Submitted:
20 October 2023
Posted:
08 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Construction of Mutant Strains
2.3. Nucleic Acid Manipulation andAnalyses
2.4. PhenotypeAnalyses
2.5. Enzyme Assay and Western Blot Analysis
2.6. Virulence and Phagocytosis Assay
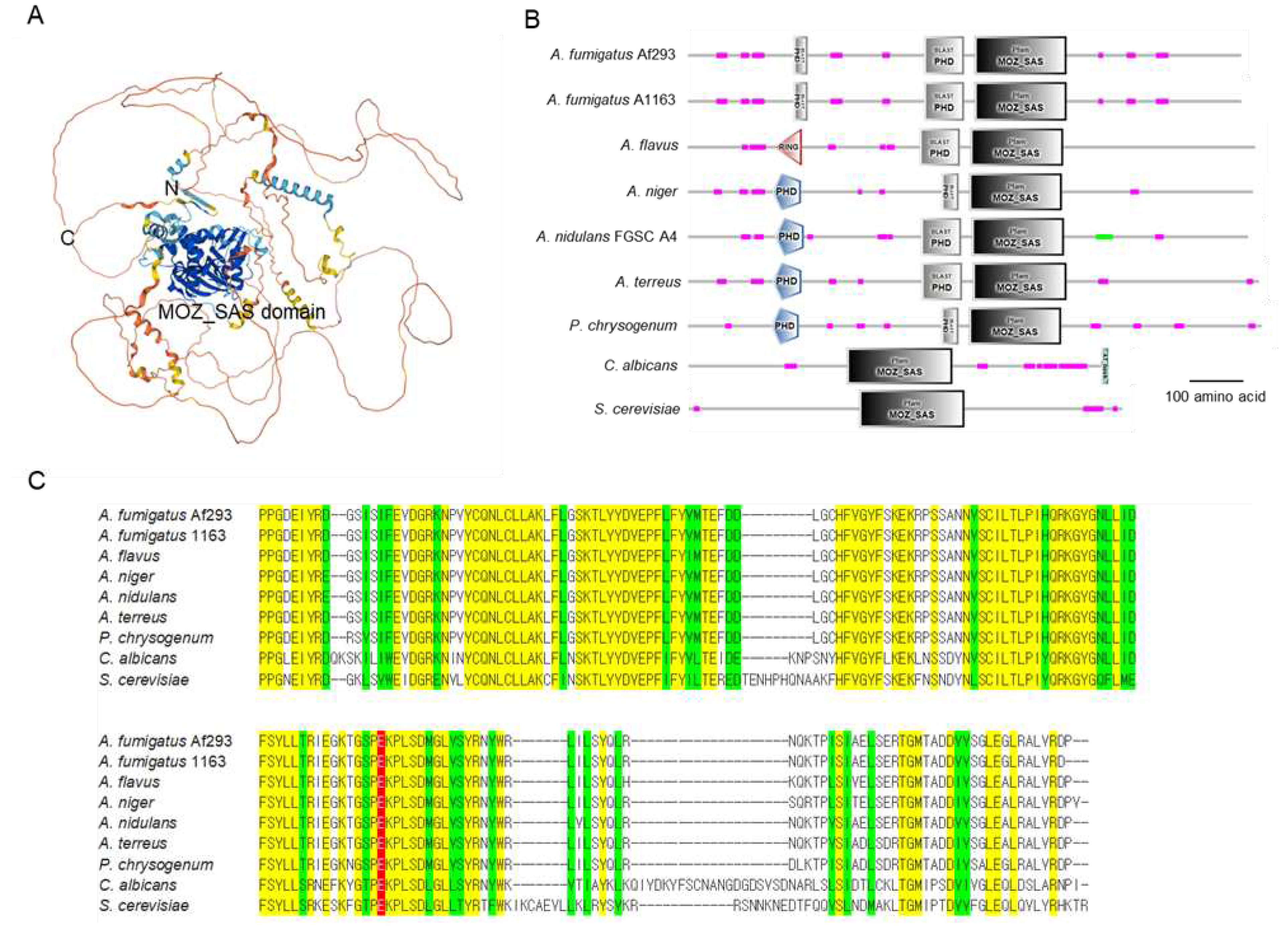
2.7. Bioinformatic Analyses
2.8. Statistics
3. Results
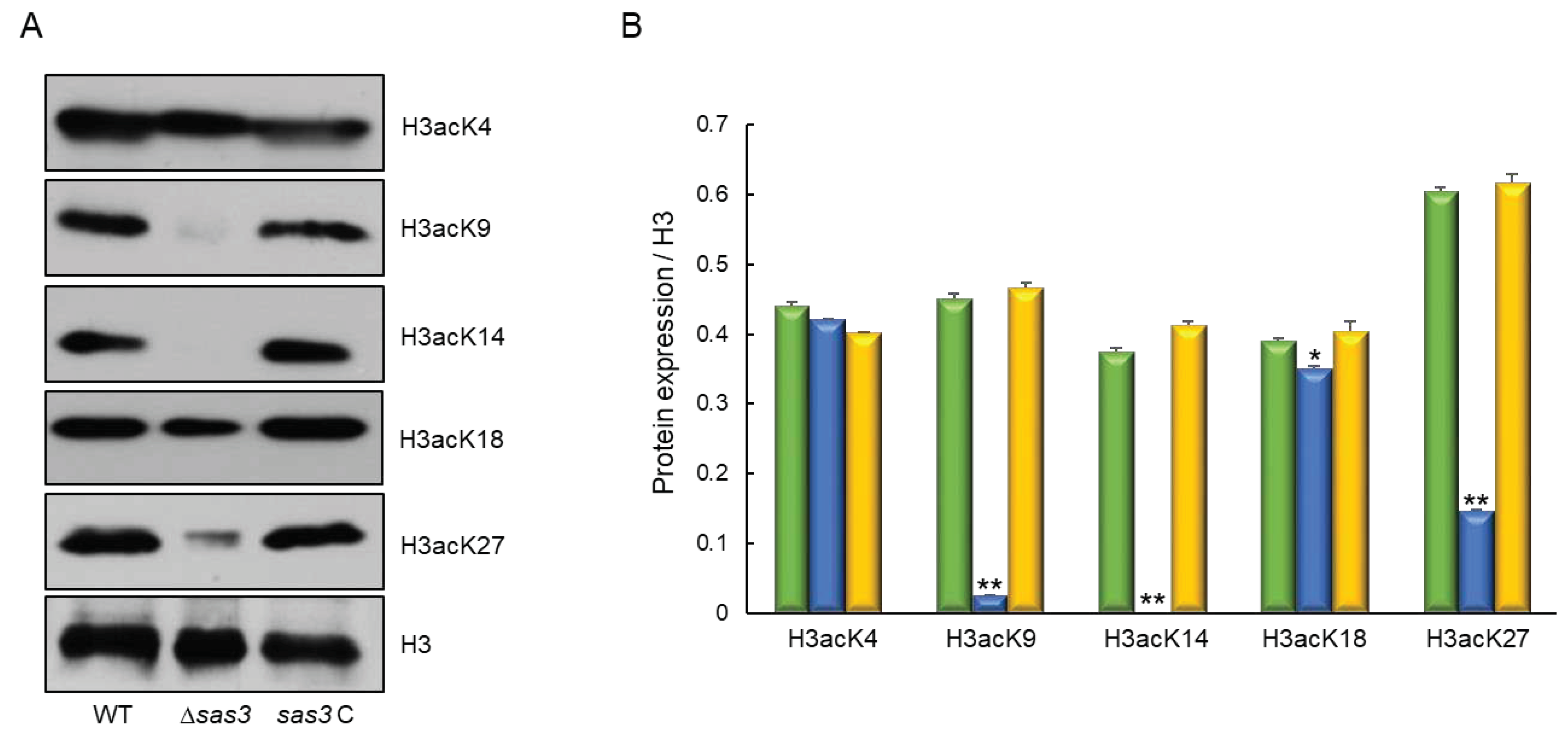
3.1. Summary of A. fumigatus Sas3
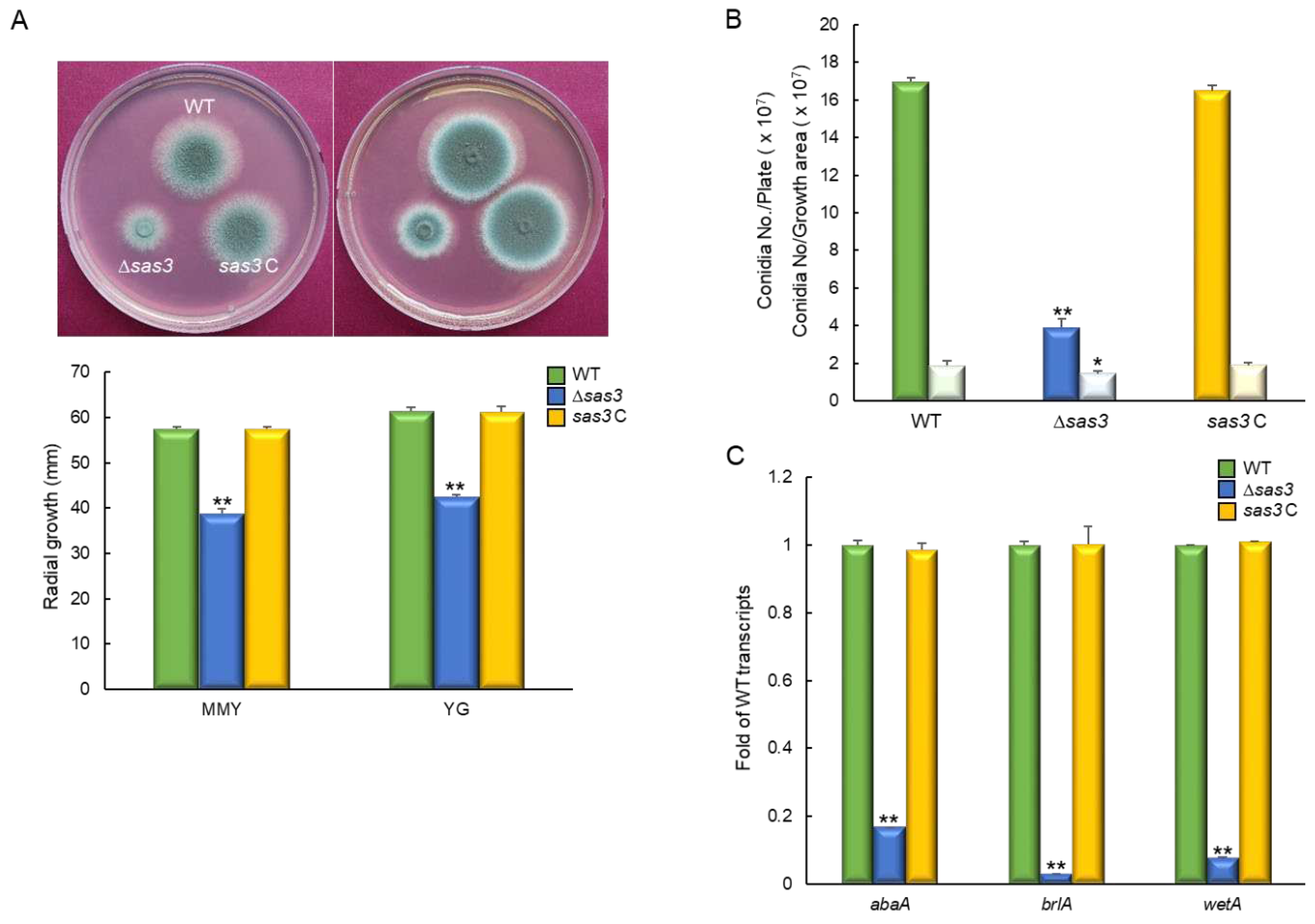
3.2. Sas3 is Required for Proper Growth and Development
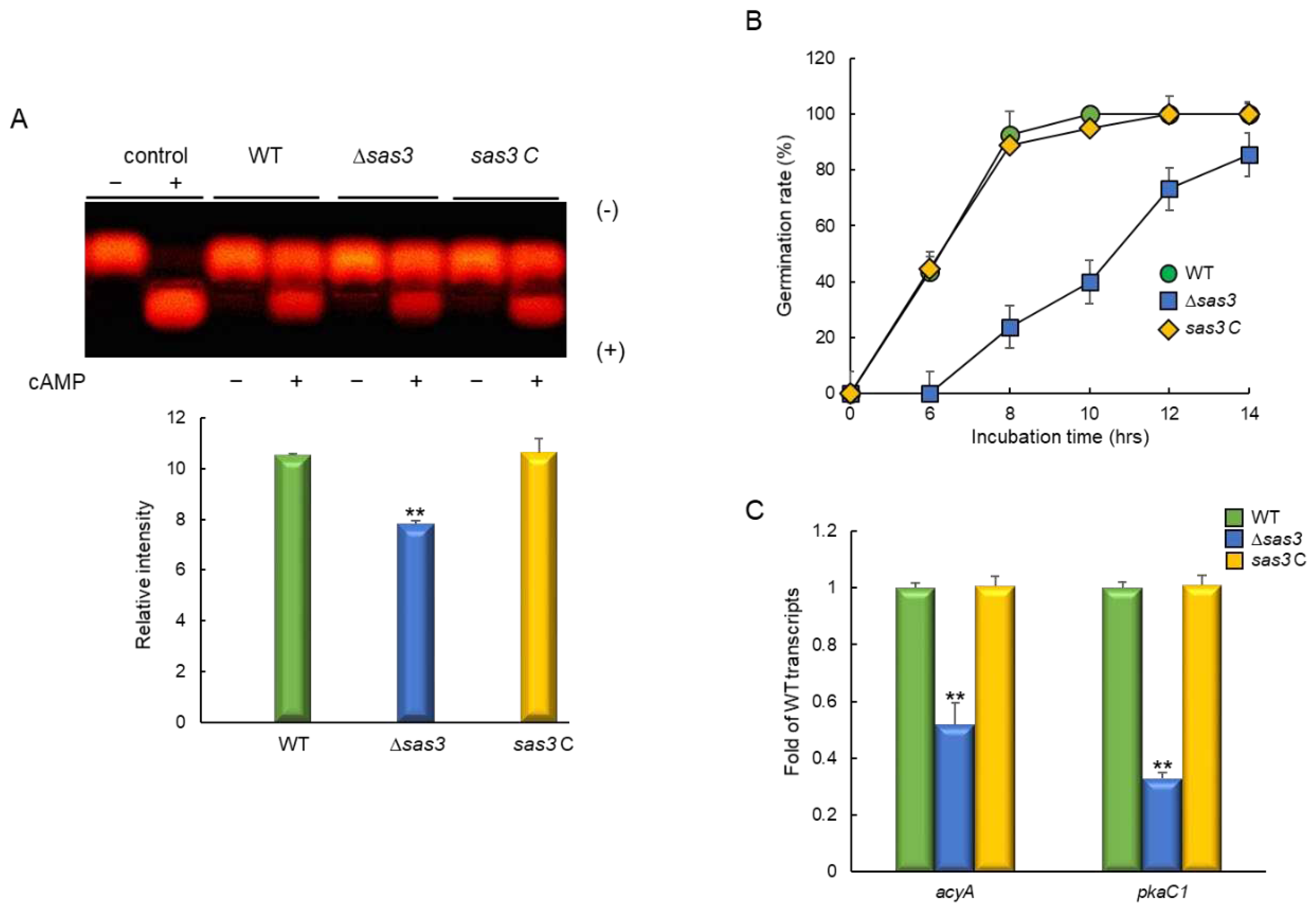
3.3. Sas3 Positively Affects PKA Signaling Pathway and Spore Germination
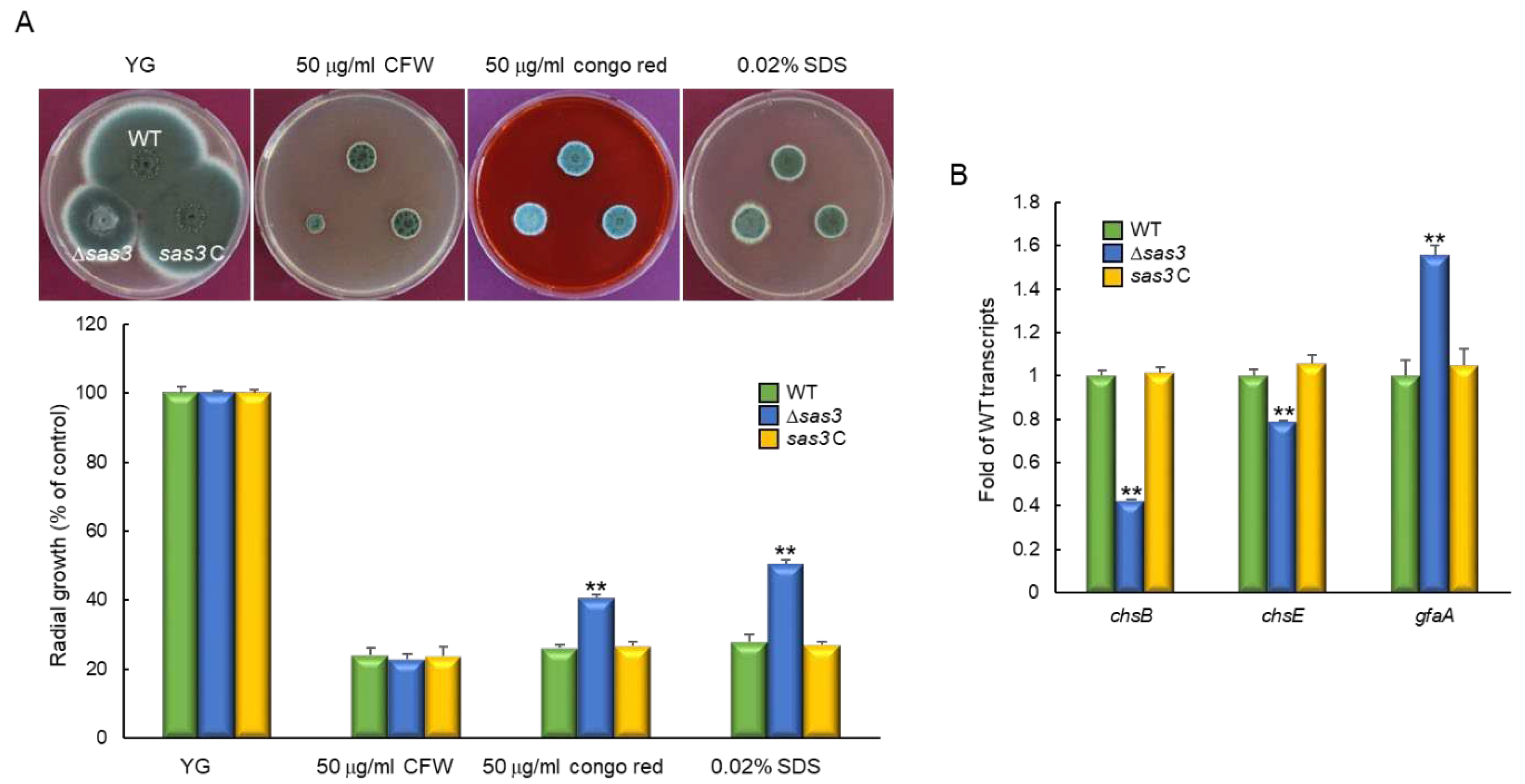
3.4. Sas3 is Involved in Cell Wall Stress Response
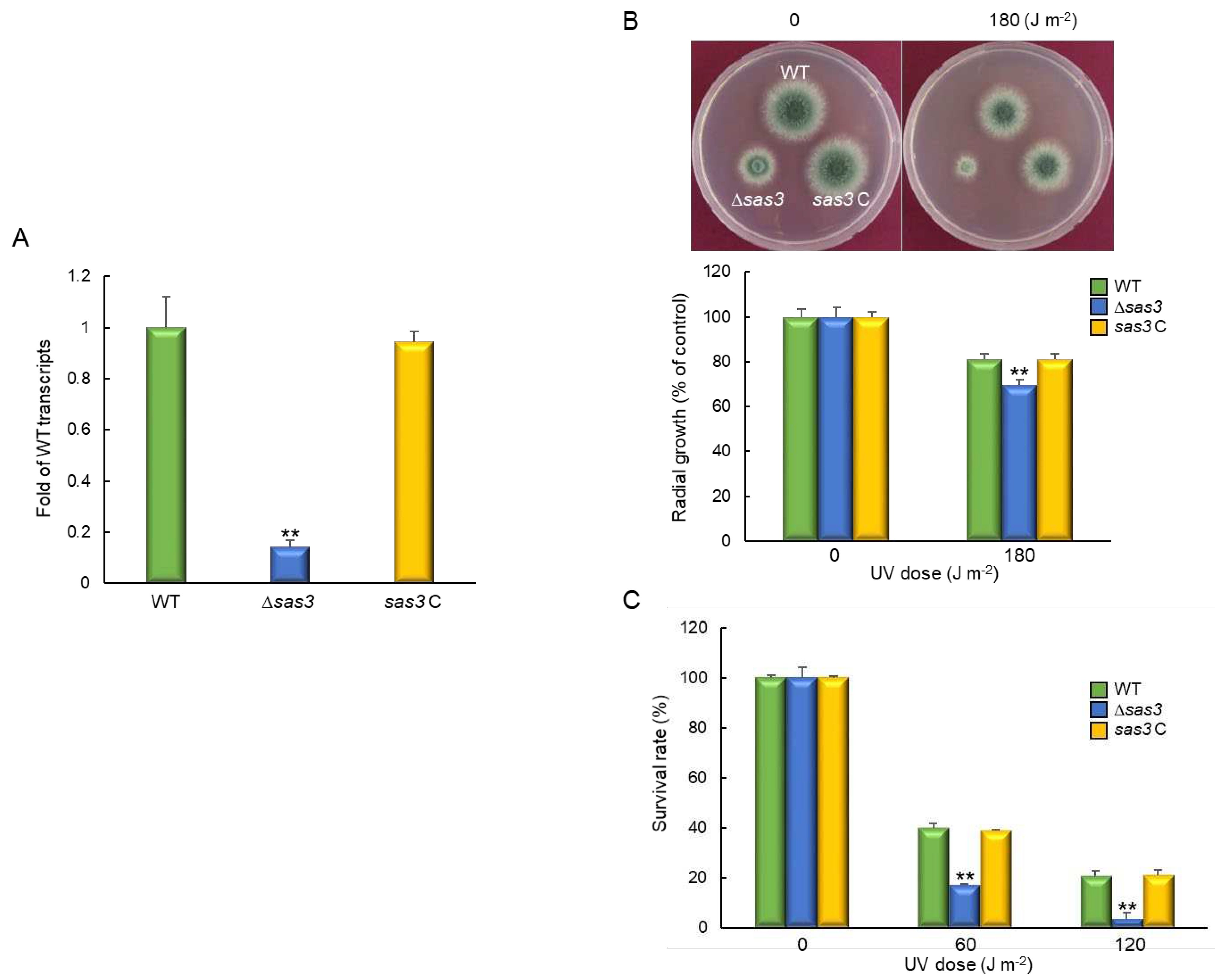
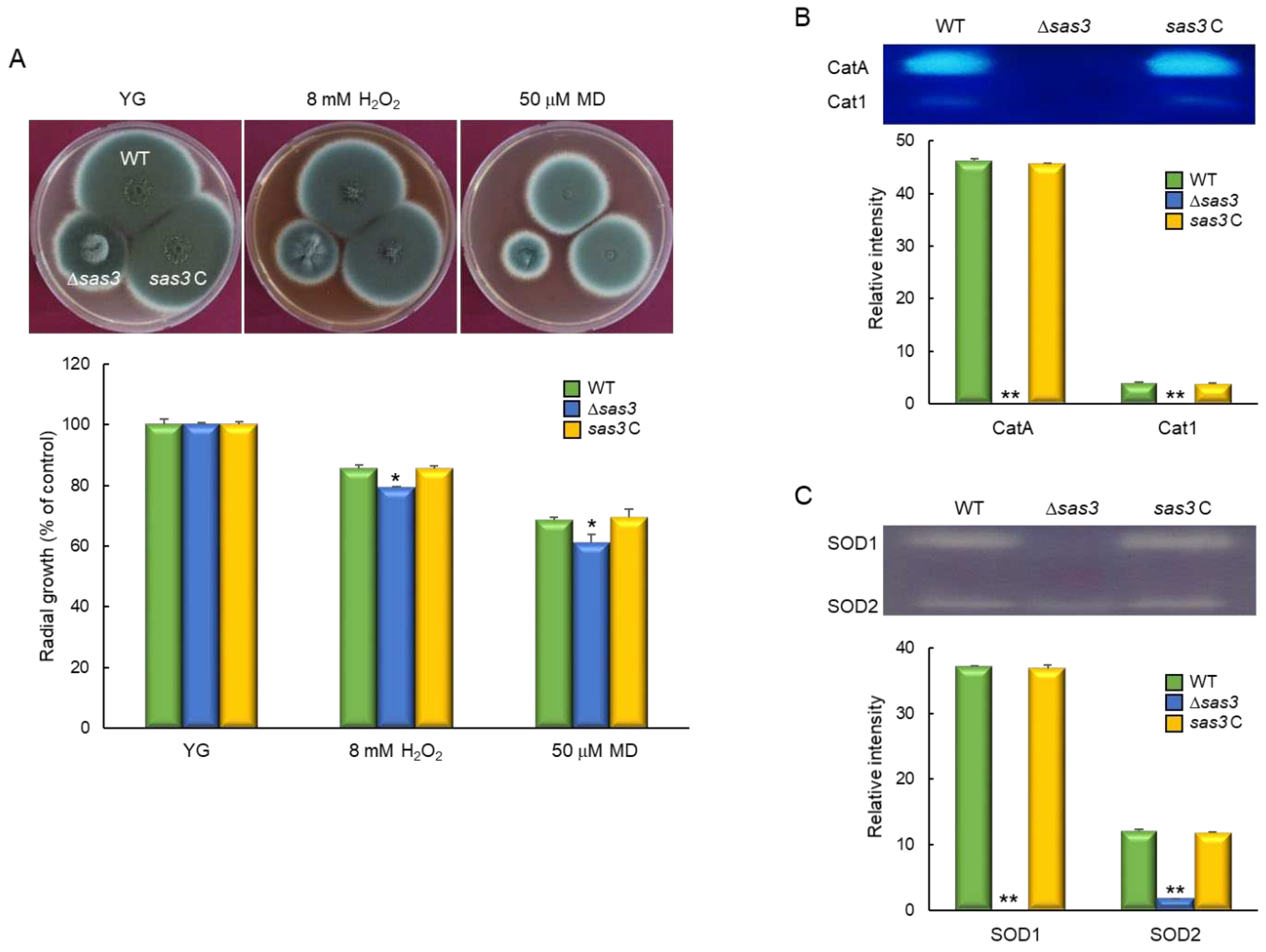
3.5. Sas3 Functions in Oxidative Stress Response
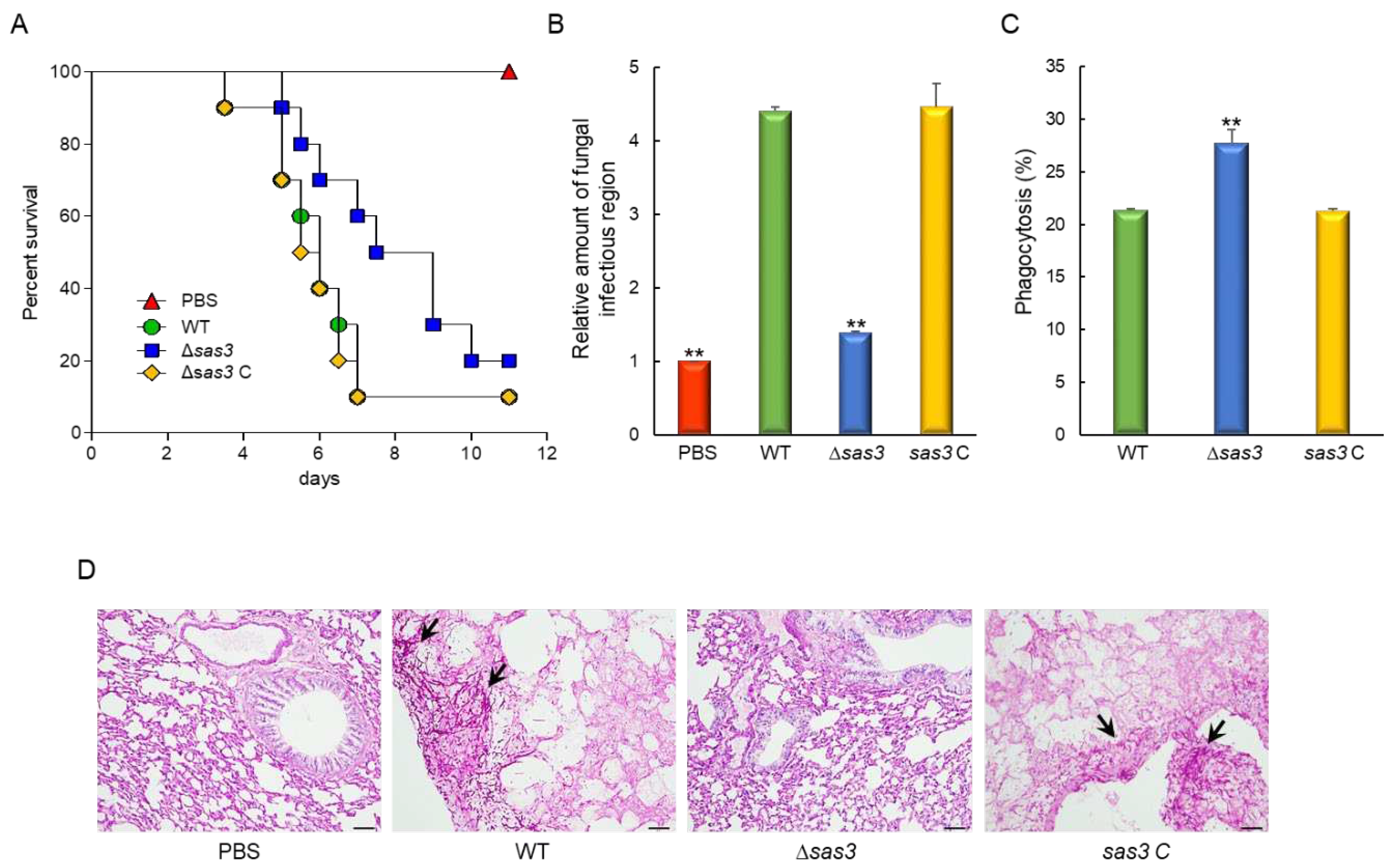
3.6. The Role of Sas3 in Fungal Virulence
3.7. Transcriptome Analysis
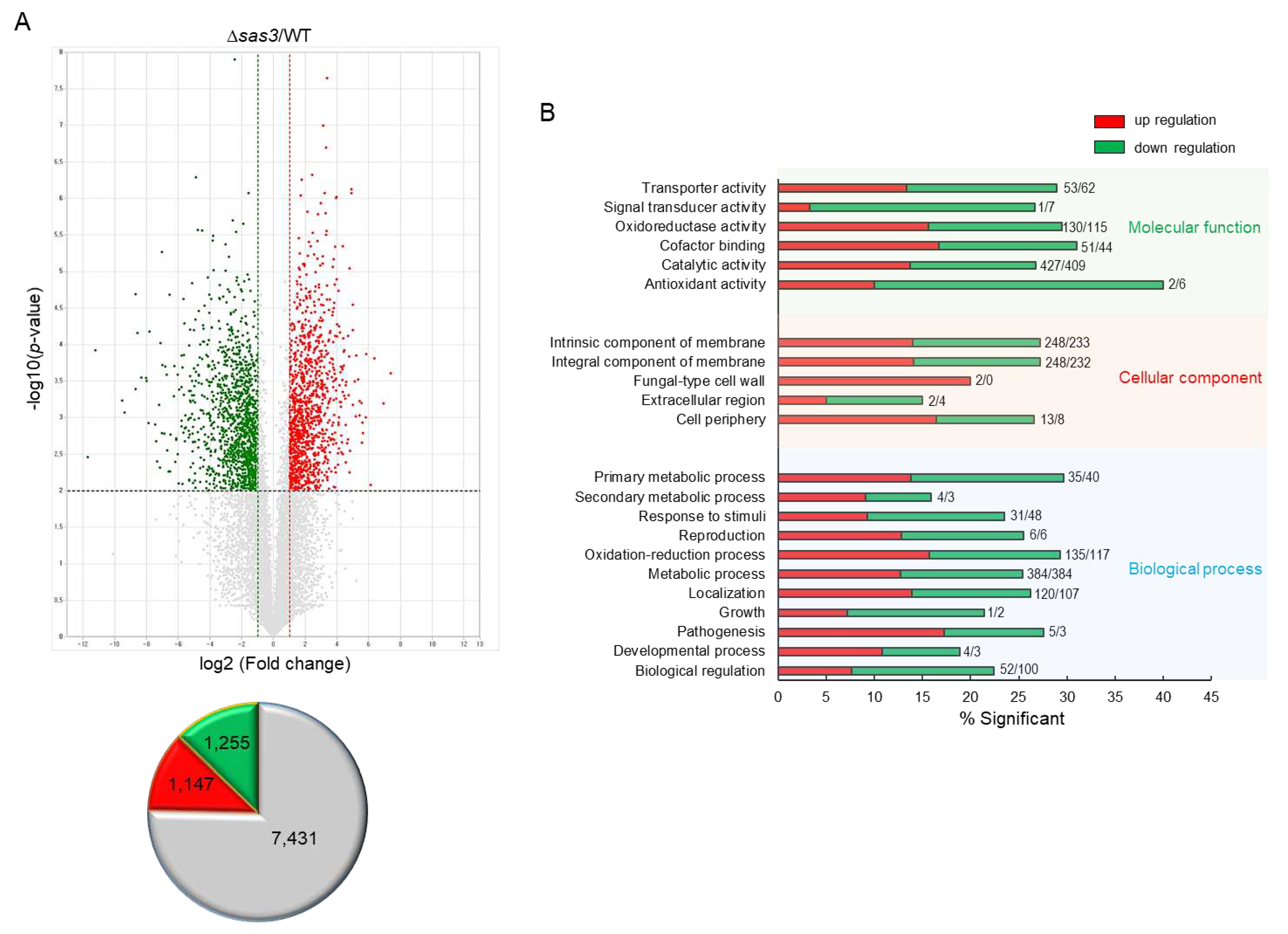
3.8. Potential Targets of Sas3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Phillips, D.M. The presence of acetyl groups of histones. Biochem J 1963, 87, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu Rev Biochem 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Jeon, J.; Kwon, S.; Lee, Y.H. Histone acetylation in fungal pathogens of plants. Plant Pathol J 2014, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sapountzi, V.; Cote, J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci 2011, 68, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Osada, S.; Sutton, A.; Muster, N.; Brown, C.E.; Yates, J.R., 3rd; Sternglanz, R.; Workman, J.L. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev 2001, 15, 3155–3168. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Howe, L.; Tafrov, S.T.; Grant, P.A.; Sternglanz, R.; Workman, J.L. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev 2000, 14, 1196–1208. [Google Scholar] [CrossRef]
- Chen, X.; Wu, L.; Lan, H.; Sun, R.; Wen, M.; Ruan, D.; Zhang, M.; Wang, S. Histone acetyltransferases MystA and MystB contribute to morphogenesis and aflatoxin biosynthesis by regulating acetylation in fungus Aspergillus flavus. Environ Microbiol 2022, 24, 1340–1361. [Google Scholar] [CrossRef]
- Kong, X.; van Diepeningen, A.D.; van der Lee, T.A.J.; Waalwijk, C.; Xu, J.; Xu, J.; Zhang, H.; Chen, W.; Feng, J. The Fusarium graminearum Histone Acetyltransferases Are Important for Morphogenesis, DON Biosynthesis, and Pathogenicity. Front Microbiol 2018, 9, 654. [Google Scholar] [CrossRef]
- Dubey, A.; Lee, J.; Kwon, S.; Lee, Y.H.; Jeon, J. A MYST family histone acetyltransferase, MoSAS3, is required for development and pathogenicity in the rice blast fungus. Mol Plant Pathol 2019, 20, 1491–1505. [Google Scholar] [CrossRef]
- Fan, A.; Mi, W.; Liu, Z.; Zeng, G.; Zhang, P.; Hu, Y.; Fang, W.; Yin, W.B. Deletion of a Histone Acetyltransferase Leads to the Pleiotropic Activation of Natural Products in Metarhizium robertsii. Org Lett 2017, 19, 1686–1689. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Cai, Q.; Qiu, L.; Ying, S.H.; Feng, M.G. The histone acetyltransferase Mst2 sustains the biological control potential of a fungal insect pathogen through transcriptional regulation. Appl Microbiol Biotechnol 2018, 102, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Fang, W.; Deng, W.; Yu, Z.; Li, J.; Chen, M.; Liao, W.; Xie, J.; Pan, W. Global profiling of lysine acetylation in human histoplasmosis pathogen Histoplasma capsulatum. Int J Biochem Cell Biol 2016, 73, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brandao, F.; Esher, S.K.; Ost, K.S.; Pianalto, K.; Nichols, C.B.; Fernandes, L.; Bocca, A.L.; Pocas-Fonseca, M.J.; Alspaugh, J.A. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci Rep 2018, 8, 5209. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Hou, Y.H.; Chen, Y.L. The histone acetyltransferase GcnE regulates conidiation and biofilm formation in Aspergillus fumigatus. Med Mycol 2020, 58, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Park, S.H.; Kim, S.S.; Lee, M.W.; Yu, J.H.; Shin, K.S. Functional Characterization of the GNAT Family Histone Acetyltransferase Elp3 and GcnE in Aspergillus fumigatus. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Brookman, J.L.; Denning, D.W. Molecular genetics in Aspergillus fumigatus. Curr Opin Microbiol 2000, 3, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Kafer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet 1977, 19, 33–131. [Google Scholar] [PubMed]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Dominguez, Y.; Scazzocchio, C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Han, K.H.; Seo, J.A.; Yu, J.H. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol Microbiol 2004, 51, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Huan, P.; Wang, H.; Liu, B. Assessment of housekeeping genes as internal references in quantitative expression analysis during early development of oyster. Genes Genet Syst 2017, 91, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Dang, X.; He, Y.Q.; Zhang, T.; Wang, H.Y. Selection of housekeeping genes as internal controls for quantitative RT-PCR analysis of the veined rapa whelk (Rapana venosa). PeerJ 2017, 5, e3398. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Jun, S.C.; Lee, M.W.; Yu, J.H.; Shin, K.S. Characterization of the mbsA Gene Encoding a Putative APSES Transcription Factor in Aspergillus fumigatus. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.F.; Malavazi, I.; von Zeska Kress Fagundes, M.R.; Savoldi, M.; Goldman, M.H.; Schwier, E.; Braus, G.H.; Goldman, G.H. The csnD/csnE signalosome genes are involved in the Aspergillus nidulans DNA damage response. Genetics 2005, 171, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Lwin, H.P.; Choi, Y.H.; Lee, M.W.; Yu, J.H.; Shin, K.S. RgsA Attenuates the PKA Signaling, Stress Response, and Virulence in the Human Opportunistic Pathogen Aspergillus fumigatus. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G.; Diaz, G.A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal Biochem 1986, 157, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Weydert, C.J.; Cullen, J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc 2010, 5, 51–66. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Rocha, M.C.; Fabri, J.H.; Franco de Godoy, K.; Alves de Castro, P.; Hori, J.I.; Ferreira da Cunha, A.; Arentshorst, M.; Ram, A.F.; van den Hondel, C.A.; Goldman, G.H. Aspergillus fumigatus MADS-Box Transcription Factor rlmA Is Required for Regulation of the Cell Wall Integrity and Virulence. G3 (Bethesda) 2016, 6, 2983–3002. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim-Granet, O.; Philippe, B.; Boleti, H.; Boisvieux-Ulrich, E.; Grenet, D.; Stern, M.; Latge, J.P. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect Immun 2003, 71, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Ram, A.F.; Arentshorst, M.; Damveld, R.A.; vanKuyk, P.A.; Klis, F.M.; van den Hondel, C.A. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine : fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology (Reading) 2004, 150, 3315–3326. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: one size doesn't fit all. Nature reviews. Molecular cell biology 2007, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.; Auston, D.; Grant, P.; John, S.; Cook, R.G.; Workman, J.L.; Pillus, L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev 2001, 15, 3144–3154. [Google Scholar] [CrossRef] [PubMed]
- Grosse, C.; Heinekamp, T.; Kniemeyer, O.; Gehrke, A.; Brakhage, A.A. Protein kinase A regulates growth, sporulation, and pigment formation in Aspergillus fumigatus. Appl Environ Microbiol 2008, 74, 4923–4933. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos, T.; Thireos, G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J 1992, 11, 4145–4152. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol 2010, 76, 1376–1386. [Google Scholar] [CrossRef]
- Reyes-Dominguez, Y.; Boedi, S.; Sulyok, M.; Wiesenberger, G.; Stoppacher, N.; Krska, R.; Strauss, J. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet Biol 2012, 49, 39–47. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Nutzmann, H.W.; Reyes-Dominguez, Y.; Scherlach, K.; Schroeckh, V.; Horn, F.; Gacek, A.; Schumann, J.; Hertweck, C.; Strauss, J.; Brakhage, A.A. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci U S A 2011, 108, 14282–14287. [Google Scholar] [CrossRef] [PubMed]
- Nutzmann, H.W.; Fischer, J.; Scherlach, K.; Hertweck, C.; Brakhage, A.A. Distinct amino acids of histone H3 control secondary metabolism in Aspergillus nidulans. Appl Environ Microbiol 2013, 79, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Soukup, A.A.; Chadwick, E.; Chiang, Y.M.; Wang, C.C.; Keller, N.P. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol Microbiol 2013, 89, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Zavala, B.; Kruger, I.; Dunker, C.; Jacobsen, I.D.; Morschhauser, J. The protein kinase Ire1 has a Hac1-independent essential role in iron uptake and virulence of Candida albicans. PLoS Pathog 2022, 18, e1010283. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Montero, A.; Goity, A.; Larrondo, L.F. The bZIP Transcription Factor HAC-1 Is Involved in the Unfolded Protein Response and Is Necessary for Growth on Cellulose in Neurospora crassa. PLoS One 2015, 10, e0131415. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Idnurm, A. The Uve1 endonuclease is regulated by the white collar complex to protect Cryptococcus neoformans from UV damage. PLoS Genet 2013, 9, e1003769. [Google Scholar] [CrossRef] [PubMed]
- Eberharter, A.; Becker, P.B. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO reports 2002, 3, 224–229. [Google Scholar] [CrossRef]
- Kalkhoven, E.; Teunissen, H.; Houweling, A.; Verrijzer, C.P.; Zantema, A. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol Cell Biol 2002, 22, 1961–1970. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




