Submitted:
01 November 2023
Posted:
02 November 2023
You are already at the latest version
Abstract
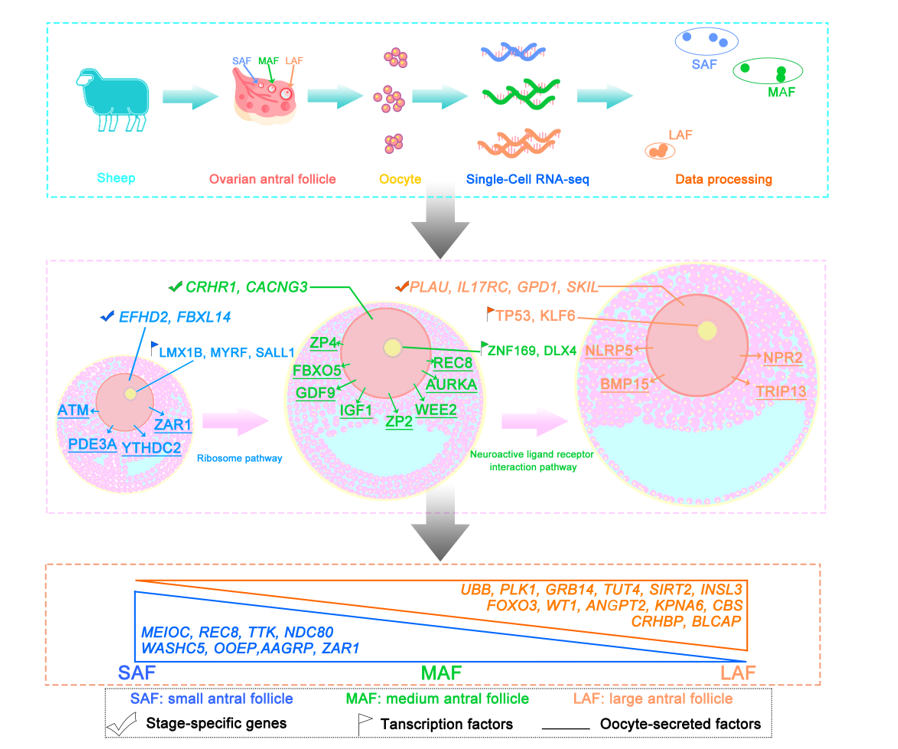
Keywords:
1. Introduction
2. Results
2.1. Overview of single-oocyte sequence data
2.2. Gene expression dynamics and transcriptional characteristics of oocytes during antral follicle development
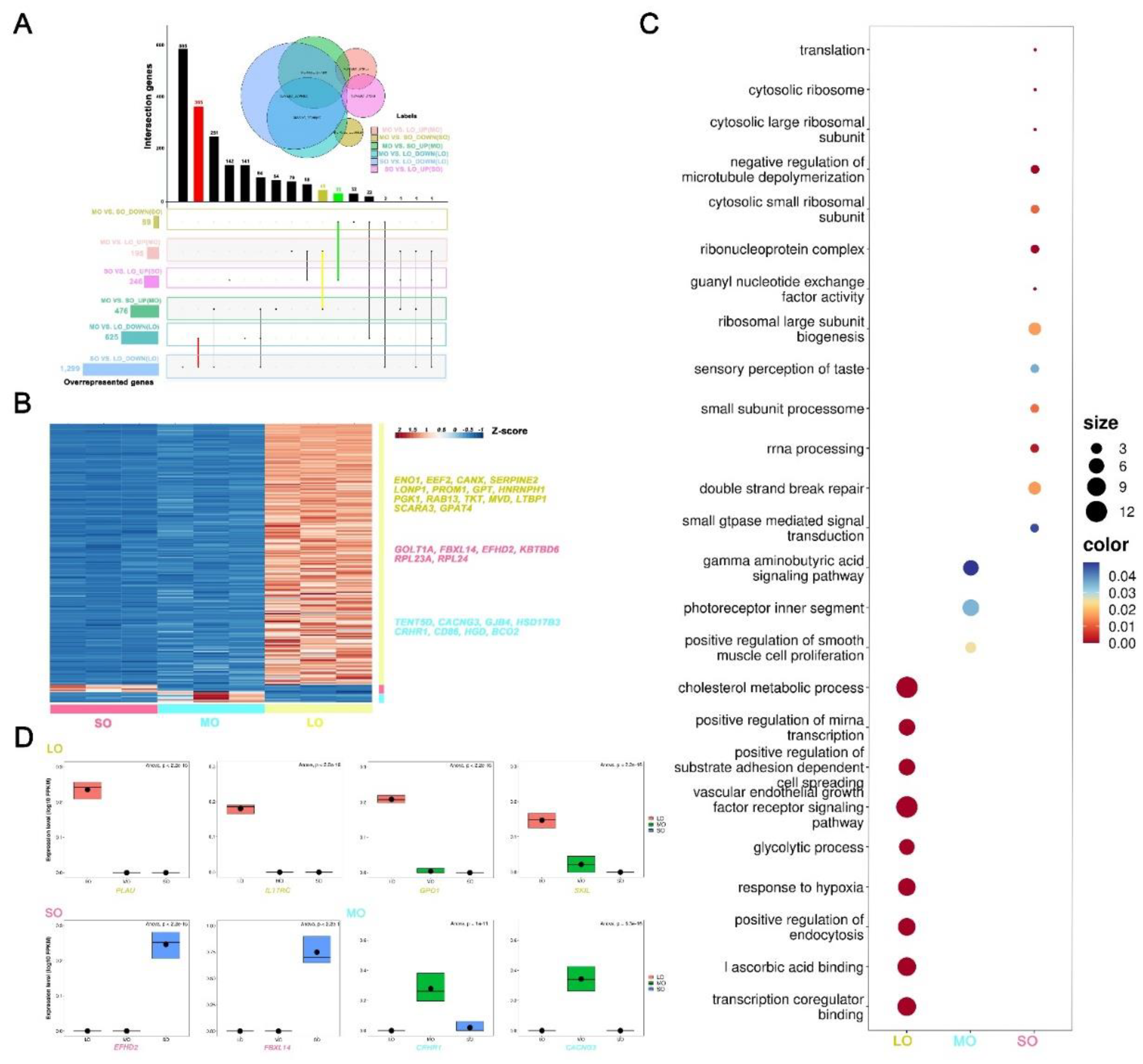
2.3. Gene expression signatures of oocytes at different antral follicle stages
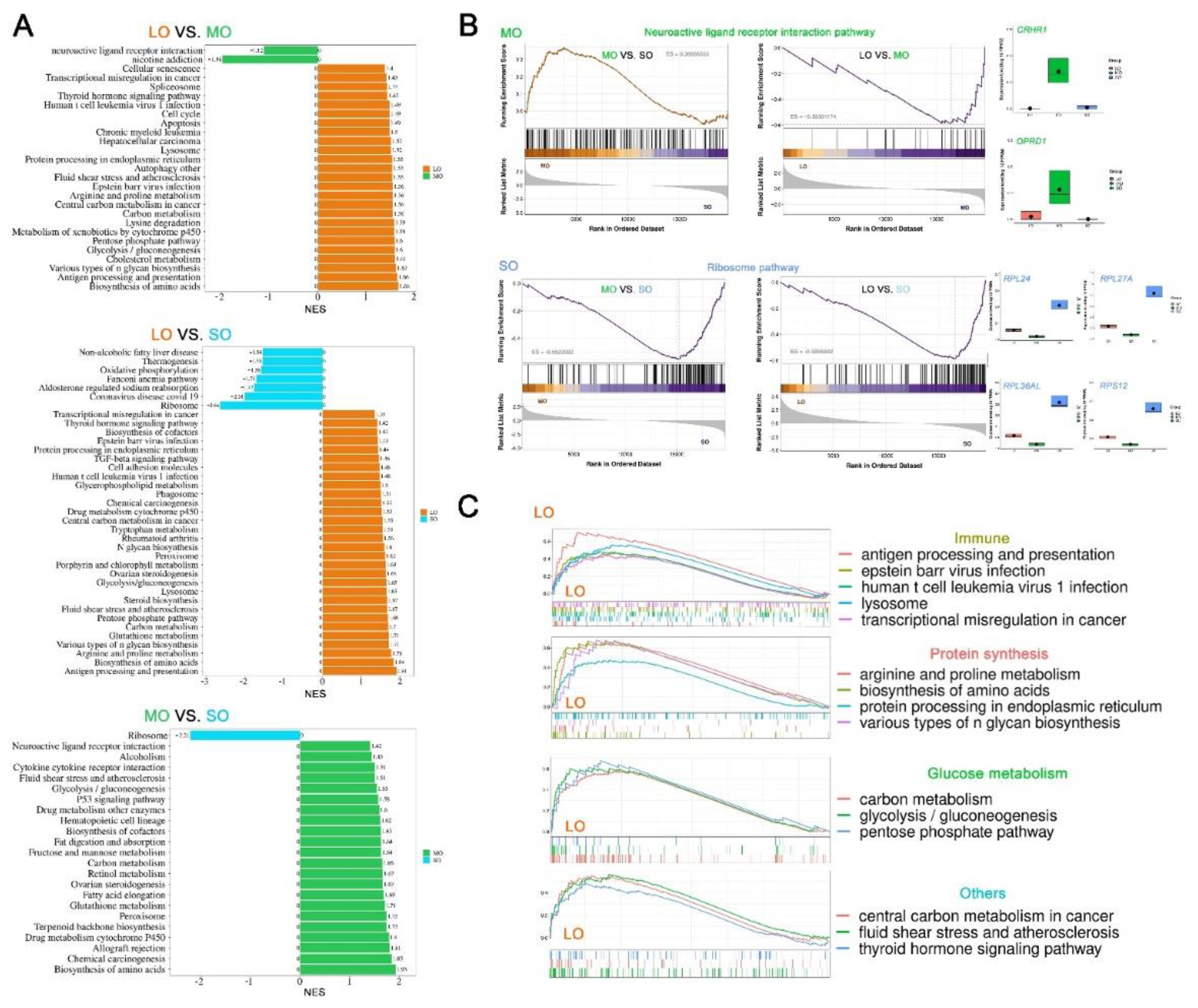
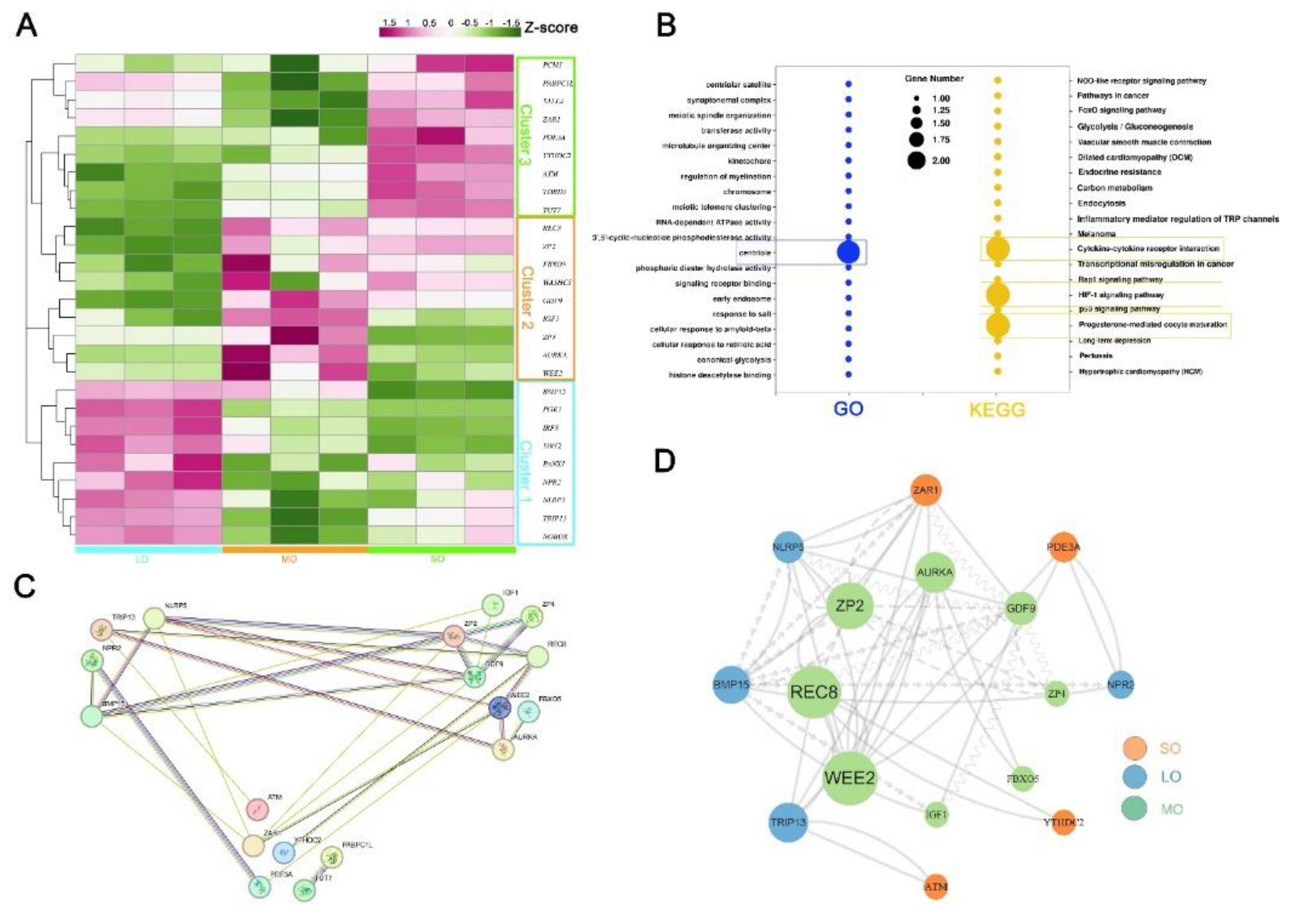
2.4. Characterization of key pathways throughout antral follicle development
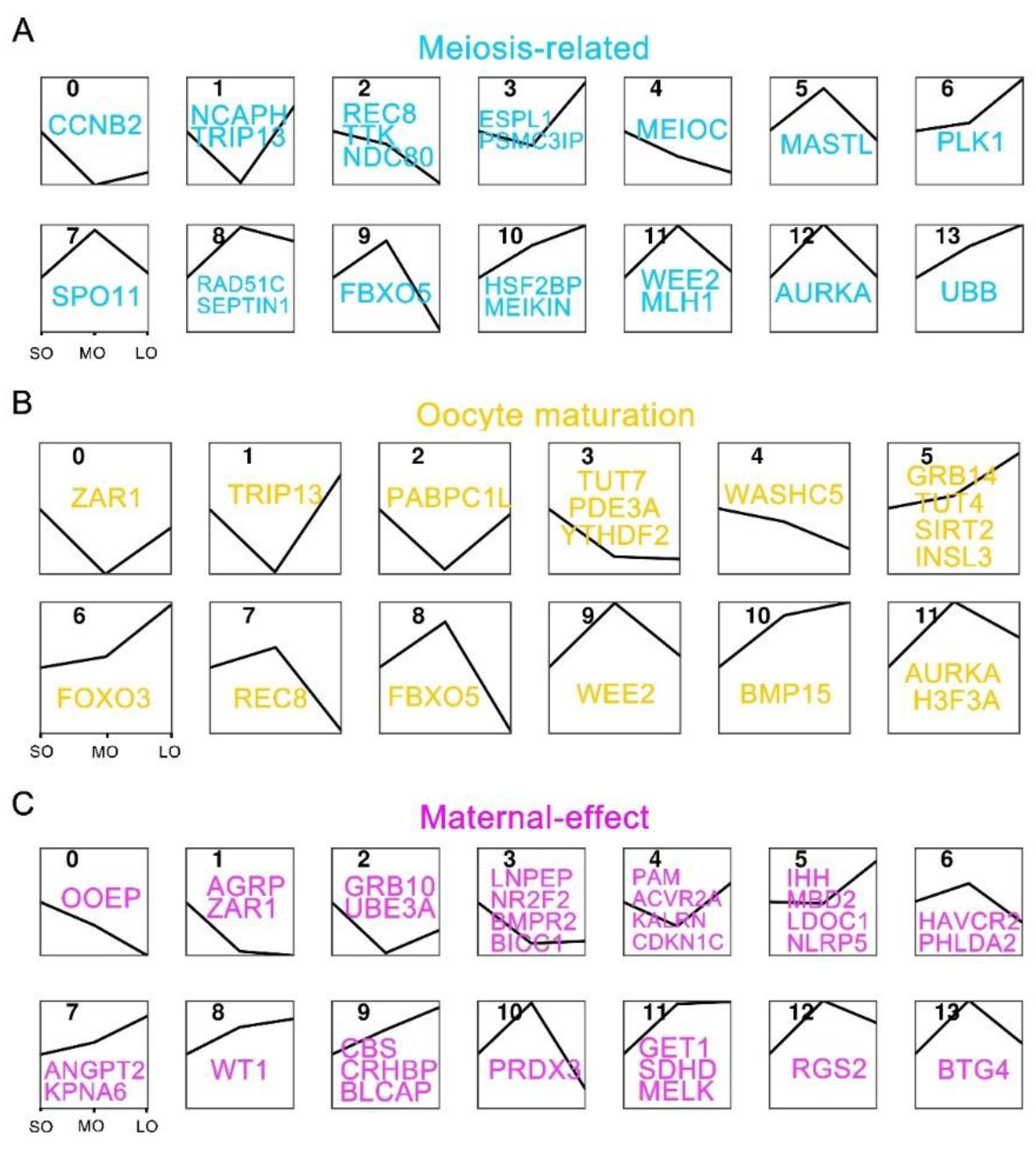
2.5. Expression patterns of oocyte-specific genes at antral follicle developmental stages
2.6. Identification of oocyte-secreted factors throughout antral follicle development
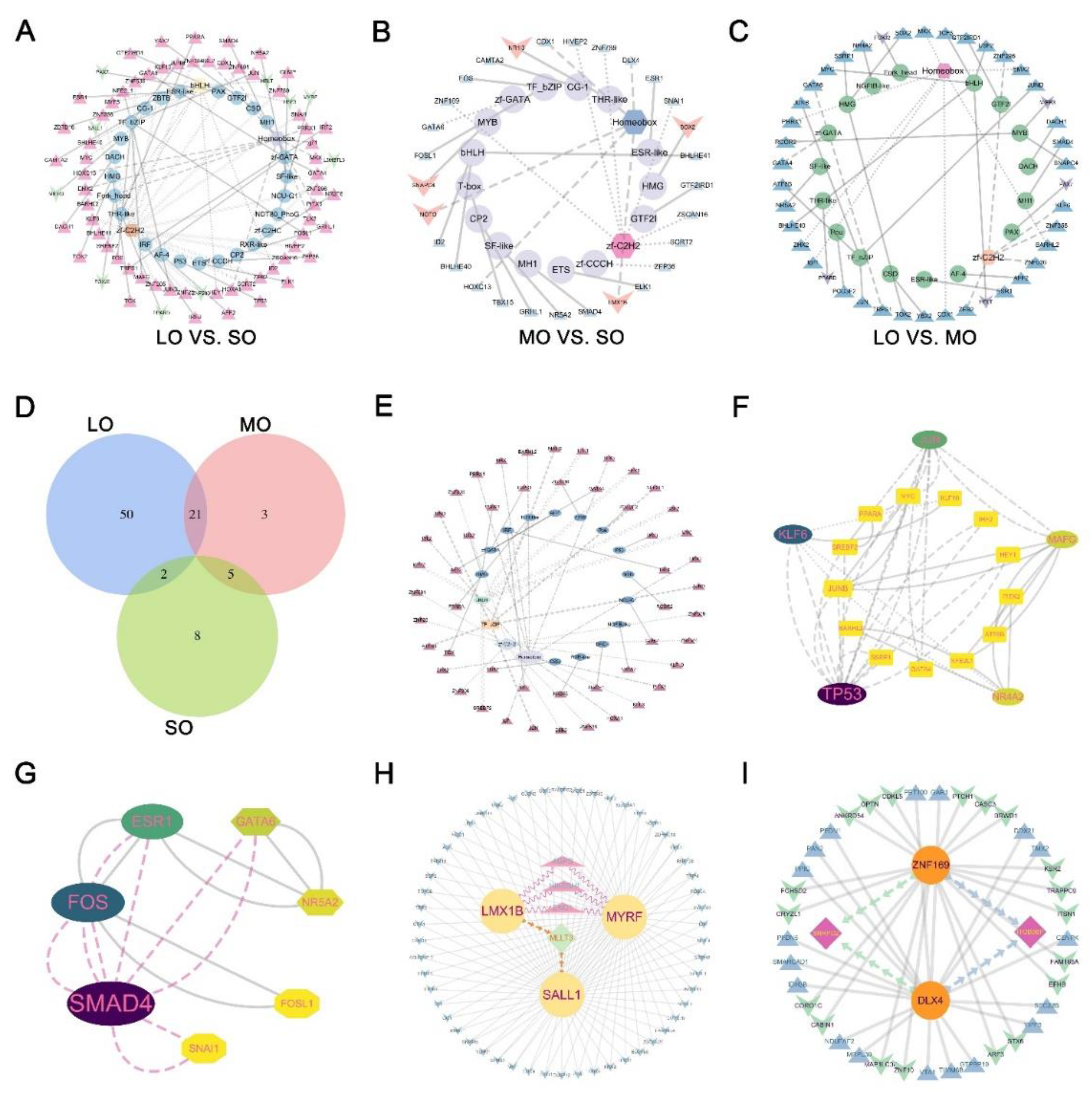
2.7. TF regulatory networks in the oocytes
3. Discussion
4. Materials and Methods
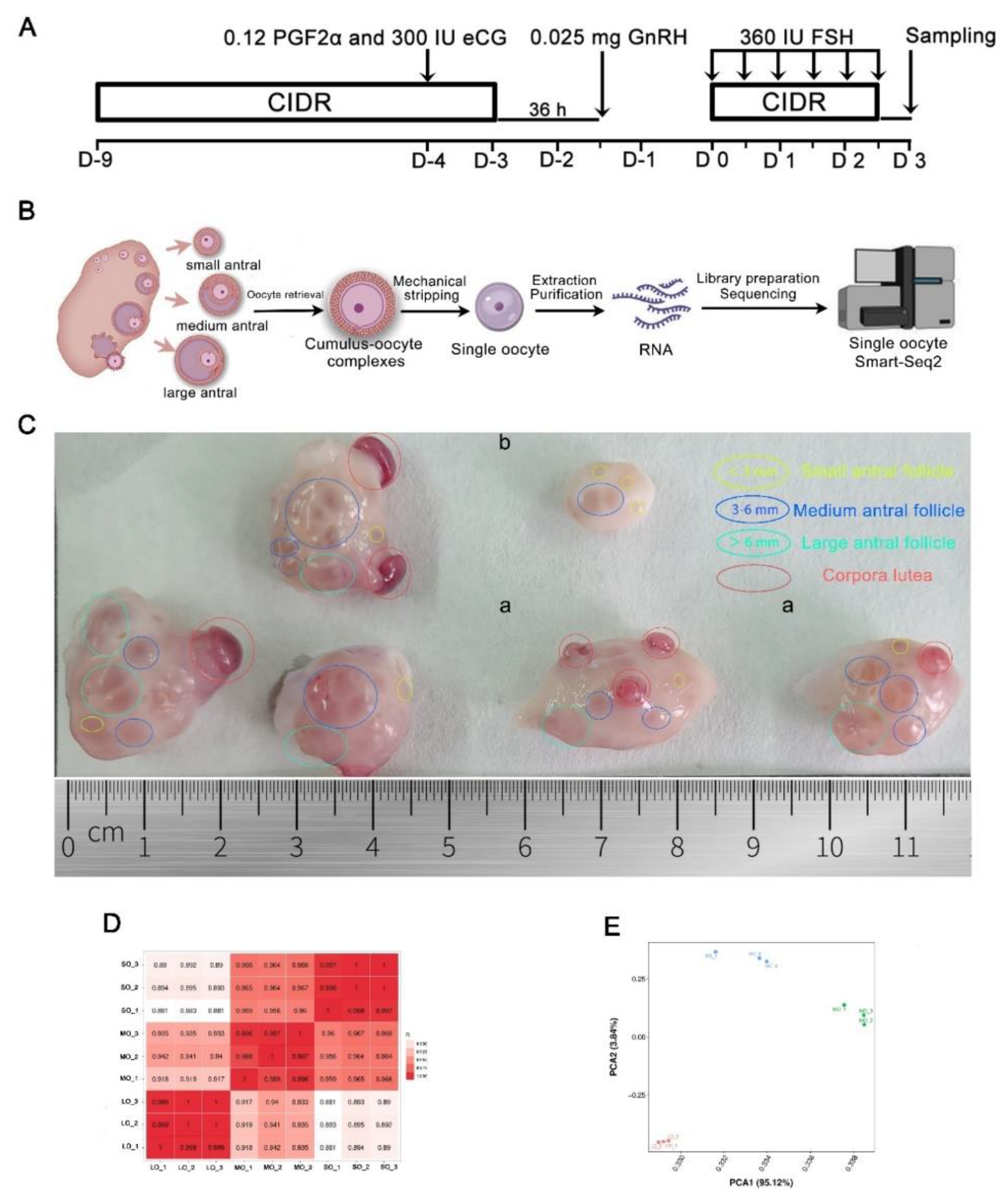
4.1. Ovarian stimulation and oocyte collection
4.2. Single-cell RNA sequencing and data analysis
4.3. PCA and identification of DEGs
4.4. Identification of stage-specific DEGs
4.5. Identifying expression patterns of oocyte-specific genes from DEGs
4.6. Identifying OSFs at sequential antral follicle developmental stages
4.7. Construction of transcriptional regulatory factor network in oocytes
4.8. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Gong, X.; Zhang, Y.; Ai, J.; Li, K. , Application of Single-Cell RNA Sequencing in Ovarian Development. Biomolecules 2022, 13, (1), 47. [Google Scholar] [CrossRef] [PubMed]
- Jozkowiak, M.; Hutchings, G.; Jankowski, M.; Kulcenty, K.; Mozdziak, P.; Kempisty, B.; Spaczynski, R. Z.; Piotrowska-Kempisty, H. , The Stemness of Human Ovarian Granulosa Cells and the Role of Resveratrol in the Differentiation of MSCs-A Review Based on Cellular and Molecular Knowledge. Cells 2020, 9, (6), 1418. [Google Scholar] [CrossRef]
- Eppig, J. J.; Wigglesworth, K.; Pendola, F. L. , The mammalian oocyte orchestrates the rate of ovarian follicular development. Proc Natl Acad Sci U S A 2002, 99, (5), 2890–2894. [Google Scholar] [CrossRef]
- Wang, S.; Gong, Y.; Wang, Z.; Greenbaum, J.; Xiao, H. M.; Deng, H. W. , Cell-specific network analysis of human folliculogenesis reveals network rewiring in antral stage oocytes. J Cell Mol Med 2021, 25, (6), 2851–2860. [Google Scholar] [CrossRef]
- Evans, A. C. , Ovarian follicle growth and consequences for fertility in sheep. Anim Reprod Sci 2003, 78, (3–4), 289-306. [Google Scholar] [CrossRef] [PubMed]
- Bartlewski, P. M.; Beard, A. P.; Rawlings, N. C. , An ultrasound-aided study of temporal relationships between the patterns of LH/FSH secretion, development of ovulatory-sized antral follicles and formation of corpora lutea in ewes. Theriogenology 2000, 54, (2), 229–245. [Google Scholar] [CrossRef]
- Keefe, D.; Kumar, M.; Kalmbach, K. , Oocyte competency is the key to embryo potential. Fertil Steril 2015, 103, (2), 317–322. [Google Scholar] [CrossRef]
- Leroy, J. L.; Valckx, S. D.; Jordaens, L.; De Bie, J.; Desmet, K. L.; Van Hoeck, V.; Britt, J. H.; Marei, W. F.; Bols, P. E. , Nutrition and maternal metabolic health in relation to oocyte and embryo quality: critical views on what we learned from the dairy cow model. Reprod Fertil Dev 2015, 27, (4), 693–703. [Google Scholar] [CrossRef]
- Lequarre, A. S.; Vigneron, C.; Ribaucour, F.; Holm, P.; Donnay, I.; Dalbiès-Tran, R.; Callesen, H.; Mermillod, P. , Influence of antral follicle size on oocyte characteristics and embryo development in the bovine. Theriogenology 2005, 63, (3), 841–859. [Google Scholar] [CrossRef]
- Ntostis, P.; Iles, D.; Kokkali, G.; Vaxevanoglou, T.; Kanavakis, E.; Pantou, A.; Huntriss, J.; Pantos, K.; Picton, H. M. , The impact of maternal age on gene expression during the GV to MII transition in euploid human oocytes. Hum Reprod 2021, 37, (1), 80–92. [Google Scholar] [CrossRef]
- Sorensen, R. A.; Wassarman, P. M. , Relationship between growth and meiotic maturation of the mouse oocyte. Dev Biol 1976, 50, (2), 531–536. [Google Scholar] [CrossRef]
- Eppig, J. J.; Schultz, R. M.; O'Brien, M.; Chesnel, F. , Relationship between the developmental programs controlling nuclear and cytoplasmic maturation of mouse oocytes. Dev Biol 1994, 164, (1), 1–9. [Google Scholar] [CrossRef]
- Kirillova, A.; Smitz, J. E. J.; Sukhikh, G. T.; Mazunin, I. , The Role of Mitochondria in Oocyte Maturation. Cells 2021, 10, (9), 2484. [Google Scholar] [CrossRef]
- Bernabé, B. P.; Woodruff, T.; Broadbelt, L. J.; Shea, L. D. , Ligands, Receptors, and Transcription Factors that Mediate Inter-Cellular and Intra-Cellular Communication during Ovarian Follicle Development. Reprod Sci 2020, 27, (2), 690–703. [Google Scholar] [CrossRef]
- Guo, Y.; Cai, L.; Liu, X.; Ma, L.; Zhang, H.; Wang, B.; Qi, Y.; Liu, J.; Diao, F.; Sha, J.; Guo, X. , Single-Cell Quantitative Proteomic Analysis of Human Oocyte Maturation Revealed High Heterogeneity in In Vitro-Matured Oocytes. Mol Cell Proteomics 2022, 21, (8), 100267. [Google Scholar] [CrossRef]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S. A. , Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc 2018, 13, (4), 599–604. [Google Scholar] [CrossRef]
- Freour, T.; Vassena, R. , Transcriptomics analysis and human preimplantation development. J Proteomics 2017, 162, 135–140. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, Q.; Dai, J.; Cui, Y.; Zhang, C.; Wen, X.; Li, J.; Xiao, Y.; Peng, X.; Liu, M.; Shen, B.; Sha, J.; Hu, Z.; Li, J.; Shu, W. , Single-cell RNA-Seq reveals a highly coordinated transcriptional program in mouse germ cells during primordial follicle formation. Aging Cell 2021, 20, (7), e13424. [Google Scholar] [CrossRef]
- Wang, J. J.; Ge, W.; Zhai, Q. Y.; Liu, J. C.; Sun, X. W.; Liu, W. X.; Li, L.; Lei, C. Z.; Dyce, P. W.; De Felici, M.; Shen, W. , Single-cell transcriptome landscape of ovarian cells during primordial follicle assembly in mice. PLoS Biol 2020, 18, (12), e3001025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Z.; Qin, Q.; Nisenblat, V.; Chang, H. M.; Yu, Y.; Wang, T.; Lu, C.; Yang, M.; Yang, S.; Yao, Y.; Zhu, X.; Xia, X.; Dang, Y.; Ren, Y.; Yuan, P.; Li, R.; Liu, P.; Guo, H.; Han, J.; He, H.; Zhang, K.; Wang, Y.; Wu, Y.; Li, M.; Qiao, J.; Yan, J.; Yan, L. , Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol Cell 2018, 72, (6), 1021–1034.e4. [Google Scholar] [CrossRef]
- Llonch, S.; Barragán, M.; Nieto, P.; Mallol, A.; Elosua-Bayes, M.; Lorden, P.; Ruiz, S.; Zambelli, F.; Heyn, H.; Vassena, R.; Payer, B. , Single human oocyte transcriptome analysis reveals distinct maturation stage-dependent pathways impacted by age. Aging Cell 2021, 20, (5), e13360. [Google Scholar] [CrossRef]
- Hu, W.; Zeng, H.; Shi, Y.; Zhou, C.; Huang, J.; Jia, L.; Xu, S.; Feng, X.; Zeng, Y.; Xiong, T.; Huang, W.; Sun, P.; Chang, Y.; Li, T.; Fang, C.; Wu, K.; Cai, L.; Ni, W.; Li, Y.; Yang, Z.; Zhang, Q. C.; Chian, R.; Chen, Z.; Liang, X.; Kee, K. , Single-cell transcriptome and translatome dual-omics reveals potential mechanisms of human oocyte maturation. Nat Commun 2022, 13, (1), 5114. [Google Scholar] [CrossRef]
- Maside, C.; Sánchez-Ajofrín, I.; Medina-Chávez, D.; Alves, B.; Garde, J. J.; Soler, A. J. , Oocyte Morphometric Assessment and Gene Expression Profiling of Oocytes and Cumulus Cells as Biomarkers of Oocyte Competence in Sheep. Animals (Basel) 2021, 11, (10),2818. [Google Scholar] [CrossRef] [PubMed]
- Mroczek, S.; Chlebowska, J.; Kuliński, T. M.; Gewartowska, O.; Gruchota, J.; Cysewski, D.; Liudkovska, V.; Borsuk, E.; Nowis, D.; Dziembowski, A. , The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat Commun 2017, 8, (1), 619. [Google Scholar] [CrossRef]
- Nemethova, M.; Radvanszky, J.; Kadasi, L.; Ascher, D. B.; Pires, D. E.; Blundell, T. L.; Porfirio, B.; Mannoni, A.; Santucci, A.; Milucci, L.; Sestini, S.; Biolcati, G.; Sorge, F.; Aurizi, C.; Aquaron, R.; Alsbou, M.; Lourenço, C. M.; Ramadevi, K.; Ranganath, L. R.; Gallagher, J. A.; van Kan, C.; Hall, A. K.; Olsson, B.; Sireau, N.; Ayoob, H.; Timmis, O. G.; Sang, K. H.; Genovese, F.; Imrich, R.; Rovensky, J.; Srinivasaraghavan, R.; Bharadwaj, S. K.; Spiegel, R.; Zatkova, A. , Twelve novel HGD gene variants identified in 99 alkaptonuria patients: focus on 'black bone disease' in Italy. Eur J Hum Genet 2016, 24, (1), 66–72. [Google Scholar] [CrossRef]
- Kwon, M.; Lee, J. H.; Yoon, Y.; Pleasure, S. J.; Yoon, K. , The CRHR1/CREB/REST signaling cascade regulates mammalian embryonic neural stem cell properties. EMBO Rep 2023, 24, (2), e55313. [Google Scholar] [CrossRef]
- Kennedy, A.; Waters, E.; Rowshanravan, B.; Hinze, C.; Williams, C.; Janman, D.; Fox, T. A.; Booth, C.; Pesenacker, A. M.; Halliday, N.; Soskic, B.; Kaur, S.; Qureshi, O. S.; Morris, E. C.; Ikemizu, S.; Paluch, C.; Huo, J.; Davis, S. J.; Boucrot, E.; Walker, L. S. K.; Sansom, D. M. , Differences in CD80 and CD86 transendocytosis reveal CD86 as a key target for CTLA-4 immune regulation. Nat Immunol 2022, 23, (9), 1365–1378. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, R.; Cheng, Y.; Wu, X.; Xi, S.; Sun, Y.; Jiang, H. , Lidocaine inhibits the proliferation of lung cancer by regulating the expression of GOLT1A. Cell Prolif. 2017, 50, (5), e12364. [Google Scholar] [CrossRef]
- Genau, H. M.; Huber, J.; Baschieri, F.; Akutsu, M.; Dötsch, V.; Farhan, H.; Rogov, V.; Behrends, C. , CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol Cell 2015, 57, (6), 995–1010. [Google Scholar] [CrossRef]
- Liu, Y. T.; Liu, F.; Cao, L.; Xue, L.; Gu, W. T.; Zheng, Y. Z.; Tang, H.; Wang, Y.; Yao, H.; Zhang, Y.; Xie, W. Q.; Ren, B. H.; Xiao, Z. H.; Nie, Y. J.; Hu, R.; Wu, Z. B. , The KBTBD6/7-DRD2 axis regulates pituitary adenoma sensitivity to dopamine agonist treatment. Acta Neuropathol 2020, 140, (3), 377–396. [Google Scholar] [CrossRef]
- Cui, Y. H.; Kim, H.; Lee, M.; Yi, J. M.; Kim, R. K.; Uddin, N.; Yoo, K. C.; Kang, J. H.; Choi, M. Y.; Cha, H. J.; Kwon, O. S.; Bae, I. H.; Kim, M. J.; Kaushik, N.; Lee, S. J. , FBXL14 abolishes breast cancer progression by targeting CDCP1 for proteasomal degradation. Oncogene 2018, 37, (43), 5794–5809. [Google Scholar] [CrossRef]
- Vega, I. E. , EFhd2, a Protein Linked to Alzheimer's Disease and Other Neurological Disorders. Front. Neurosci. 2016, 10, (2), 150. [Google Scholar] [CrossRef]
- Li, H.; You, L.; Tian, Y.; Guo, J.; Fang, X.; Zhou, C.; Shi, L.; Su, Y. Q. , DPAGT1-Mediated Protein N-Glycosylation Is Indispensable for Oocyte and Follicle Development in Mice. Adv Sci (Weinh) 2020, 7, (14), 2000531. [Google Scholar] [CrossRef]
- Tan, Z.; Lu, W.; Li, X.; Yang, G.; Guo, J.; Yu, H.; Li, Z.; Guan, F. , Altered N-Glycan expression profile in epithelial-to-mesenchymal transition of NMuMG cells revealed by an integrated strategy using mass spectrometry and glycogene and lectin microarray analysis. J Proteome Res 2014, 13, (6), 2783–2795. [Google Scholar] [CrossRef]
- Anyaogu, D. C.; Hansen, A. H.; Hoof, J. B.; Majewska, N. I.; Contesini, F. J.; Paul, J. T.; Nielsen, K. F.; Hobley, T. J.; Yang, S.; Zhang, H.; Betenbaugh, M.; Mortensen, U. H. , Glycoengineering of Aspergillus nidulans to produce precursors for humanized N-glycan structures. Metab Eng 2021, 67, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Elsharkawi, I.; Wongkittichote, P.; Daniel, E. J. P.; Starosta, R. T.; Ueda, K.; Ng, B. G.; Freeze, H. H.; He, M.; Shinawi, M. , DDOST-CDG: Clinical and molecular characterization of a third patient with a milder and a predominantly movement disorder phenotype. J Inherit Metab Dis 2023, 46, (1), 92–100. [Google Scholar] [CrossRef]
- Li, R.; Albertini, D. F. , The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol 2013, 14, (3), 141–152. [Google Scholar] [CrossRef]
- Innocenti, F.; Fiorentino, G.; Cimadomo, D.; Soscia, D.; Garagna, S.; Rienzi, L.; Ubaldi, F. M.; Zuccotti, M. , Maternal effect factors that contribute to oocytes developmental competence: an update. J Assist Reprod Genet 2022, 39, (4), 861–871. [Google Scholar] [CrossRef]
- Hobeika, E.; Armouti, M.; Kala, H.; Fierro, M. A.; Winston, N. J.; Scoccia, B.; Zamah, A. M.; Stocco, C. , Oocyte-Secreted Factors Synergize With FSH to Promote Aromatase Expression in Primary Human Cumulus Cells. J Clin Endocrinol Metab 2019, 104, (5), 1667–1676. [Google Scholar] [CrossRef]
- Fernandes, R.; Tsuda, C.; Perumalsamy, A. L.; Naranian, T.; Chong, J.; Acton, B. M.; Tong, Z. B.; Nelson, L. M.; Jurisicova, A. , NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod 2012, 86, (5), 138, 1–10. [Google Scholar] [CrossRef]
- Rankin, T. L.; O'Brien, M.; Lee, E.; Wigglesworth, K.; Eppig, J.; Dean, J. , Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development 2001, 128, (7), 1119–1126. [Google Scholar] [CrossRef]
- Wu, X.; Viveiros, M. M.; Eppig, J. J.; Bai, Y.; Fitzpatrick, S. L.; Matzuk, M. M. , Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet 2003, 33, (2), 187–191. [Google Scholar] [CrossRef]
- Tachibana-Konwalski, K.; Godwin, J.; van der Weyden, L.; Champion, L.; Kudo, N. R.; Adams, D. J.; Nasmyth, K. , Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev 2010, 24, (22), 2505–2516. [Google Scholar] [CrossRef]
- Nasmyth, K.; Haering, C. H. , Cohesin: its roles and mechanisms. Annu Rev Genet 2009, 43, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chen, T.; Yu, M.; Bian, Y.; Cao, Y.; Ning, Y.; Su, S.; Zhang, J.; Zhao, S. , Novel WEE2 gene variants identified in patients with fertilization failure and female infertility. Fertil Steril 2019, 111, (3), 519–526. [Google Scholar] [CrossRef]
- Sun, R.; Li, M.; He, N.; Wen, X.; Zhang, J. , Molecular Characterization, Expression Profiles of SMAD4, SMAD5 and SMAD7 Genes and Lack of Association with Litter Size in Tibetan Sheep. Animals (Basel) 2022, 12, (17), 2232. [Google Scholar] [CrossRef]
- Wang, S.; Yang, H.; Fu, Y.; Teng, X.; Wang, C.; Xu, W. , The Key Role of Peroxisomes in Follicular Growth, Oocyte Maturation, Ovulation, and Steroid Biosynthesis. Oxid. Med. Cell. Longev. 2022, 2022, (4), 7982344. [Google Scholar] [CrossRef]
- Yang, C. X.; Song, Z. Q.; Pei, S.; Yu, X. X.; Miao, J. K.; Liang, H.; Miao, Y. L.; Du, Z. Q. , Single cell RNA-seq reveals molecular pathways altered by 7, 12-dimethylbenz[a]anthracene treatment on pig oocytes. Theriogenology 2020, 157, (1), 449–457. [Google Scholar] [CrossRef]
- Kim, S. G.; Jang, S. J.; Soh, J.; Lee, K.; Park, J. K.; Chang, W. K.; Park, E. W.; Chun, S. Y. , Expression of ectodermal neural cortex 1 and its association with actin during the ovulatory process in the rat. Endocrinology 2009, 150, (8), 3800–3806. [Google Scholar] [CrossRef]
- Conti, M.; Franciosi, F. , Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum Reprod Update 2018, 24, (3), 245–266. [Google Scholar] [CrossRef]
- Mercer, M.; Jang, S.; Ni, C.; Buszczak, M. , The Dynamic Regulation of mRNA Translation and Ribosome Biogenesis During Germ Cell Development and Reproductive Aging. Front Cell Dev Biol 2021, 9, 710186. [Google Scholar] [CrossRef] [PubMed]
- Biase, F. H.; Kimble, K. M. , Functional signaling and gene regulatory networks between the oocyte and the surrounding cumulus cells. BMC Genomics 2018, 19, (1), 351. [Google Scholar] [CrossRef] [PubMed]
- Rae, M. T.; Gubbay, O.; Kostogiannou, A.; Price, D.; Critchley, H. O.; Hillier, S. G. , Thyroid hormone signaling in human ovarian surface epithelial cells. J Clin Endocrinol Metab 2007, 92, (1), 322–327. [Google Scholar] [CrossRef]
- Sánchez, F.; Romero, S.; De Vos, M.; Verheyen, G.; Smitz, J. , Human cumulus-enclosed germinal vesicle oocytes from early antral follicles reveal heterogeneous cellular and molecular features associated with in vitro maturation capacity. Hum Reprod 2015, 30, (6), 1396–1409. [Google Scholar] [CrossRef]
- Singh, P.; Fragoza, R.; Blengini, C. S.; Tran, T. N.; Pannafino, G.; Al-Sweel, N.; Schimenti, K. J.; Schindler, K.; Alani, E. A.; Yu, H.; Schimenti, J. C. , Human MLH1/3 variants causing aneuploidy, pregnancy loss, and premature reproductive aging. Nat Commun 2021, 12, (1), 5005. [Google Scholar] [CrossRef]
- Lodde, V.; Modina, S.; Maddox-Hyttel, P.; Franciosi, F.; Lauria, A.; Luciano, A. M. , Oocyte morphology and transcriptional silencing in relation to chromatin remodeling during the final phases of bovine oocyte growth. Mol Reprod Dev 2008, 75, (5), 915–924. [Google Scholar] [CrossRef]
- Soh, Y. Q. S.; Mikedis, M. M.; Kojima, M.; Godfrey, A. K.; de Rooij, D. G.; Page, D. C. , Meioc maintains an extended meiotic prophase I in mice. PLoS Genet 2017, 13, (4), e1006704. [Google Scholar] [CrossRef] [PubMed]
- Tora, L.; Vincent, S. D. , What defines the maternal transcriptome? Biochem Soc Trans 2021, 49, (5), 2051–2062. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, X.; Zhen, P.; Tian, Y.; Ji, D.; Xue, K.; Yan, W.; Chai, J.; Liu, H.; Wang, W. , Cystathionine β-synthase is required for oocyte quality by ensuring proper meiotic spindle assembly. Cell Prolif 2022, 55, (12), e13322. [Google Scholar] [CrossRef]
- Kona, S. S.; Praveen Chakravarthi, V.; Siva Kumar, A. V.; Srividya, D.; Padmaja, K.; Rao, V. H. , Quantitative expression patterns of GDF9 and BMP15 genes in sheep ovarian follicles grown in vivo or cultured in vitro. Theriogenology 2016, 85, (2), 315–322. [Google Scholar] [CrossRef]
- Xi, G.; An, L.; Jia, Z.; Tan, K.; Zhang, J.; Wang, Z.; Zhang, C.; Miao, K.; Wu, Z.; Tian, J. , Natriuretic peptide receptor 2 (NPR2) localized in bovine oocyte underlies a unique mechanism for C-type natriuretic peptide (CNP)-induced meiotic arrest. Theriogenology 2018, 106, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Blengini, C. S.; Ibrahimian, P.; Vaskovicova, M.; Drutovic, D.; Solc, P.; Schindler, K. , Aurora kinase A is essential for meiosis in mouse oocytes. PLoS Genet 2021, 17, (4), e1009327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, B.; Fu, J.; Li, R.; Diao, F.; Li, C.; Chen, B.; Du, J.; Zhou, Z.; Mu, J.; Yan, Z.; Wu, L.; Liu, S.; Wang, W.; Zhao, L.; Dong, J.; He, L.; Liang, X.; Kuang, Y.; Sun, X.; Sang, Q.; Wang, L. , Bi-allelic Missense Pathogenic Variants in TRIP13 Cause Female Infertility Characterized by Oocyte Maturation Arrest. Am J Hum Genet 2020, 107, (1), 15–23. [Google Scholar] [CrossRef]
- Lambert, S. A.; Jolma, A.; Campitelli, L. F.; Das, P. K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T. R.; Weirauch, M. T. , The Human Transcription Factors. Cell 2018, 172, (4), 650–665. [Google Scholar] [CrossRef]
- Wu, S.; Tong, X.; Li, C.; Lu, K.; Tan, D.; Hu, H.; Liu, H.; Dai, F. , Genome-wide identification and expression profiling of the C2H2-type zinc finger protein genes in the silkworm Bombyx mori. PeerJ 2019, 7, e7222. [Google Scholar] [CrossRef] [PubMed]
- Torres-Machorro, A. L. , Homodimeric and Heterodimeric Interactions among Vertebrate Basic Helix-Loop-Helix Transcription Factors. Int. J. Mol. Sci. 2021, 22, (23), 12855. [Google Scholar] [CrossRef]
- Bragança, G. M.; Souza-Fabjan, J. M. G.; Ribeiro, L. S.; Brair, V. L.; Côrtes, L. R.; Souza, C. V.; Batista, R.; Fonseca, J. F.; Menchaca, A.; Brandão, F. Z. , Exogenous progestogen does not affect first-wave follicle populations and oocyte quality during ovarian stimulation with FSH in sheep. Domest Anim Endocrinol 2020, 72, 106369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Fan, X.; Li, R.; Zhang, C.; Zhang, J. , Effects of pre-incubation with C-type natriuretic peptide on nuclear maturation, mitochondrial behavior, and developmental competence of sheep oocytes. Biochem Biophys Res Commun 2018, 497, (1), 200–206. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, P.; Ge, W.; Feng, Y.; Li, L.; Sun, Z.; Zhang, H.; Shen, W. , Alginate oligosaccharides improve germ cell development and testicular microenvironment to rescue busulfan disrupted spermatogenesis. Theranostics 2020, 10, (7), 3308–3324. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund Å, K.; Faridani, O. R.; Sagasser, S.; Winberg, G.; Sandberg, R. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013, 10, (11), 1096–1098. [Google Scholar] [CrossRef]
- Macosko, E. Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A. R.; Kamitaki, N.; Martersteck, E. M.; Trombetta, J. J.; Weitz, D. A.; Sanes, J. R.; Shalek, A. K.; Regev, A.; McCarroll, S. A. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, (5), 1202–1214. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; Fu, X.; Liu, S.; Bo, X.; Yu, G. , clusterProfiler 4. 0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021, 2, (3), 100141. [Google Scholar]
- Yevshin, I.; Sharipov, R.; Kolmykov, S.; Kondrakhin, Y.; Kolpakov, F. , GTRD: a database on gene transcription regulation-2019 update. Nucleic Acids Res 2019, 47, (D1), D100–D105. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, W.; Zhang, H. M.; Xie, G. Y.; Miao, Y. R.; Xia, M.; Guo, A. Y. , hTFtarget: A Comprehensive Database for Regulations of Human Transcription Factors and Their Targets. Genomics Proteomics Bioinformatics 2020, 18, (2), 120–128. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



