Submitted:
03 November 2023
Posted:
03 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Peptides
2.2. ELISA
2.3. Flow cytometriy
2.4. Immunohistochemical analysis
3. Results
3.1. Epitope mapping of C44Mab-108 with alanine (or glycine)-substituted CD44v4 peptide
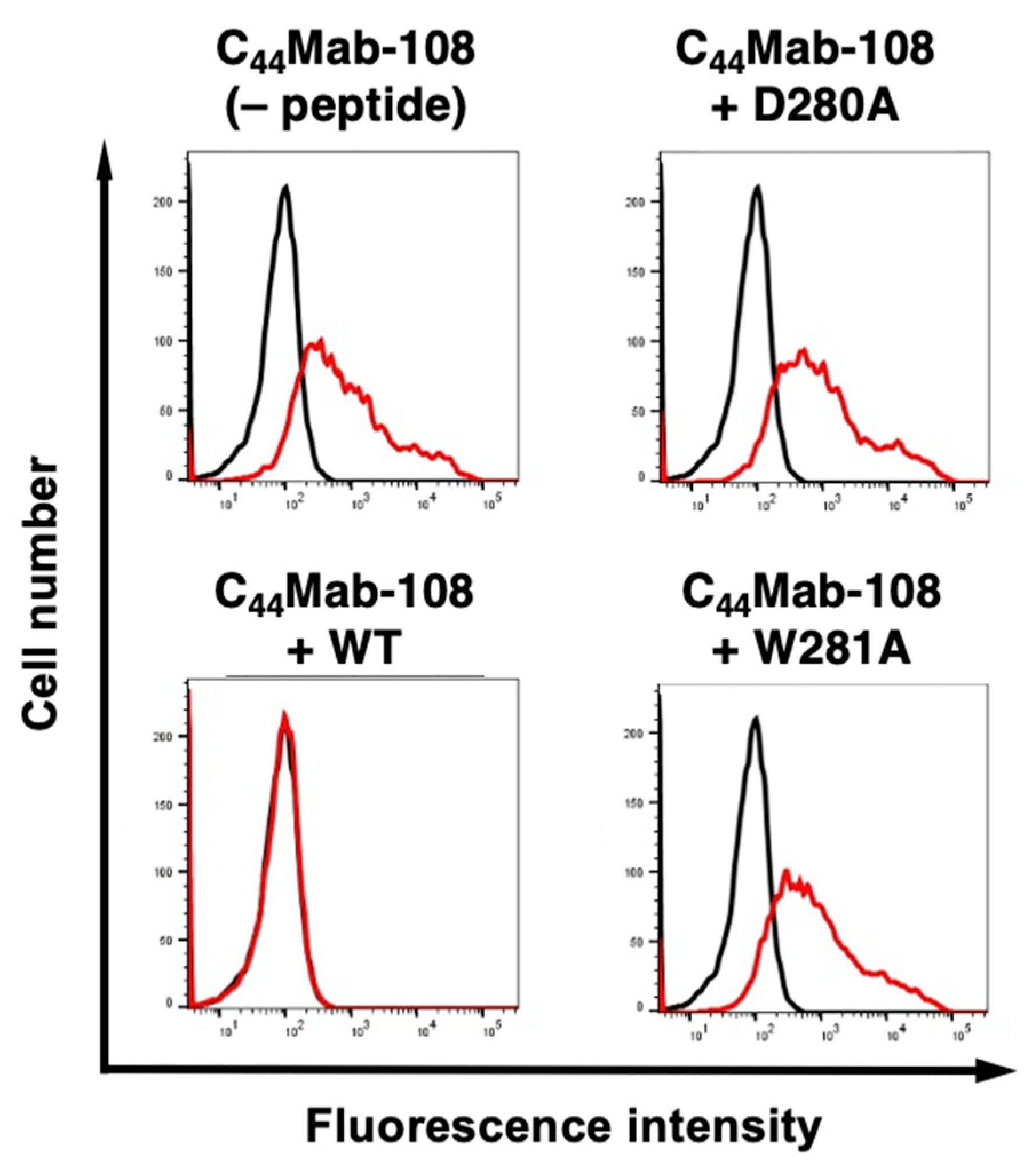
3.2. Flow cytometry using C44Mab-108 with alanine-substituted CD44v4 peptides
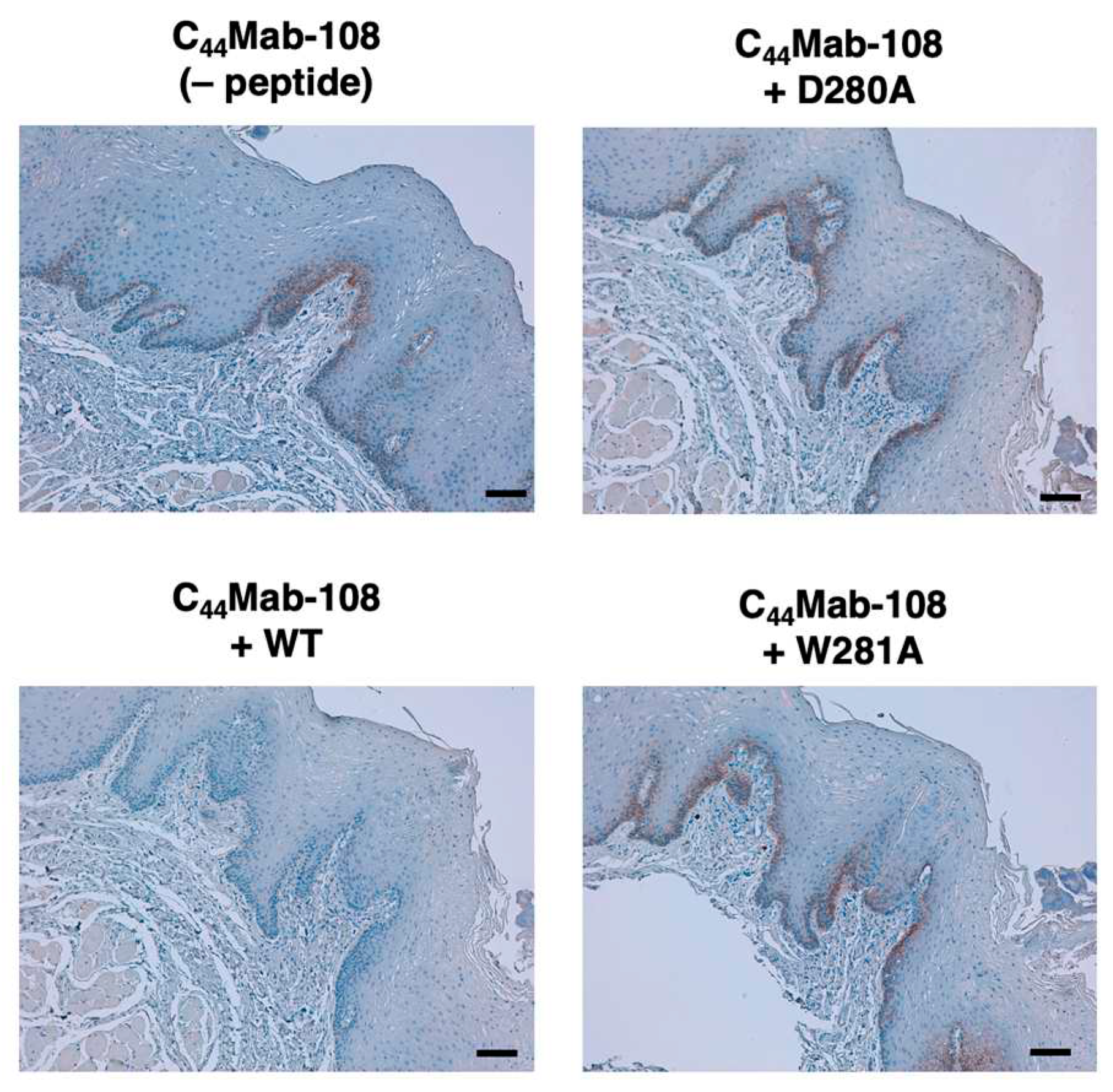
3.3. Immunohistochemistry using C44Mab-108 with alanine-substituted CD44v4 peptides
4. Discussion
Funding
Conflicts of Interest
References
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl Med 2015, 4, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Wang, Q.; An, J.; Chen, J.; Li, X.; Long, Q.; Xiao, L.; Guan, X.; Liu, J. CD44 Glycosylation as a Therapeutic Target in Oncology. Front Oncol 2022, 12, 883831. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.N.; Chandavarkar, V.; Sharma, R.; Bhargava, D. Structure, function and role of CD44 in neoplasia. J Oral Maxillofac Pathol 2019, 23, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Zöller, M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Jackson, D.G.; Simon, J.C.; Tanczos, E.; Peach, R.; Modrell, B.; Stamenkovic, I.; Plowman, G.; Aruffo, A. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J Cell Biol 1995, 128, 687–698. [Google Scholar] [CrossRef]
- Jackson, D.G.; Bell, J.I.; Dickinson, R.; Timans, J.; Shields, J.; Whittle, N. Proteoglycan forms of the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J Cell Biol 1995, 128, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Matzke, A.; Sargsyan, V.; Holtmann, B.; Aramuni, G.; Asan, E.; Sendtner, M.; Pace, G.; Howells, N.; Zhang, W.; Ponta, H.; et al. Haploinsufficiency of c-Met in cd44-/- mice identifies a collaboration of CD44 and c-Met in vivo. Mol Cell Biol 2007, 27, 8797–8806. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V.; Chen, L.; Sleeman, J.P.; Herrlich, P.; Ponta, H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002, 16, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Itai, S.; Nakamura, T.; Yanaka, M.; Kaneko, M.K.; Kato, Y. Detection of high CD44 expression in oral cancers using the novel monoclonal antibody, C(44)Mab-5. Biochem Biophys Rep 2018, 14, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Monoclonal Antibody for Multiple Applications against Esophageal Squamous Cell Carcinomas. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Ejima, R.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 6 Monoclonal Antibody C(44)Mab-9 for Multiple Applications against Colorectal Carcinomas. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Establishment of a Novel Anti-CD44 Variant 10 Monoclonal Antibody C(44)Mab-18 for Immunohistochemical Analysis against Oral Squamous Cell Carcinomas. Curr Issues Mol Biol 2023, 45, 5248–5262. [Google Scholar] [CrossRef] [PubMed]
- Kudo, Y.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 5 Monoclonal Antibody C(44)Mab-3 for Multiple Applications against Pancreatic Carcinomas. Antibodies (Basel) 2023, 12. [Google Scholar] [CrossRef]
- Suzuki, H.; Goto, N.; Tanaka, T.; Ouchida, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 8 Monoclonal Antibody C(44)Mab-94 against Gastric Carcinomas. Antibodies (Basel) 2023, 12. [Google Scholar] [CrossRef]
- Suzuki, H.; Kitamura, K.; Goto, N.; Ishikawa, K.; Ouchida, T.; Tanaka, T.; Kaneko, M.K.; Kato, Y. A Novel Anti-CD44 Variant 3 Monoclonal Antibody C(44)Mab-6 Was Established for Multiple Applications. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Ozawa, K.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 7/8 Monoclonal Antibody, C(44)Mab-34, for Multiple Applications against Oral Carcinomas. Biomedicines 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tanaka, T.; Goto, N.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Variant 4 Monoclonal Antibody C(44)Mab-108 for Immunohistochemistry. Curr Issues Mol Biol 2023, 45, 1875–1888. [Google Scholar] [CrossRef] [PubMed]
- Tawara, M.; Suzuki, H.; Goto, N.; Tanaka, T.; Kaneko, M.K.; Kato, Y. A Novel Anti-CD44 Variant 9 Monoclonal Antibody C(44)Mab-1 Was Developed for Immunohistochemical Analyses against Colorectal Cancers. Curr Issues Mol Biol 2023, 45, 3658–3673. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Epitope Mapping System: RIEDL Insertion for Epitope Mapping Method. Monoclon Antib Immunodiagn Immunother 2021, 40, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Kaneko, M.K.; Takei, J.; Tateyama, N.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C(44)Mab-46) Using the REMAP Method. Monoclon Antib Immunodiagn Immunother 2021, 40, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Takei, J.; Asano, T.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C(44)Mab-46) Using Alanine-Scanning Mutagenesis and Surface Plasmon Resonance. Monoclon Antib Immunodiagn Immunother 2021, 40, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Itai, S.; Ohishi, T.; Kaneko, M.K.; Yamada, S.; Abe, S.; Nakamura, T.; Yanaka, M.; Chang, Y.W.; Ohba, S.I.; Nishioka, Y.; et al. Anti-podocalyxin antibody exerts antitumor effects via antibody-dependent cellular cytotoxicity in mouse xenograft models of oral squamous cell carcinoma. Oncotarget 2018, 9, 22480–22497. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. Embo j 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Martins Á, M.; Gomes, C.; Balmaña, M.; Macedo, J.A.; Polom, K.; Roviello, F.; Magalhães, A.; Reis, C.A. O-glycan truncation enhances cancer-related functions of CD44 in gastric cancer. FEBS Lett 2019, 593, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- Arimori, T.; Mihara, E.; Suzuki, H.; Ohishi, T.; Tanaka, T.; Kaneko, M.K.; Takagi, J.; Kato, Y. Locally misfolded HER2 expressed on cancer cells is a promising target for development of cancer-specific antibodies Cell Press Community Review 2023. [CrossRef]
- Suzuki, H.; Kaneko, M.K.; Kato, Y. A cancer-specific monoclonal antibody against HER2 for breast cancers Preprint 2023. [CrossRef]
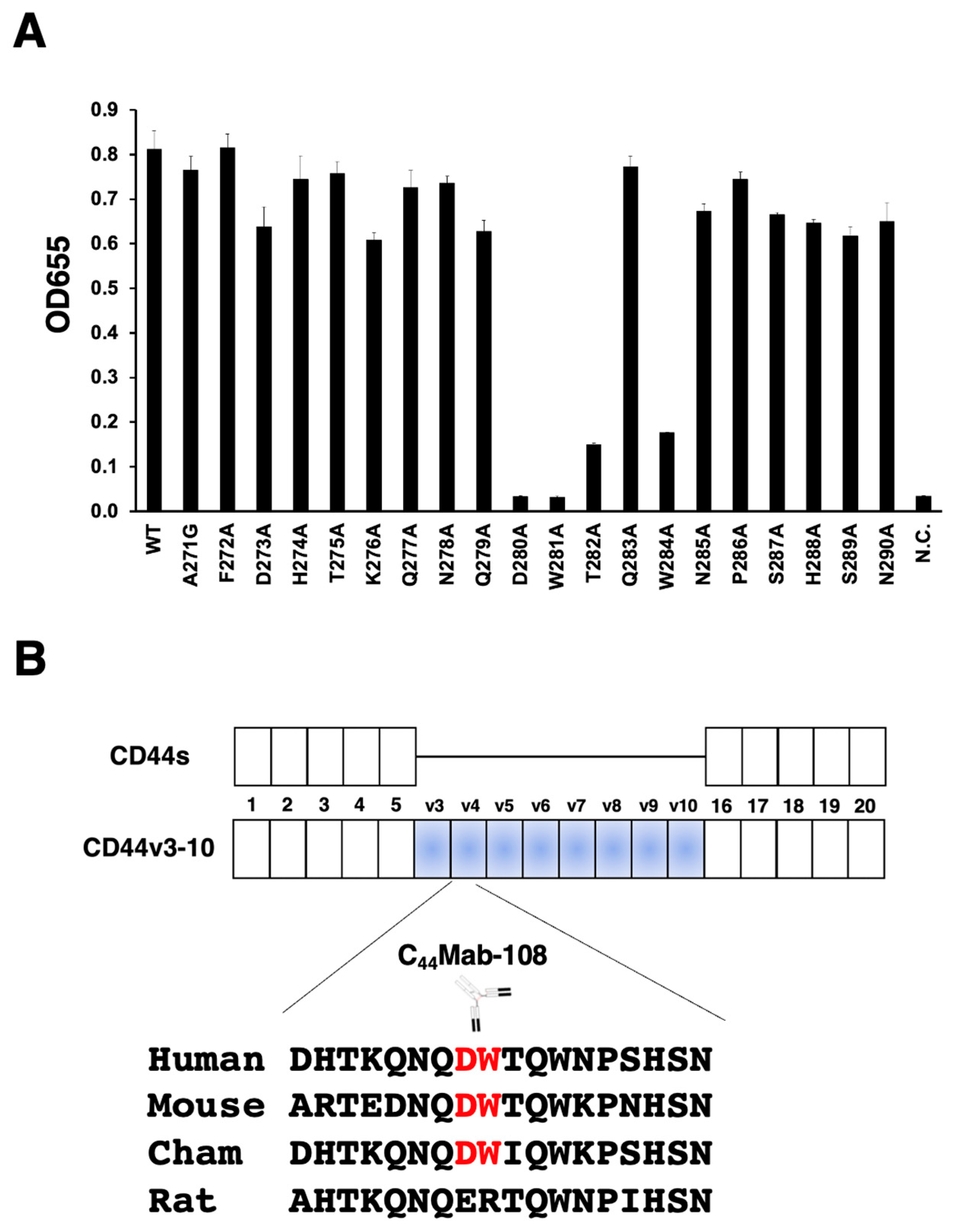
| Peptides | Sequences | C44Mab-108 |
| WT (271-290) | AFDHTKQNQDWTQWNPSHSN | +++ |
| A271G | GFDHTKQNQDWTQWNPSHSN | +++ |
| F272A | AADHTKQNQDWTQWNPSHSN | +++ |
| D273A | AFAHTKQNQDWTQWNPSHSN | +++ |
| H274A | AFDATKQNQDWTQWNPSHSN | +++ |
| T275A | AFDHAKQNQDWTQWNPSHSN | +++ |
| K276A | AFDHTAQNQDWTQWNPSHSN | +++ |
| Q277A | AFDHTKANQDWTQWNPSHSN | +++ |
| N278A | AFDHTKQAQDWTQWNPSHSN | +++ |
| Q279A | AFDHTKQNADWTQWNPSHSN | +++ |
| D280A | AFDHTKQNQAWTQWNPSHSN | – |
| W281A | AFDHTKQNQDATQWNPSHSN | – |
| T282A | AFDHTKQNQDWAQWNPSHSN | + |
| Q283A | AFDHTKQNQDWTAWNPSHSN | +++ |
| W284A | AFDHTKQNQDWTQANPSHSN | + |
| N285A | AFDHTKQNQDWTQWAPSHSN | +++ |
| P286A | AFDHTKQNQDWTQWNASHSN | +++ |
| S287A | AFDHTKQNQDWTQWNPAHSN | +++ |
| H288A | AFDHTKQNQDWTQWNPSASN | +++ |
| S289A | AFDHTKQNQDWTQWNPSHAN | +++ |
| N290A | AFDHTKQNQDWTQWNPSHSA | +++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





