1. Introduction
The human immunodeficiency virus (HIV), which causes AIDS, is spread differently from country to country[1-4]. According to the World Health Organisation (WHO), around 40.1 million people have died from HIV/AIDS by 2021 and around 38.4 million people are infected with HIV worldwide [
5].This study investigates the relationship between HIV-1 Protease and or HIV-2 Protease with the natural compound Polydatin ( called Piceid), a natural precursor and glycoside form of resveratrol with a monocrystalline structure [
6].
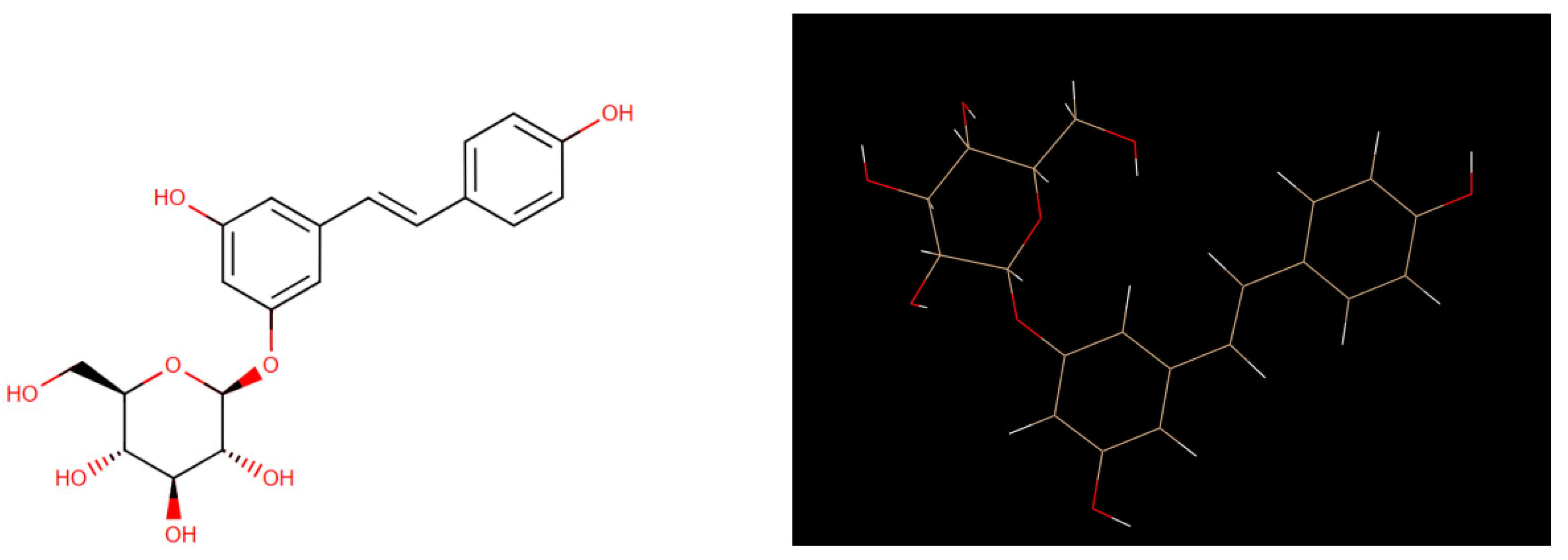
Figure 1 shows its 2D and its 3D crystal structure. It is isolated from the bark of Picea sitchensis or Polygonum cuspidatum, polydatin may be detected in grape, peanut, hop cones, red wines, hop pellets, cocoa-containing products, chocolate products, and many daily diets [
6].The detailed structure of the HIV protease has been used to design effective drugs for the treatment of AIDS [1;3]. HIV-2 is a virus of the Retroviridae family, Lentivirus genus; it represents one of the two HIV serotypes, together with the much more widespread HIV-1, which is responsible for the HIV pandemic worldwide. HIV-2, on the other hand, has a more limited geographic spread [
7].
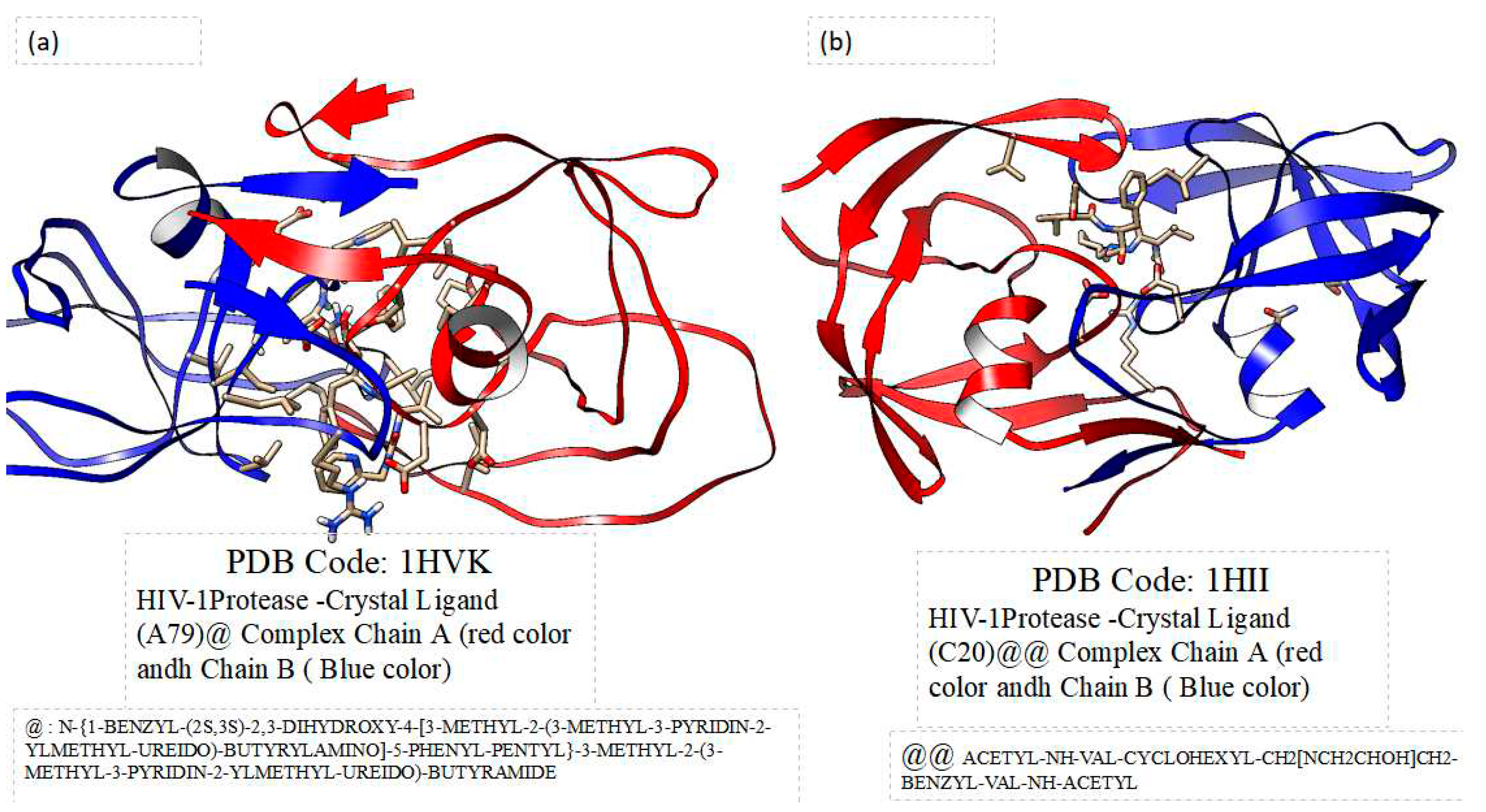
Figure 2 shows the comparison of the 3D crystal structures of HIV-1 protease and HIV-2 protease with their crystal inhibitors in the active site. The focus of this work was to evaluate, using computational methods, the potential biological role of these HIV proteases with polydatin. We chose to investigate Piceid because many scientific articles report not only that it has low side effects compared to other natural substances but also because it has different biological properties, for example, antioxidant [
8], anti-inflammatory [
9], and anticancer properties [
10]. In addition, polydatin has powerful anti-free radical activity [
8], and it reduces oxidative stress on cells and tissues [
11].
Crystal structures were taken by Protein Data Bank ( PDB code: 1hvk and PDB code: 1hii).
2. Material and Methods
SwissDock, a web service for predicting molecular interactions that might occur between a target protein and a small molecule [
12], was used to predict the potential role of Polydatin with HIV-1 protease and HIV-2 protease. SwissDock and S3DB are developed by Aurélien Grosdidier, Vincent Zoete and Olivier Michielin, from the Molecular Modeling Group of the Swiss Institute of Bioinformatics in Lausanne, Switzerland[
12]. SwissDock is based on the docking software EADock DSS, whose algorithm consists of the following steps[
12]:
- many binding modes are generated either in a box (local docking) or in the vicinity of all target cavities (blind docking).
-simultaneously, their CHARMM energies are estimated on a grid.
-the binding modes with the most favorable energies are evaluated with FACTS, and clustered.
-the most favorable clusters can be visualized online and downloaded on your computer.
In this wok, 3D Crystal structures of HIV-1 protease and HIV-2 protease with their crystal inhibithor in the Active site and saved in PDB format .Crystal structures were taken by Protein Data Bank ( PDB code: 1hvk and PDB code: 1hii). Polydatin was downloaded from Pubchem Database and saved in SDF format.
3. Results and Discussion
For the first time, this theoretical study focused on the relation between HIV-1 protease and HIV-2 protease with a natural compound named polydatin (or Piceid, 3,4,5-trihydroxystilbene-3-beta-monoglucoside). Several papers have reported the excellent biological role of this compound in the literature [
8,
9,
10,
11]. However, there are few recent works that have studied the role of polydatin in the HIV virus [13, 14]. Heredia et al. 2020 reported synergistic inhibition of HIV-1 in activated and remaining peripheral blood mononuclear cells, monocyte-derived macrophages, and selected drug-resistant isolates with nucleoside analogs combined with a natural product, resveratrol [
13]. Pflieger et al. (2013 studied natural stilbenoids isolated from grapevine exhibiting inhibitory effects against HIV-1 integration and eukaryote MOS1 transposase in vitro activities [
14]. Our main contribution was to investigate polydatin by molecular docking with HIV-1 protease and HIV-2 protease, respectively. The methods were performed by SwissDock Server, a web service for predicting molecular interactions that might occur between a target protein and a small molecule
Table 1. shows the comparison docking results evaluated by SwissDock. As can be seen from this Table, Polydatin has been shown to have a higher estimated ΔG score than the ligand crystal both in the case of HIV-2 protease and HIV-1 : ( Estimated ΔG of Polydatin was -9.89 kcal/mol VS Estimated ΔG of Crystal Ligand C20 [ACETYL-NH-VAL-CYCLOHEXYL-CH2[NCH2CHOH]CH2-BENZYL-VAL-NH-ACETYL) was -12.14 kcal/mol, when they binds in the acitive site of HIV-2, while Estimated ΔG of Polydatin was -9.47 kcal/mol VS Estimated ΔG of Crystal ligand A79 ( N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE) was -12.54 kcal/mol, when they binds in the acitive site of HIV-1 protease. However, a very important aspect that we noticed was the binding ability of Polydatin when it binds to HIV-1 protease or HIV-2 protease is not the same. In fact, if we observe the respective estimated ΔG scores , it can be seen that Polydatin shows a greater ability to bind with HIV-2 protease compared to when it binds with HIV-1 protease ( excellent low ΔG score of -9.89 kcal/mol of ΔG when Polydatin binds with HIV-2 protease ; VS a good ΔG score of -9.47 kcal/mol of ΔG , when Polydatin binds with HIV-1 protease.
These results are significant not only because they demonstrate the ability of polydatin to bind better with HIV-2 protease compared to when it binds with HIV-1 protease, even though they have a 54% similar sequence between them [
15], but also because the polydatin in the active site of these proteases could bind to two key amino acids of the catalytic triad sequence common to aspartic proteases (Asp25 and Gly27; see below
Figure 4 and
Figure 6). In
Figure 3 and
Figure 4, we reported the 3D structure of HIV-1 protease with crystal ligand A79 and polydatin complex, respectively, and the 2D diagrams that occur between HIV-1 protease amino acid residues and atoms in crystal ligand A79 and polydatin, respectively. In Figs. 5 and 6, we reported the 3D structure of HIV-2 protease with crystal ligand C20 and polydatin complex, respectively, and the 2D diagrams that occur between HIV-2 protease amino acid residues and atoms in crystal ligand C20 and polydatin, respectively.
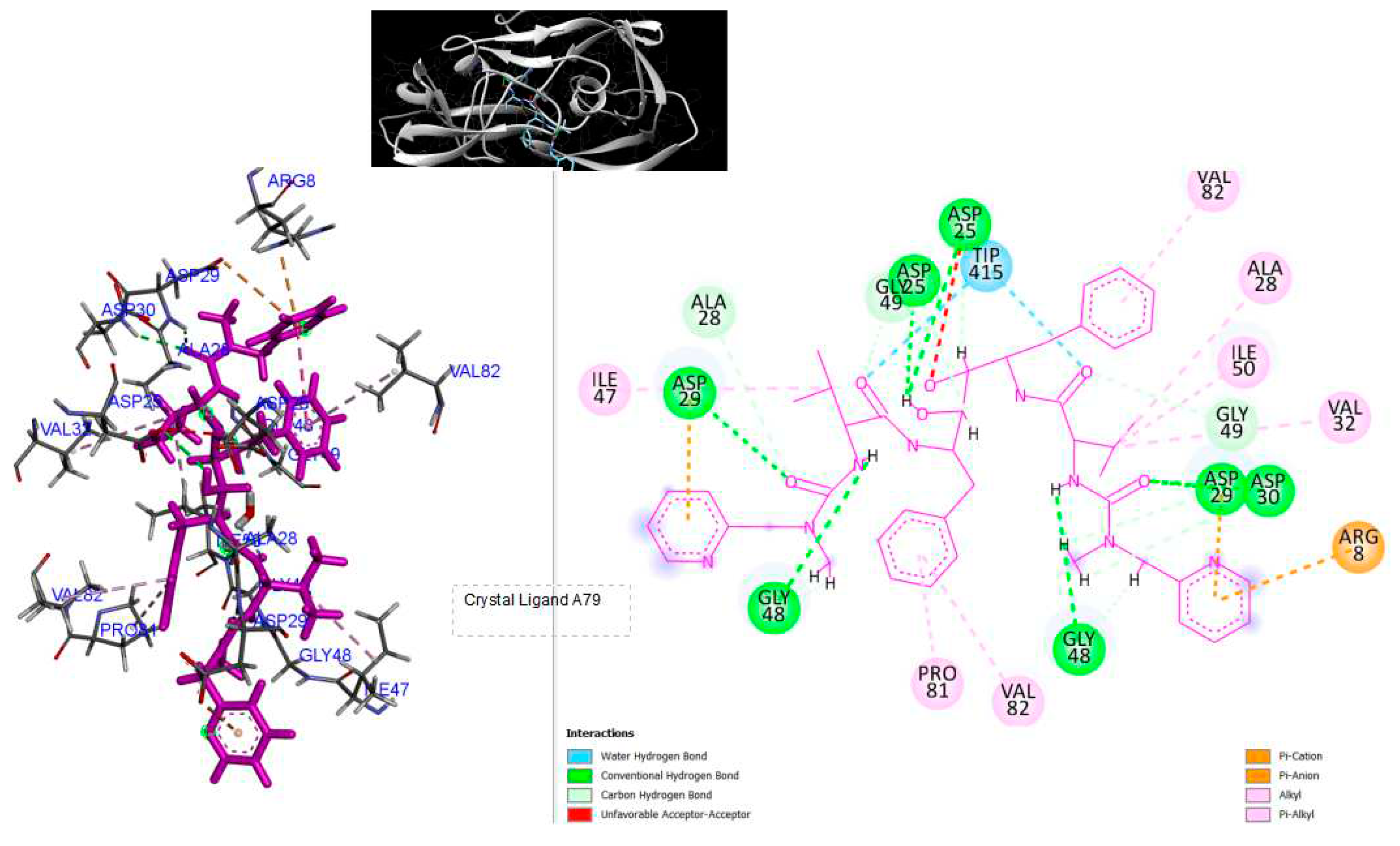
Figure 3.
3D Crystal structures of HIV-1 protease and 2D Diagrams residues interactions that occur between HIV-1 protease amino acid residues and atoms in crystal ligand A79: [N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE ] in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1hvk).
Figure 3.
3D Crystal structures of HIV-1 protease and 2D Diagrams residues interactions that occur between HIV-1 protease amino acid residues and atoms in crystal ligand A79: [N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE ] in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1hvk).
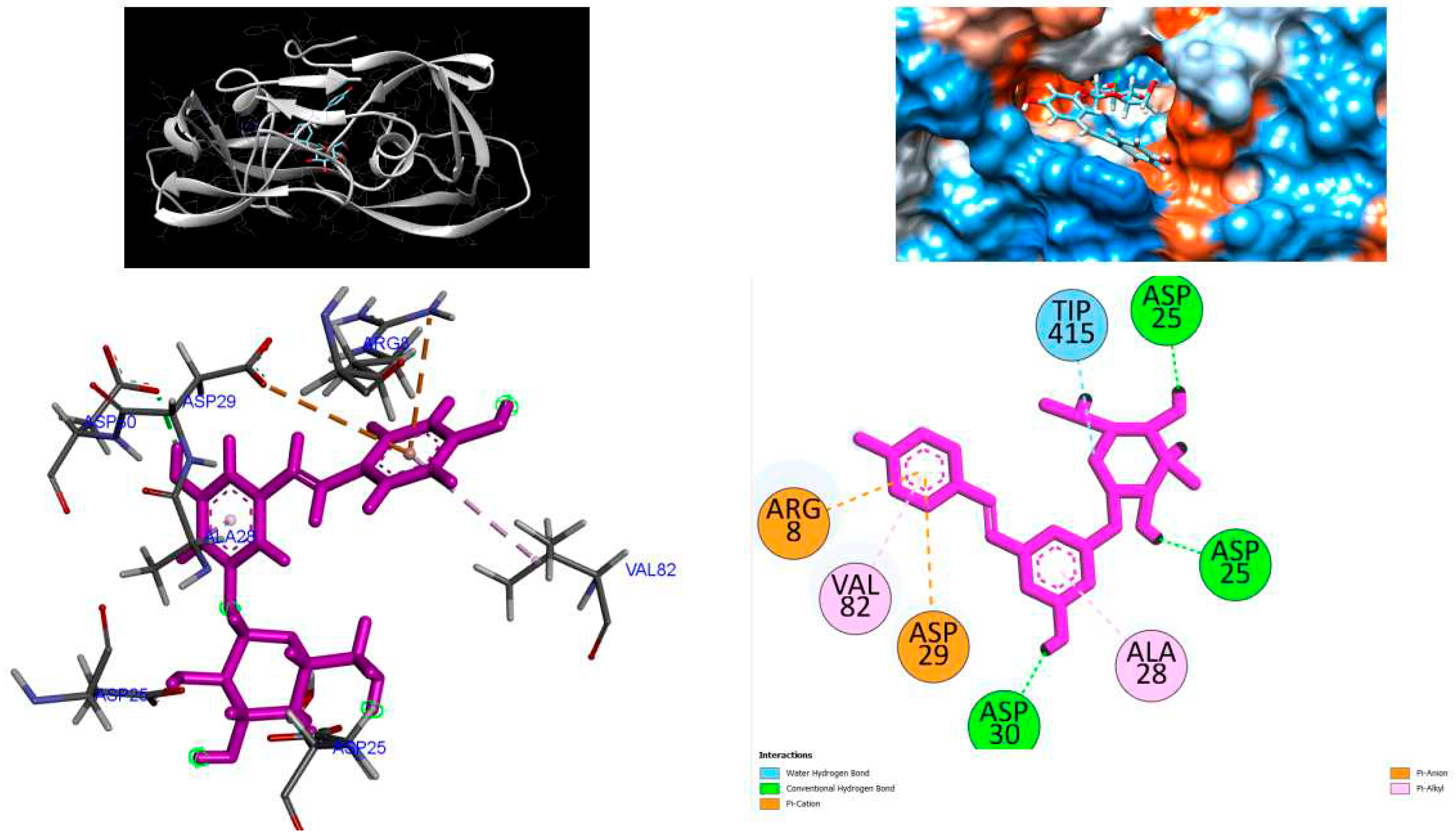
Figure 4.
3D Crystal structures of HIV-1 protease and 2D Diagrams residues interactions that occur between HIV-1 protease amino acid residues and atoms in Polydatin in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1hvk).
Figure 4.
3D Crystal structures of HIV-1 protease and 2D Diagrams residues interactions that occur between HIV-1 protease amino acid residues and atoms in Polydatin in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1hvk).
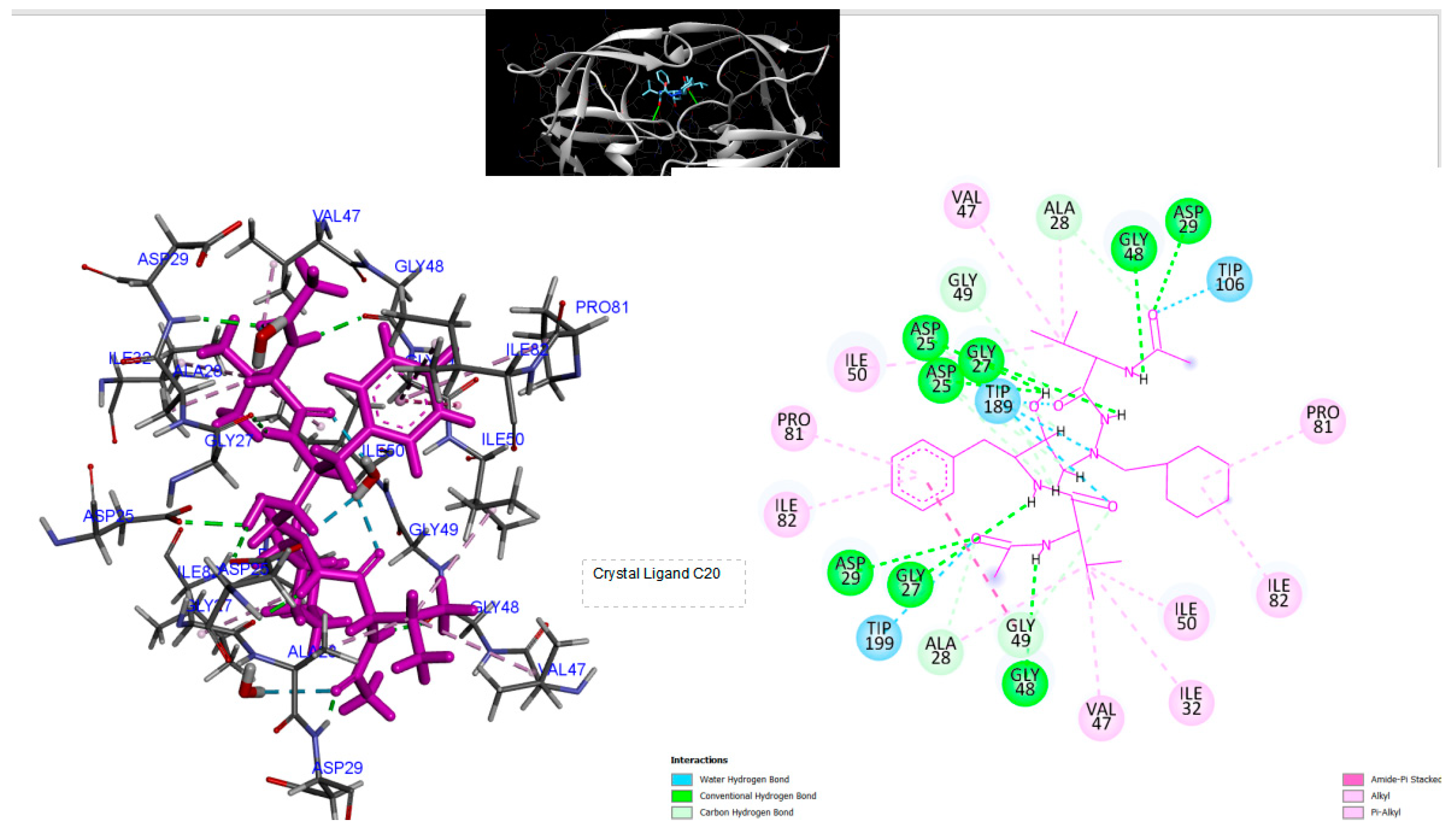
Figure 5.
3D Crystal structures of HIV-2 protease and 2D Diagrams residues interactions that occur between HIV-2 protease amino acid residues and atoms in Cystal Ligand C20 [ACETYL-NH-VAL-CYCLOHEXYL-CH2[NCH2CHOH]CH2-BENZYL-VAL-NH-ACETYL)] in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1HII).
Figure 5.
3D Crystal structures of HIV-2 protease and 2D Diagrams residues interactions that occur between HIV-2 protease amino acid residues and atoms in Cystal Ligand C20 [ACETYL-NH-VAL-CYCLOHEXYL-CH2[NCH2CHOH]CH2-BENZYL-VAL-NH-ACETYL)] in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1HII).
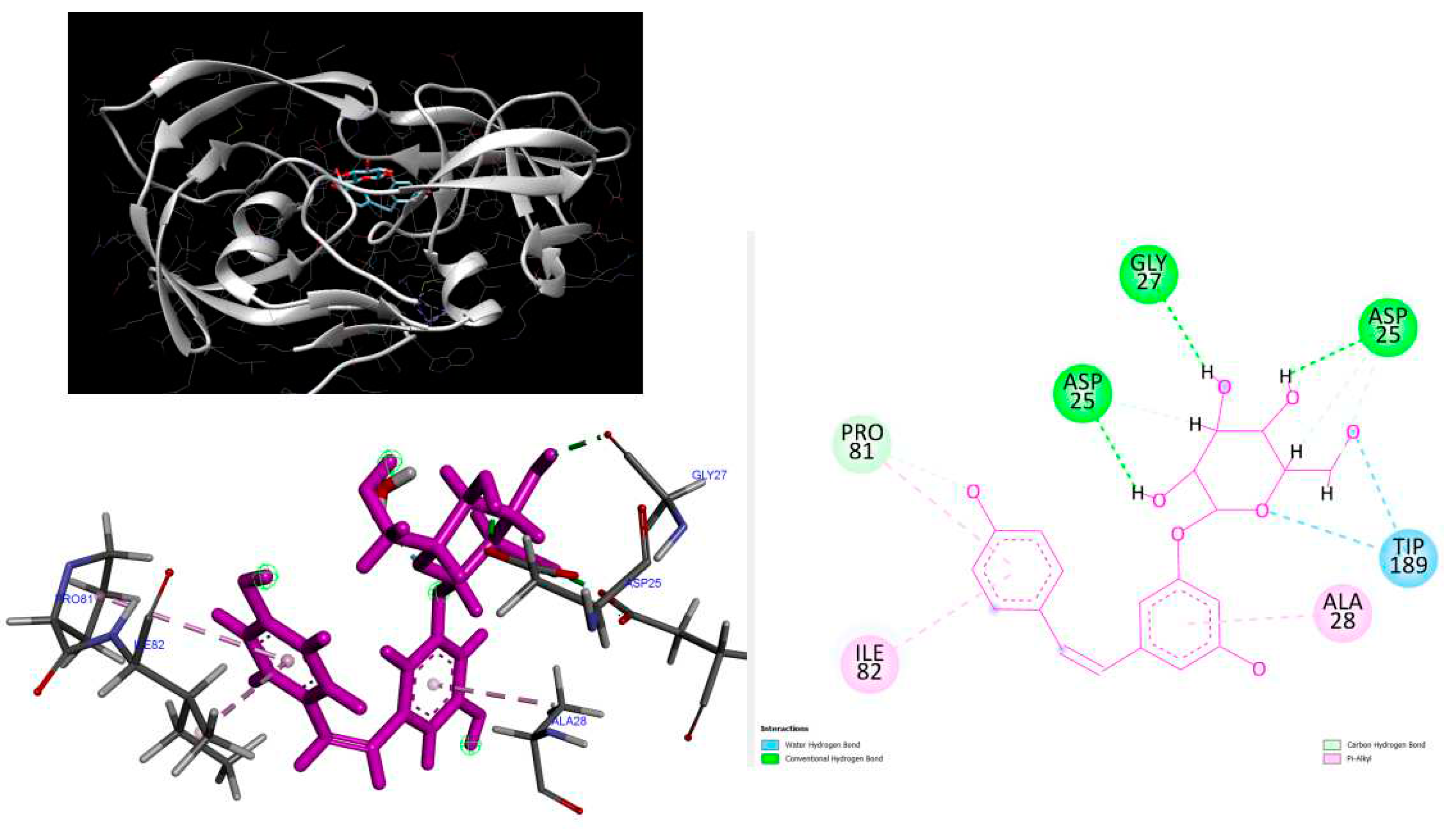
Figure 6.
3D Crystal structures of HIV-2 protease and 2D Diagrams residues interactions that occur between HIV-2 protease amino acid residues and atoms in Polydatin in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1HII).
Figure 6.
3D Crystal structures of HIV-2 protease and 2D Diagrams residues interactions that occur between HIV-2 protease amino acid residues and atoms in Polydatin in the Active site. The figure was reproduced by Discovery Studio Biovia Visualizer program. Crystal structure was taken by Protein Data Bank ( PDB code: 1HII).
Table 1.
Comparison Docking results of Polydatin and crystal ligands respectively, when they are complexed in HIV-1 protease or in HIV-2 protease respectively. The results were performed by SWISS Dock Service.
Table 1.
Comparison Docking results of Polydatin and crystal ligands respectively, when they are complexed in HIV-1 protease or in HIV-2 protease respectively. The results were performed by SWISS Dock Service.
| PDB |
Target |
Compound |
FullFitness (kcal/mol) |
Estimated ΔG (kcal/mol) |
| 1HII |
HIV-2 PROTEASE |
Polydatin |
-780.51 |
-9.89 |
| 1HVK |
HIV-1 PROTEASE |
Polydatin |
-770.76 |
-9.47 |
| |
|
|
|
|
| 1HII |
HIV-2 PROTEASE |
Crystal Ligand C20 |
-814.83 |
-12.14 |
| 1HVK |
HIV-1 PROTEASE |
Crystal Ligand A79 |
-822.14 |
-12.54 |
4. Conclussion
This short theoretical communication focused on the detection of the biological compound polydatin with HIV-1 protease or HIV-2 protease. The method evaluated for this goal was the molecular docking approach, investigated by SWISS Dock. From these results, polydatin seems to demonstrate a greater ability to bond with HIV-2 protease than it does with HIV-1 protease. Indeed, comparing the estimated ΔG of polydatin with HIV-2, it was reported at -9.9 kcal/mol versus -9.5 kcal/mol.
These computational results are very promising, although in vitro and in vivo studies are necessary to confirm our findings.
Author Contributions
Protocol designed by IVF . All authors read and approved the final manuscript.
Conflict of Interest
Authors declare that they do not have any conflict of interest
References
- Levy, J.A. (1993). Pathogenesis of human immunodeficiency virus infection. Microbiological reviews, 57(1), 183-289.
- Pantaleo, G.; Graziosi, C.; Fauci, A.S. (1993). The immunopathogenesis of human immunodeficiency virus infection. New England Journal of Medicine, 328(5), 327-335. [CrossRef]
- Pantaleo, G., Menzo, S., Vaccarezza, M., Graziosi, C., Cohen, O.J., Demarest, J.F., ... & Fauci, A.S. (1995). Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. New England Journal of Medicine, 332(4), 209-216. [CrossRef]
- Parekh, B.S., Ou, C.Y., Fonjungo, P.N., Kalou, M.B., Rottinghaus, E., Puren, A., ... & Nkengasong, J.N. (2018). Diagnosis of human immunodeficiency virus infection. Clinical microbiology reviews, 32(1), 10-1128. [CrossRef] [PubMed]
- World Health Organization. Retrieved 6 March 2022.
- Du, Q.H.; Peng, C.; Zhang, H. (2013). Polydatin: a review of pharmacology and pharmacokinetics. Pharmaceutical biology, 51(11), 1347-1354. [CrossRef]
- Gottlieb, G.S.; Raugi, D.N.; Smith, R.A. (2018). 90-90-90 for HIV-2? Ending the HIV-2 epidemic by enhancing care and clinical management of patients infected with HIV-2. The lancet HIV, 5(7), e390-e399. [CrossRef]
- Jin, J.; Li, Y.; Zhang, X.; Chen, T.; Wang, Y.; Wang, Z. (2016). Evaluation of both free radical scavenging capacity and antioxidative damage effect of polydatin. In Oxygen Transport to Tissue XXXVIII (pp. 57-62). Springer International Publishing.
- Lanzilli, G.; Cottarelli, A.; Nicotera, G.; Guida, S.; Ravagnan, G.; Fuggetta, M.P. (2012). Anti-inflammatory effect of resveratrol and polydatin by in vitro IL-17 modulation. Inflammation, 35, 240-248. [CrossRef]
- Zhang, Y.; Zhuang, Z.; Meng, Q.; Jiao, Y.; Xu, J.; Fan, S. (2014). Polydatin inhibits growth of lung cancer cells by inducing apoptosis and causing cell cycle arrest. Oncology letters, 7(1), 295-301. [CrossRef]
- Qiao, H.; Chen, H.; Dong, Y.; Ma, H.; Zhao, G.; Tang, F.; Li, Z. (2016). Polydatin attenuates H 2 O 2-induced oxidative stress via PKC pathway. Oxidative medicine and cellular longevity, 2016.
- Grosdidier, A.; Zoete, V.; Michielin, O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic acids research, 39(suppl_2), W270-W277. [CrossRef]
- Heredia, A.; Davis, C.; Redfield, R. (2000). Synergistic inhibition of HIV-1 in activated and resting peripheral blood mononuclear cells, monocyte-derived macrophages, and selected drug-resistant isolates with nucleoside analogues combined with a natural product, resveratrol. JAIDS Journal of Acquired Immune Deficiency Syndromes, 25(3), 246-255.
- Pflieger, A.; Waffo Teguo, P.; Papastamoulis, Y.; Chaignepain, S.; Subra, F.; Munir, S.; Parissi, V. (2013). Natural stilbenoids isolated from grapevine exhibiting inhibitory effects against HIV-1 integrase and eukaryote MOS1 transposase in vitro activities. PLoS One, 8(11), e81184. [CrossRef]
- Motomura, K.; Chen, J.; Hu, W.S. (2008). Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. Journal of virology, 82(4), 1923-1933. [CrossRef]
- Bragina, M.E.; Daina, A.; Perez, M.A.S.; Michielin, O.; Zoete, V. SwissSimilarity 2021 Web Tool: Novel Chemical Libraries and Additional Methods for an Enhanced Ligand-Based Virtual Screening Experience., Int. J. Mol. Sci, 2022, 23(2), 811. [CrossRef]
- Zoete, V.; Daina, A.; Bovigny, C.; Michielin, O. SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening., J. Chem. Inf. Model., 2016, 56(8), 1399. [CrossRef]
- SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. (2017) 7:42717.
- SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules, Nucl. Acids Res. (2019).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).