulgaris (order
Lamiales, family
Plantaginaceae), although encoding SP24, differs significantly from other plant negev-like viruses in terms of RdRp phylogeny (
Supplementary Figure S1) and gene organization. Unlike FVaV-1 and CKLV, which have four genes,
Linaria vulgaris VLRA encodes six potential proteins (
Figure 4). Replicase protein of more than 1865 residues is potentially encoded by two overlapping ORFs: ORF1a protein has single annotated domain of superfamily 1 viral RNA helicase, while ORF1b overlaps ORF1 by 555 nucleotides and encodes a protein part with RdRp domain. ORF2 protein (637 amino acids) is distantly related to the predicted DiSB domain-containing glycoprotein of astegopteryx formosana nege-like virus 1. ORF3 is completely embedded in ORF2 and encodes a hydrophobic protein with no similarity to known viral proteins. ORF4 protein (329 amino acids) includes a cysteine-rich region, while overlapping ORF5 coding for protein possessing the SP24 domain. ORF6 encodes a small protein of 69 amino acids (
Figure 4). Our extensive search of the plant NCBI TSA database revealed that SP24-encoding virus of
Linaria vulgaris can infect several other monocot and dicot plants, namely,
Hordeum vulgare,
Alloteropsis semialata,
Papaver nudicaule,
Papaver armeniacum,
Papaver californicum,
Papaver atlanticum and
Silene latifolia (
Table 4). The RdRp domain of
Linaria vulgaris VLRA and its isolates specific for the listed above hosts forms a common cluster with virga-like viruses of brown algae in the PdRp phylogenetic tree (
Supplementary Figure S1).
Figure 4.
Gene organization of the selected VLRAs. See
Figure 1 for details of ORF naming and shading.
Figure 4.
Gene organization of the selected VLRAs. See
Figure 1 for details of ORF naming and shading.
Strikingly, one of brown algae virga-like viruses infecting
Sargassum vulgare (class
Phaeophyceae, order
Fucales) encodes SP24 (
Table 4). This VLRA encodes 3 proteins typical for nege-like viruses (
Figure 4). Incomplete ORF1 codes for replicase C-terminal part with superfamily 1 viral RNA helicase domain (pfam01443, e-value—2.74e-38) and RdRp domain (pfam00978, e-value—2.27e-85). ORF2 protein (490 residues) contains DiSB-like domain but has no significant sequence similarity to negevirus proteins. ORF3 encodes protein of 185 amino acids which possesses SP24 domain (e-value—1.06e-07).
3.5. NCBI conserved domain database search reveals horizontal transfer of SP24 protein genes to viruses of order Hepelivirales and insect genomes
The NCBI-conserved domain search tool [
30] was used to find potential SP24 genes encoded by cellular genomes. Among the four SP24-producing genes, three belong to insects, namely,
Macrosiphum euphorbiae, order
Hemiptera (locus CAI6355448, protein size—206 amino acids),
Drosophila melanogaster, order
Diptera (locus ABC86319, protein size—202 amino acids), and
Glossina morsitans morsitans, order
Diptera (locus ADD20599, protein size—271 amino acids). Another gene is found in the nematode
Nippostrongylus brasiliensis, order
Strongylida (locus VDL74358, protein size—184 amino acids). Thus, insect and nematode genomes may acquire viral SP24 genes by putative horizontal transfer from negev-like viruses known to infect species of both taxa (see above).
Another result of our NCBI CDD search is the pioneering discovery of SP24 genes in the genomes of two insect RNA viruses belonging to the order
Hepelivirales. The genome of hubei hepe-like virus 1 [
5] encodes three proteins (accession KX883803). ORF1 encodes a replicase (1661 amino acids) with the C-terminal RdRp domain showing obvious similarity to RdRp of hepeviruses (
Table 5). ORF2 encodes a protein (206 amino acids) with SP24 domain, whereas ORF3 encodes a longer protein (560 amino acids) with DiSB-chro domain. Thus, the general genome organization of this virus is different from that of negeviruses because of the inverse order of DiSB-chro and SP24 genes. Interestingly, although the replicase of sogatella furcifera hepe-like virus (1844 amino acids) is also related to viruses belonging to the order
Hepelivirales [
13] (
Table 5), the genome cistron order in this case is identical to that of negev virus (
Figure 1). ORF2 encodes a protein (622 amino acids) with DiSB-chro domain, and ORF3 encodes the SP24 protein (209 amino acids).
Table 5.
Similarity of Hubei hepe-like virus 1 RdRp to sequences in NCBI protein database.
Table 5.
Similarity of Hubei hepe-like virus 1 RdRp to sequences in NCBI protein database.
| Virus and accession number |
E-value |
Identity to HHLV1 RdRp (%) |
|
Astroviridae sp.; QYJ54474
|
2e-48 |
34.54 |
|
Hepelivirales sp.; WAY16503
|
9e-48 |
34.08 |
| Sogatella furcifera hepe-like virus; YP_009553211 |
3e-47 |
34.53 |
| Flumine bastrovirus 3; UQB75993 |
9e-47 |
32.84 |
| Hubei sediment bastro-like virus; QYF50028 |
2e-45 |
33.80 |
|
Hepelivirales sp.; WAY16393
|
2e-44 |
32.34 |
| Bastrovirus-like_virus/VietNam; YP_009333174 |
5e-44 |
33.42 |
| Microbat bastrovirus; UBK24595 |
1e-42 |
31.91 |
4. Discussion
The present and previously published studies show that there are several important aspects to the evolutionary history of the SP24 protein genes. The high evolutionary impact of the virion SP24 protein is evident because this protein is encoded by many members of at least three orders of the phylum
Kitrinoviricota (orders
Martellivirales,
Tolivirales and
Hepelivirales) as well as the family
Permutotetraviridae. Moreover, SP24 appears to be capable of forming virions of different morphologies, from pleiomorphic and elliptical shape to filamentous. In addition, some negev-like viruses of the order
Martellivirales encode two or even three copies of moderately related SP24 proteins. We propose that these variants of SP24 may have different functions, similar to the tandem duplicated capsid protein genes in members of
Closteriviridae [
31,
32] and novel virga-like viruses infecting
Hevea brasiliensis (rubber tree latent virus 1 and rubber tree latent virus 2) [
33]. In this context, it is important to note that one of the SP24 variants in RdFV plays a role in inter-tissue transport through the insect body [
19].
We hypothesize that the extremely broad host specificity of SP24-encoded viruses is related to an adaptive role of SP24. These viruses (mainly negev-like viruses) infect most animals (from primitive members of the phyla
Porifera and
Cnidaria to arthropods and vertebrates) as well as green plants and brown algae. Such a wide range of host organisms may result from the ability of viruses to rapidly adapt to new hosts upon occasional virus transmission between close and distant eukaryotic taxa. An illustrative example of this phenomenon is SINV-17 and its isolates, which can infect dozens of arthropods (mostly spiders) and some vertebrates. In addition to SINV-17, many insect RNA viruses are also known to be multi-host species [
34]. In this paper, similar wide host range covering both monocots and dicots is found for the unique plant virus isolated from
Linaria vulgaris and showing quite unusual genome organization. Some general properties of RNA viruses that allow high rates of adaptive evolution and facilitate species barriers crossing have been discussed [
34]. From an ecological perspective, previous studies have suggested that food-borne transmission is a common route for arthropod predators or scavengers, including spiders and ants. A specific route for arthropods to become infected with viruses is the ingestion of contaminated pollen or feces [
35,
36].
Our previous data have shown that in virgavirus-like insect viruses, the coding region of replicative RNA helicase is often integrated into insect retrotransposons [
37,
38]. It is currently clear that horizontal transfer of individual viral genes, including SP24 gene or genome fragments, into insect genomes is a fairly common phenomenon. Such endogenous viral elements (EVE) have been annotated in at least eight insect orders and can be assigned to at least 22 families of positive- and negative-sense RNA viruses [
14,
39,
40,
41]. Studies of EVE have already provided important insights into insect-virus interactions, including the discovery of novel forms of adaptive antiviral immunity [
38,
39,
40,
41,
42]. Thus, future studies of SP24 genes may shed a light on the functions of the protein in the context of viral genomes and genomes of insects and other eukaryotes.