Submitted:
31 October 2023
Posted:
01 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Culture and Morphological Analyses
2.3. Molecular Identification of Free-Living amoeba DNA Sequencing
2.4. Assessment of Viability
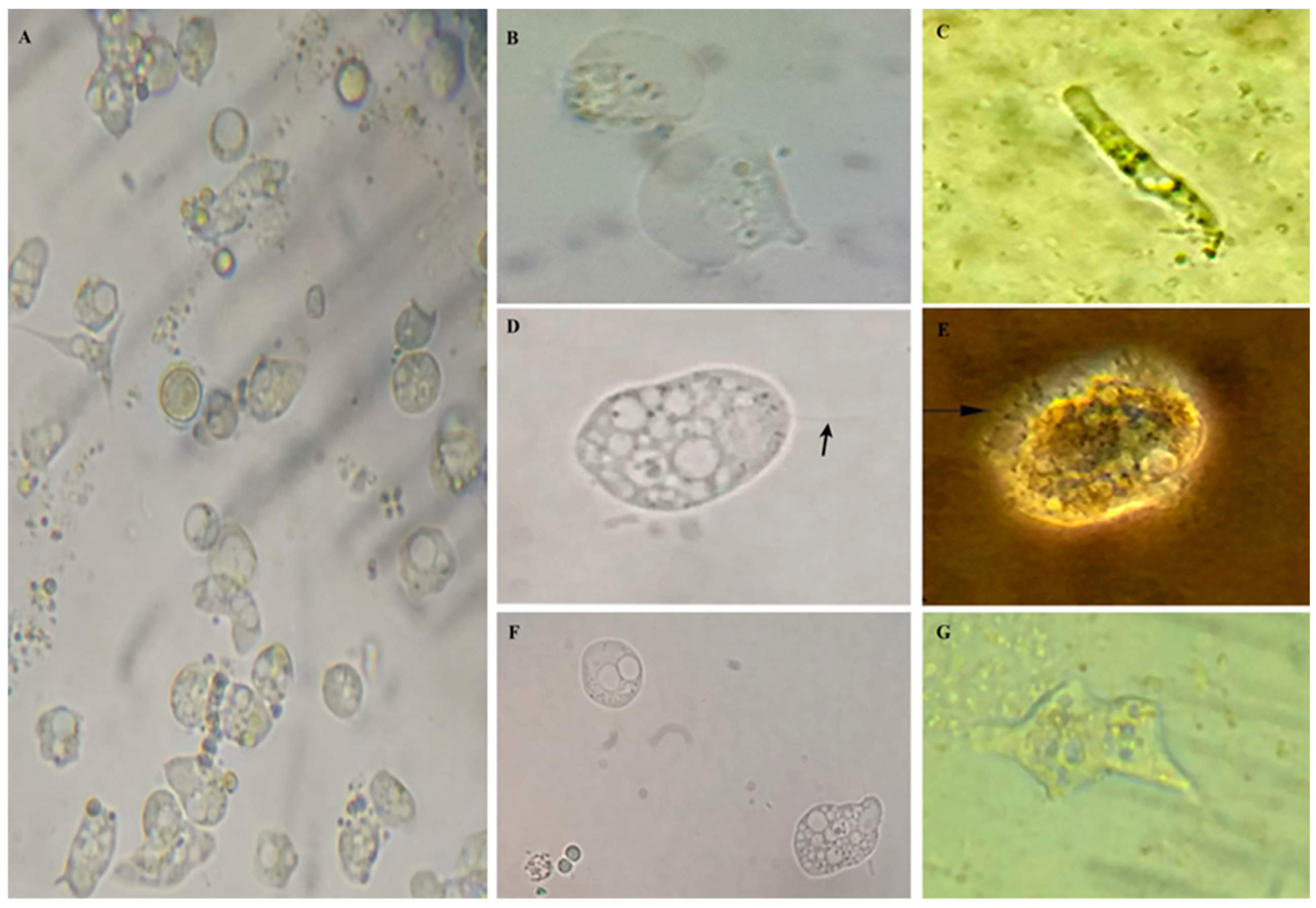
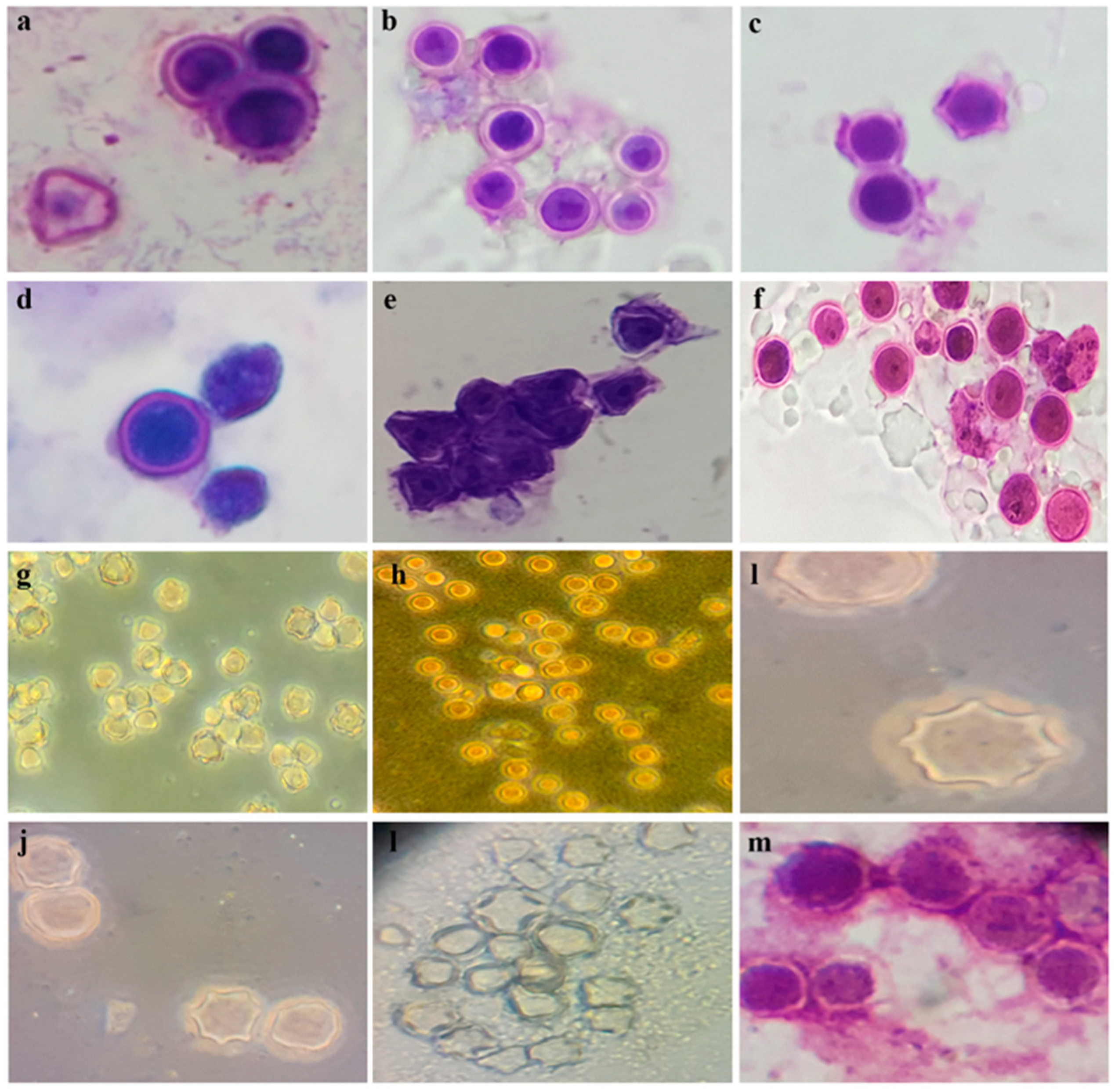
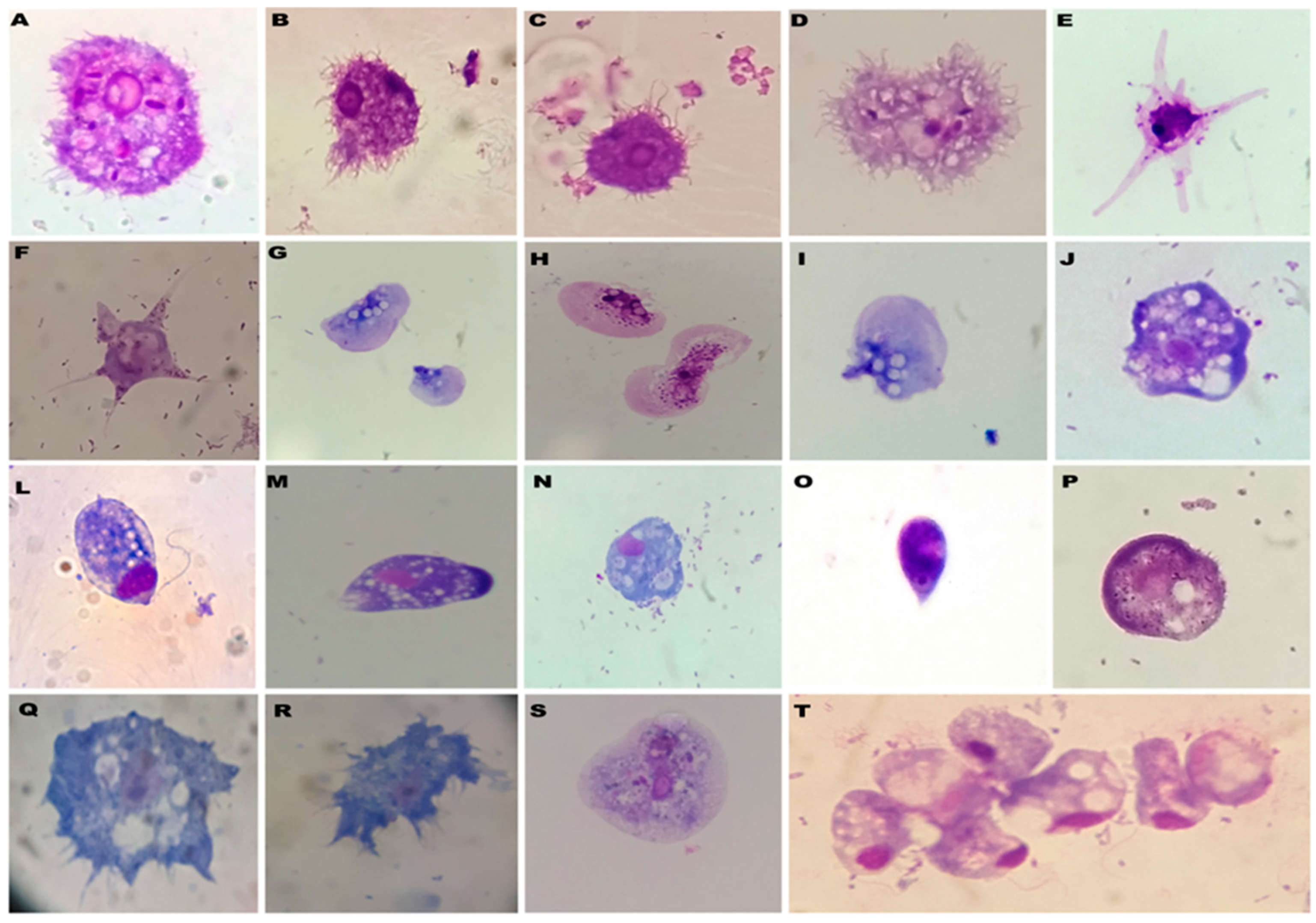
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berrilli F, Di Cave D, Cavallero S, D’Amelio S. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol. 2012, 2, 141. [Google Scholar] [CrossRef]
- Rastelli M, Cani PD, Knauf C. The Gut Microbiome Influences Host Endocrine Functions. Endocr Rev. 2019, 40, 1271–1284. [Google Scholar] [CrossRef] [PubMed]
- Thomas AM, Segata N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019, 17, 48. [Google Scholar] [CrossRef]
- Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019, 568, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Hooks KB, O’Malley MA. Contrasting Strategies: Human Eukaryotic Versus Bacterial Microbiome Research. J Eukaryot Microbiol. 2020, 67, 279–295. [Google Scholar] [CrossRef]
- Lind AL, Pollard KS. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome. 2021, 9, 58. [Google Scholar] [CrossRef]
- Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Manara S, Asnicar F, Beghini F, Bazzani D, Cumbo F, Zolfo M, et al. Microbial genomes from non-human primate gut metagenomes expand the primate-associated bacterial tree of life with over 1000 novel species. Genome Biol. 2019, 20, 299. [Google Scholar] [CrossRef]
- Ashford, R.W. Reid G.D.F., Butynski T.M. The intestinal faunas of man and mountain gorillas in a shared habitat. Ann. Trop. Med. Parasitol. 1990, 84, 337–340. [Google Scholar] [CrossRef]
- Muriuki, S.M.K. Murugu R.K., Munene E., Karere G.M., Chai D.C. Some gastro-intestinal parasites of zoonotic (public health) importance commonly observed in old world non-human primates in Kenya. Acta Trop. 1998, 71, 73–82. [Google Scholar] [CrossRef]
- Gómez, J.M. Nunn C.L., Verdú M. Centrality in primate-parasite networks reveals the potential for the transmission of emerging infectious diseases to humans. Proc. Natl. Acad. Sci. USA. 2013, 110, 7738–7741. [Google Scholar] [CrossRef] [PubMed]
- Gogarten JF, Calvignac-Spencer S, Nunn CL, Ulrich M, Saiepour N, Nielsen HV, et al. Metabarcoding of eukaryotic parasite communities describes diverse parasite assemblages spanning the primate phylogeny. Mol Ecol Resour. 2020, 20, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Mehta AP, Guirges SY. Acute amoebic dysentery due to free-living amoebae treated with methronidazole. J Trop Med Hyg. 1979, 82, 134–136. [Google Scholar]
- de Moura H, Salazar HC, Fernandes O, Lisboa DC, de Carvalho FG. [Free-living amoeba in the human intestine. Evidences of parasitism]. Rev Inst Med Trop Sao Paulo. 1985, 27, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, RS. Free-living amoebae recovered from human stool samples in Strongyloides agar culture. J Clin Microbiol. 2014, 52, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Chavatte N, Lambrecht E, Van Damme I, Sabbe K, Houf K. Free-living protozoa in the gastrointestinal tract and feces of pigs: Exploration of an unknown world and towards a protocol for the recovery of free-living protozoa. Vet Parasitol. 2016, 225, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Mulec, J., Dietersdorfer, E., Ustunturk-Onan, M., and Walochnik, J. Acanthamoeba and other free-living amoebae in bat guano, an extreme habitat. Parasitol Res. 2016, 115, 1375–1383. [CrossRef]
- Angelici MC, Walochnik J, Calderaro A, Saxinger L, Dacks JB. Free-living amoebae and other neglected protistan pathogens: Health emergency signals? Eur J Protistol. 2021, 77, 125760. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef]
- Carlesso AM, Simonetti AB, Artuso GL, Rott MB. [Isolation and identification of potentially pathogenic free-living amoebae in samples from environments in a public hospital in the city of Porto Alegre, Rio Grande do Sul]. Rev Soc Bras Med Trop. 2007, 40, 316–320. [Google Scholar] [CrossRef]
- Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol. 2004, 34, 1001–1027. [Google Scholar] [CrossRef] [PubMed]
- Pimentel LA, Dantas AF, Uzal F, Riet-Correa F. Meningoencephalitis caused by Naegleria fowleri in cattle of northeast Brazil. Res Vet Sci. 2012, 93, 811–812. [Google Scholar] [CrossRef] [PubMed]
- Henker LC, Lorenzett MP, Dos Santos DL, Virginio VG, Driemeier D, Rott MB, et al. Naegleria fowleri-associated meningoencephalitis in a cow in Southern Brazil-first molecular detection of N. fowleri in Brazil. Parasitol Res. 2021, 120, 2873–2879. [Google Scholar] [CrossRef] [PubMed]
- Madrigal Sesma MJ, Santillana Lopez I. Isolation of free-living amoebas from samples of respiratory origin]. Rev Sanid Hig Publica (Madr). 1989, 63, 63–72. [Google Scholar]
- Rivera F, Medina F, Ramirez P, Alcocer J, Vilaclara G, Robles E. Pathogenic and free-living protozoa cultured from the nasopharyngeal and oral regions of dental patients. Environ Res. 1984, 33, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Santos LC, Oliveira MS, Lobo RD, Higashino HR, Costa SF, van der Heijden IM, et al. Acanthamoeba spp. in urine of critically ill patients. Emerg Infect Dis. 2009, 15, 1144–1146. [Google Scholar] [CrossRef]
- Parija SC, Dinoop K, Venugopal H. Management of granulomatous amebic encephalitis: Laboratory diagnosis and treatment. Trop Parasitol. 2015, 5, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Arab-Mazar Z, Niyyati M, Lasjerdi Z, Spotin A, Alavi Darzam I, Gachkar L. Isolation, identification, and phylogenetic analysis of potentially pathogenic free-living amoebae isolated from nasal and oral mucosa of HIV/AIDS patients in Iran. Parasitol Res. 2019, 118, 3061–3066. [Google Scholar] [CrossRef] [PubMed]
- Kot K, Lanocha-Arendarczyk N, Kosik-Bogacka D. Immunopathogenicity of Acanthamoeba spp. in the Brain and Lungs. Int J Mol Sci. 2021, 22, 3. [Google Scholar] [CrossRef]
- Van der Henst C, Vanhove AS, Drebes Dorr NC, Stutzmann S, Stoudmann C, Clerc S, et al. Molecular insights into Vibrio cholerae’s intra-amoebal host-pathogen interactions. Nat Commun. 2018, 9, 3460. [Google Scholar] [CrossRef]
- Goni P, Fernandez MT, Rubio E. Identifying endosymbiont bacteria associated with free-living amoebae. Environ Microbiol. 2014, 16, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Barker J, Brown MR. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology (Reading). 1994, 140 Pt 6, 1253–1259. [Google Scholar] [CrossRef]
- Balczun C, Scheid PL. Free-Living Amoebae as Hosts for and Vectors of Intracellular Microorganisms with Public Health Significance. Viruses. 2017, 9, 65. [Google Scholar] [CrossRef]
- Coulon C, Collignon A, McDonnell G, Thomas V. Resistance of Acanthamoeba cysts to disinfection treatments used in health care settings. J Clin Microbiol. 2010, 48, 2689–2697. [Google Scholar] [CrossRef]
- Dey R, Rieger AM, Stephens C et al. Interactions of Pseudomonas aeruginosa with Acanthamoeba polyphaga Observed by Imaging Flow Cytometry. Cytometry A 2019, 95, 555–564. [Google Scholar] [CrossRef]
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980, 33, 1179–1183. [Google Scholar] [CrossRef]
- Dey R, Rieger A, Banting, G, Ashbolt N J, Role of amoebae for survival and recovery of ‘non-culturable’ Helicobacter pylori cells in aquatic environments. FEMS Microbiology Ecology 2020, 96, fiaa182.
- De Carli, G.A. Diagnóstico Laboratorial das Parasitoses Humanas - Métodos e Técnicas, 2 edn; Atheneu: São Paulo, Brazil, 2007. [Google Scholar]
- Liu, W. A simplified cytologic staining technic. Am J Clin Pathol. 1970, 54, 767–768. [Google Scholar] [CrossRef] [PubMed]
- Gatti S, Rama P, Matuska S, Berrilli F, Cavallero A, Carletti S, et al. Isolation and genotyping of Acanthamoeba strains from corneal infections in Italy. J Med Microbiol. 2010, 59, 1324–1330. [Google Scholar] [CrossRef]
- Nazar M, Haghighi A, Taghipour N, Ortega-Rivas A, Tahvildar-Biderouni F, Nazemalhosseini Mojarad E, et al. Molecular identification of Hartmannella vermiformis and Vannella persistens from man-made recreational water environments, Tehran, Iran. Parasitol Res. 2012, 111, 835–839. [Google Scholar] [CrossRef]
- Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Zanzani SA, Gazzonis AL, Epis S, Manfredi MT. Study of the gastrointestinal parasitic fauna of captive non-human primates (Macaca fascicularis). Parasitol Res. 2016, 115, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Drakulovski P, Bertout S, Locatelli S, Butel C, Pion S, Mpoudi-Ngole E, Delaporte E, Peeters M, Mallié M. Assessment of gastrointestinal parasites in wild chimpanzees (Pan troglodytes troglodytes) in southeast Cameroon. Parasitol Res. 2014, 113, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- Zhang H, Cheng X. Various brain-eating amoebae: the protozoa, the pathogenesis, and the disease. Front Med. 2021, 15, 842–866. [Google Scholar] [CrossRef] [PubMed]
- Schuster FL, De Jonckheere JF, Moura H, Sriram R, Garner MM, Visvesvara GS. Isolation of a thermotolerant Paravahlkampfia sp. from lizard intestine: biology and molecular identification. J Eukaryot Microbiol. 2003, 50, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Westmoreland SV, Rosen J, MacKey J, Romsey C, Xia DL, Visvesvera GS, et al. Necrotizing meningoencephalitis and pneumonitis in a simian immunodeficiency virus-infected rhesus macaque due to Acanthamoeba. Vet pathol. 2004, 41, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara GS, Martinez AJ, Schuster FL, Leitch GJ, Wallace SV, Sawyer TK, et al. Leptomyxid ameba, a new agent of amebic meningoencephalitis in humans and animals. J Clin Microbiol. 1990, 28, 2750–2756. [Google Scholar] [CrossRef] [PubMed]
- Anderson MP, Oosterhuis JE, Kennedy S, et al. Pneumonia and meningoencephalitis due to amoeba in a lowland gorilla. J Zoo Anim Med 1976, 17, 87–91. [Google Scholar]
- Canfield PJ, Vogelnest L, Cunningham ML, et al. Amoebic meningoencephalitis caused by Balamuthia mandrillaris in an orangutan. Aust Vet J 1997, 75, 97–100. [Google Scholar] [CrossRef]
- Rideout BA, Gardiner CH, Stalis IH, et al. Fatal infections with Balamuthia mandrillaris (a free-living amoeba) in gorillas and other Old World primates. Vet Pathol 1997, 34, 15–22. [Google Scholar] [CrossRef]
- Vellosa SAG, Mangini ACS, Nunes LR, Schlodtmann AG. Frequência de amebas de vida livre em fezes de indivíduos de uma creche da cidade de Säo Paulo / Frequency free-living amebas in the feces of persons from a nursery in São Paulo city. Rev Inst Adolfo Lutz. 1984, 44, 61–65. [Google Scholar] [CrossRef]
- Zaman, V. Acanthamoeba in human faeces from Karachi. Ann Trop Med Parasitol. 1999, 93, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Michel R, Schmid EN, Boker T, Hager DG, Muller KD, Hoffmann R, et al. Vannella sp. harboring Microsporidia-like organisms isolated from the contact lens and inflamed eye of a female keratitis patient. Parasitol Res. 2000, 86, 514–520.
- Montalbano Di Filippo M, Novelletto A, Di Cave D, Berrilli F. Identification and phylogenetic position of Naegleria spp. from geothermal springs in Italy. Exp Parasitol. 2017, 183, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Esboei BR, Fakhar M, Saberi R, Barati M, Moslemi M, Hassannia H, Dadimoghadam Y, Jalallou N. Genotyping and phylogenic study of Acanthamoeba isolates from human keratitis and swimming pool water samples in Iran. Parasite Epidemiol Control. 2020, 11, e00164. [Google Scholar] [CrossRef] [PubMed]
- Fuerst PA, Booton GC. Species, Sequence Types and Alleles: Dissecting Genetic Variation in Acanthamoeba. Pathogens 2020, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Memari F, Niyyati M, Lorenzo-Morales J, Jonaydi Z. Isolation and molecular characterization of Acanthamoeba strains isolated from the oral cavity of immunosuppressed individuals in Tehran, Iran. Acta parasitol. 2016, 61, 451–455. [Google Scholar]
- Niyyati M, Arab-Mazar Z, Lasjerdi Z, Lorenzo-Morales J, Espotin A, Yadegarynia D, et al. Molecular characterization of Acanthamoeba strains isolated from the oral cavity of hemodialysis patients in Iran. Parasitol Res. 2017, 116, 2965–2969. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D. Weissbrod, O., Barkan, E. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA. The Impact of Helicobacter pylori Infection on the Gastric Microbiota of the Rhesus Macaque. PLoS ONE. 2013, 8, e76375. [Google Scholar]
- Lam C, He L, Marciano-Cabral F. The Effect of Different Environmental Conditions on the Viability of Naegleria fowleri Amoebae. J Eukaryot Microbiol. 2019, 66, 752–756. [Google Scholar] [CrossRef]
- Sriram R, Shoff M, Booton G, Fuerst P, Visvesvara GS. Survival of Acanthamoeba cysts after desiccation for more than 20 years. J Clin Microbiol. 2008, 46, 4045–4048. [Google Scholar] [CrossRef] [PubMed]
- Ansari S, Yamaoka Y. Survival of Helicobacter pylori in gastric acidic territory. Helicobacter. 2017, 22, 10–1111. [Google Scholar] [CrossRef]
- Aqeel Y, Siddiqui R, Iftikhar H, Khan NA. The effect of different environmental conditions on the encystation of Acanthamoeba castellanii belonging to the T4 genotype. Exp Parasitol. 2013, 135, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Michel, R. Rohl, R., Schneider, H., 1982. Isolation of free-living amoebae from nasal mucosa of healthy individuals. Zentralbl. Bakteriol. Mikrobiol. Hyg. 1982, 176, 155–159. [Google Scholar]
- Lares-Jiménez LF, Borquez-Román MA, Alfaro-Sifuentes R, Meza-Montenegro MM, Casillas-Hernández R, Lares-Villa F. Detection of serum antibodies in children and adolescents against Balamuthia mandrillaris, Naegleria fowleri and Acanthamoeba T4. Exp Parasitol. 2018, 189, 28–33. [Google Scholar] [CrossRef]
- Alizadeh H, He YG, McCulley JP, et al. Successful immunization against Acanthamoeba keratitis in a pig model. Cornea 1995, 14, 180–186. [Google Scholar]
- Leher H, Zaragoza F, Taherzadeh S, Alizadeh H, Niederkorn JY. Monoclonal IgA antibodies protect against Acanthamoeba keratitis. Exp Eye Res. 1999, 69, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Garate M, Alizadeh H, Neelam S, Niederkorn JY, Panjwani N. Oral immunization with Acanthamoeba castellanii mannose-binding protein ameliorates amoebic keratitis. Infect Immun. 2006, 74, 7032–7034. [Google Scholar] [CrossRef]
- Kollars, T.M., Jr. Wilhelm, W.E. The Occurrence of Antibodies to Naegleria Species in Wild Mammals. The Journal of Parasitology. 1996, 82, 73–77. [Google Scholar] [CrossRef]
- Pietrzak B, Tomela K, Olejnik-Schmidt A, Mackiewicz A, Schmidt M. Secretory IgA in Intestinal Mucosal Secretions as an Adaptive Barrier against Microbial Cells. Int J Mol Sci. 2020, 21, 9254. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



