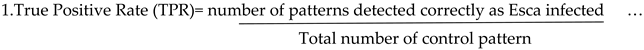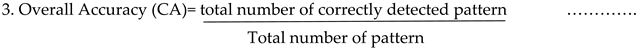Introduction
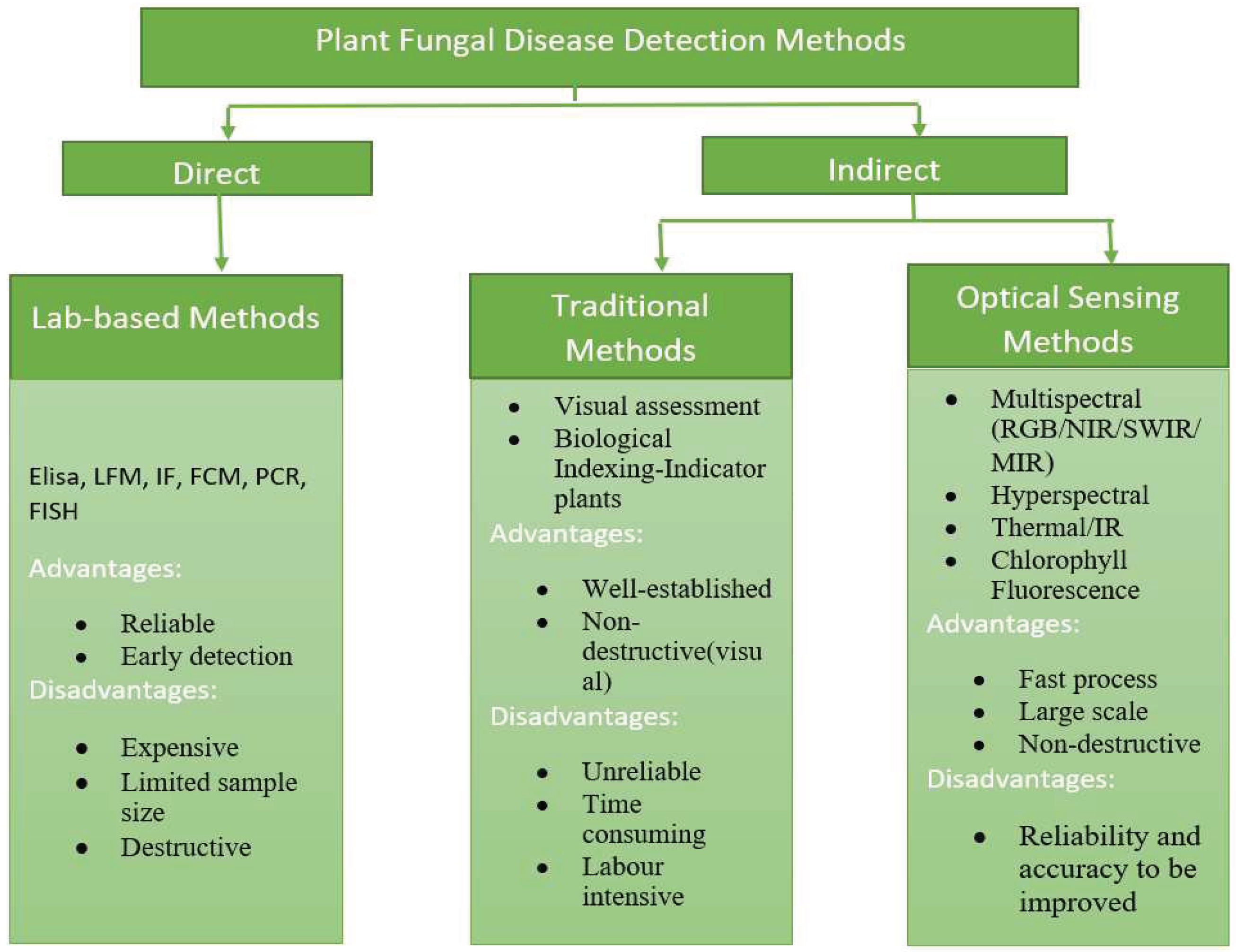
There are a variety of fungal diseases that are included in the paper mainly powdery mildew, Esca, Orchard, spider mite and black measles. Among this powdery mildew are the most harmful diseases. Methods for detecting plant diseases that are depicted in
Figure 1.
Going through the disadvantages of Lab-based methods and Traditional methods we are opting the optical based method which provide better throughput as compared to the other two. Optical methods offer the potential to manipulate living biological systems with exceptional spatial and temporal control. These methods distinguish materials whose chemical composition and crystalline structure exhibit a peculiar behavior when interacting with the full light spectrum.
In optical methods there are again different methods like Multispectral (RGB/NIR/MIR), Hyperspectral, Thermal/IR, Chlorophyll Fluorescence. But we are mainly concentrating on RGB, Multispectral and Thermal. Due to complexity and their cost of implementation we are not concentrating on Hyperspectral and Chlorophyll Fluorescence. The paper will further contain in
section 2 we will be explain about the working principle of RGB camera, existing application, various machine learning models used, different method implemented for data-preprocessing of three different papers and a comparison table with reviewers’ comments. Similarly in
section 3 we will speak about multispectral cameras and in
section 4 we will speak about thermal cameras. Finally, we will come to conclusion about various plant fungal disease detection methods, and which is the best method that we can choose by the reviewer’s comment, and we will also speak about the future scope of these cameras.
1. RGB camera
In the paper
Salvador Gutierrez et al [
1]. Proposed to use computer vision and deep learning modelling for the early identification and distinction of two of two major plant diseases, primarily downy mildew & spider mite, in grapevine leaves. The experiment was carried out in the field. Three stages make up most of the study. The initial step was classifying the three types of grapevine leaves that were collected from the fields into the RGB photos. In the second stage, the pictures that had been separated into the three classes-leaves with spider mites, and leaves without symptoms-were combined for classification models using computer vision techniques to pre-process the collected data. The next step was classifying the deep learning models that had been trained to divide the photos into three categories.
Images were captured at a working vineyard in northern Spain. Early in August, photographs were shot manually in order to capture a variety of lighting conditions, including sunshiny and cloud-masked conditions, and from both sides of the canopy. Using a Canon EOS 5D Mark IV and a Canon EF 20 mm f/2.8 USM lens in aperture-priority AE(Av) mode with the aperture set to F/6.3, leaf photos were captured from the lateral canopy side. The ISO setting was 200 in order to get a good level of light that allowed us to reduce sensor-related image noise while keeping the relatively high-speed impediment necessary for the manually captured photographs to be acceptable and to prevent movement distortion. The field of view for the camera lens employed in this experiment was around 504 mm horizontally and 360 mm vertically, which corresponded to the objective of the leaf being approached in the image frame to keep about 30 cm between them. Each leaf class has at least 250 photos captured of it.
The RGB images were cropped to remove the background noise and highlight the primary leaf in the image using GrabCut segmentation in OpenCV (version 4.0). The RGB images had widths and heights of 6720*4480 pixels, so the leaves were centered in the photographs. The color system was changed from RGB to hue-saturation-value to allow for accurate modelling & to lessen the impact of changing lighting. Since illumination primarily affects the saturation & value (or lighting or intensity) channels, hue, which is after constant, was sickness symptoms using the hue(H) component. In order to do hue thresholding, the histograms of the images were checked for any sign of a noticeable high number of pixels with H values between 1 and 40. If the H is equal to or less than 40, the pixel is left alone; if H is larger than 40, the pixel is set to 0.
Convolutional neural networks were utilized to train the classification models that were utilized in this instance. On the basis of initially utilizing all three HSV channels & then simply using the H(hue) channel, two different types of models were developed. For model validation, the hold-out approach was employed because of the execution duration each time step. Three convolutional batch-normalization & max-pooling blocks made up the experiment. Normalization and max-pooling blocks made up the experiment. A dropout layer at the end of these blocks is employed to reduce overfitting. In the second part, three completely linked blocks were employed to categorize characteristics gathered in the earlier layers in the appropriate manner. With the same architectural categorization has been used to separate all sorts of groups: healthy & sick leaves.
The training of classification models was carried out about 300 epochs. The loss functions, whose values indicates the discrepancy between the predicted & actual value for the values utilizing the Adam optimizer, is reduced by the created CNN architecture. An online data augmentation was applied progresses. The models were trained using the original images in a variety of methods, including rotation, translation, brightness adjustments, zooming & horizontal & vertical flipping, which supplied many characteristics for training the model more successfully & improving performance. Accuracy, precision, recall and F1-score were used as assessment metrics for models trained for multi-class classification. Precision, recall and F1-score were determined via macro-averaging.
The three components of the HSV colors pace & hue thresholding, not very different values were obtained, with accuracy 0.90 in training, 0.92 in validation & 0.86 in testing, slightly lower than that of without thresholding. The H component without thresholding, was acquired with an accuracy of 0.94, 0.93 in validation and 0.94 in testing. In virtually all instances, accuracy & F1-score were identical. The same CNN architecture was used to categorize the several types of data (spider mite, downy mildew & healthy) in a binary fashion. The outcomes are consistent with the metric accuracy of the tests carried out without considering the threshold for each color & utilizing the H component of the HSV color space. The highest classification accuracies across all datasets were achieved while detecting healthy & diseased leaves with a maximum accuracy of 0.91 & a minimum accuracy of 0.89 respectively.
Working within the leaf photos that were actually collected from the plants in the field presented the biggest obstacle in this. Modelling infield pictures using deep learning was a challenge for which it demonstrated its utility. Additionally, it is utilized to distinguish between two symptoms of a very distinct kind of plant disease. The use of color thresholding did not have a significant effect. It takes a lot of time gathering infield. Based on studies like Guo et al., computer vision techniques allow to create RGB pictures of grapevine leaves for classification & deep learning techniques like data augmentation & CNNs. However, further study is required if automatic identification of leaves from photos of entire canopies is needed. These techniques may be used in agriculture & vineyards to quickly identify pests & diseases in the field.
In the paper
Miaomiao Ji et al [
2] a productive automatic detection and severity examination method based on deep learning and fuzzy logics is proposed for grape black measles disease, for grape black measles disease and individual grape leaves were chosen as the research unit. The primary goal of this paper was to present a grape leaf disease segmentation model based on DeepLavbV3+ architecture with ResNet50 backbone. Plant Pathologists knowledge and experience were used to develop fuzzy rules. For grape black measles, a completely new disease severity classification method is proposed.
Proper database creation is essential for image acquisition. Plant Village which is open source, provided the raw JPG format images with 24-bit color depth. The “Labelme” tool was used to explain samples for the pixel-level semantic segmentation tasks and the interpreted data was saved in JavaScript Object Notation format. Simultaneously, areas of diseased ROI (region of interest) and POI (percentage of infection) from manually labelled images are used as the benchmark to evaluate the accuracy. PSPNet, UNet, and BiSeNet are also used for pixel-level predictions in pathological lesions segmentation tasks. ROI is a perfect value that directly reflects the size of disease lesions and is the basis for calculating POI, as shown in the equation 1.2.1 and 1.2.2 below.
DeepLabV3+ with ResNet50 backbone recognizes pathological damages while simultaneously generating high quality segmentation masks for injured and normal areas in the initial phase. The ROI and POI are extracted and fed into the fuzzy inferences systems. The sample is run through a DeepLabV3+ model with SoftMax output. DeepLabV3+ was trained using loss function-based propagation, and it is the sum of cross-entropy between references and predicted values. To reduce feature maps and capture contextual information, encoder and decoder modules were used. The Atrous spatial pyramid pooling (ASPP) mechanism was used to increase the receptive field without sacrificing information. The decoder module receives the processed combined feature map in order to capture higher semantic information. Fuzzy logic inferences system for classification input fuzzification, generating membership functions, designing fuzzy rules and output defuzzification were all included in the process. DeepLabV3+ segmentation model ROI and POI are used as inputs. When the ROI is equal to zero, the image is classified as healthy. Fuzzification rule database setup invocation and defuzzification were performed.
Metrics for semantic evaluation include mean accuracy score(mAS) mean intersection over union (mIOU). Lin’s concordance correlation coefficients (Lawrence and Lin, 1989) allow evaluating the degree to which the evaluating the degree to which falls on the line of concordance. The overall agreement(ρc) is the sum of overall bias (Cb) and correlation coefficient (r). Deviations in c, r , ρc and from 1 and zero indicate a loss of precision and agreement. Grape Black measles disease severity detection confusion matrices was used.
Fuzzy inference system was introduced so as to get the grape levels plant disease degree assessments estimated diseases severity. The mAS and mIOU metrics are calculated by analysing test images. The DeepLabV3+ model produced the highest IOU values, and the AS and IOU of the divination were highest for the background class. The leaves with small and light spots had a difficult time segmenting. DeepLabV3atrous +s convolutional layers are specially designed for capturing background information, resulting in the precise capture of information related to small targets. The Roi predictions are the most accurate values. The correlation (r=0.99) and concordance correlation(c=0.99) were both high. The experiments yielded approximately accuracy rates of 94%,100%, 97%, and 100% for Healthy, Mild Medium and severe accordingly.
The main challenge of this experiment was that the model classified medium samples as mild samples. The main reason is the disparity between the predicted values of ROI and POI. Most of the confusion were between close to sample labels which contributes a major portion to the overall error. The use of fuzzy set can reduce the impact of errors and improve classification results. The implementation of grape leaf severity detection method has proven to be useful tool with a remarkably high classification accuracy. More research is needed to fully comprehend the application of deep learning-driven semantic segmentation in automatic detection, classification and quantification for plant disease detection tasks.
In the paper
Peng Wang et al [
3]. Proposed that classical deep learning network that was used to gain high precision in recognizing grape leaf diseases. As computing resources requirements are very huge cross-channel interactive attention mechanism-based lightweight model (ECA-SNet) was chosen. When compared to commonly used lightweight methods, the performance is noticeably better. As a result, this paper can successfully use image fine-grained information to identify orchard grape leaf diseases at a low computational cost. The primary goal of the paper was to develop image enhancement techniques. Implementation of a cross-channel interaction strategy that reduces complication while maintaining model performance without depleting dimensionally.
A variety of images of downy mildew, powdery mildew, and healthy leaves are collected in the field using a MI 9 smartphone to shoot from various directions and angles. The public data set contains images of leaf blight, black measles, and black rot. An enhanced robustness test data set was formed by adding Gaussian noise, rotating right 90 degrees, rotating left 90 degrees vertically flipped and weakened sharpness. To reproduce weather inference, Gaussian blur, dissimilarity improvement of 30% and decrease of 30% and brightness improvement of 30% and decrease of 30% are applied to the remaining images in the original data set.
Because there was such a high demand for high-quality deep neural networks on mobile devices. MobileNet-v2 provides a high-quality model with exceptional feature extraction capability by stacking the inverted Residual Block Feature matrix more dimensional. This model developed the feature learning potentiality of deep separable convolution using the Squeeze-and-Excitation module on the basis of MobileNet-v2. The ShuffleNet network was designed to provide channel shuffle and point-wise group convolution. FLOPs (floating-point operations per second) were previously used to estimate the multiplication operation of convolution, and the design of this structure aims to reduce FLOPs. As there were some not to the use of point-wise group convolution. Instead of group of convolutions they used channel separation method.
The attention mechanism assigns high contribution details to larger weights while suppressing irrelevant details through weight distribution, which is a productive method for model performance optimization. This paper introduces ECA-SNet for identifying images of grape leaf diseases. The Max pooling operation was used to reduce the size of the output feature matrix. Cross-channel interaction data was recorded with Conv1D using the equation 1.3.1, taking into account each channel and its n-neighbourhoods.
The exponential function is used to fine-tune the mapping. To reduce the computational cost and time of the training process, as well as to improve the friendliness of model training, hyperparameters b and y were set to 1 and 2, respectively as shown in equation 1.3.2 and equation 1.3.2
The Python programming language is used to build a model based on the Pytorch 1.7.1 deep learning framework and the model is trained and tested on a GPU-equipped server. The numerical change of convolutional layer during the iterative process was used as the abscissa and ordinate represents the number of times the comparable numerical value appears. The model is examined with RTD and the 1.0×version of ECA-SNet as well as the 0.5 version’s confusion matrix. ECA-SNet classification output and the accuracy reached 98.86 to 96.66%. ECA-SNet 1.0× has a improved identification performance and reduced misidentification of the Black rot as Black measles.
Confusion matrix, performance measures including accuracy, precision, recall and F1-score were computed. MobileNET-v2 0.4’s RTD accuracy can reach 95.23 percent, demonstrating the bottleneck structure’s strong feature learning capabilities. ECA-SNet produces high-efficiency since the dimensionality reduction processes are not included. By enabling ECA-SNet to execute with pinpoint accuracy, the channel interaction mechanism significantly improves channel performance. The network topology that is utilised to represent the attention area is known and examined using the attention heat map visualisation technique.
Without dimensionality reduction, the basic goal of ShuffleNet is to make the model have effective channel attention through cross-channel interaction. The layer structure is reduced in several phases with fewer parameters to create an efficient ECA-SNet. One-dimensional convolution was used to reduce the calculation expenses. The invention of automatic inspection equipment for disease diagnosis in grape plants resulted from the discovery of the best recognition effect under the conditions of extremely minimal computation and parameter requirements.
Table 1.
Tabular column 1.1 summary of recent work in RGB based camera.
Table 1.
Tabular column 1.1 summary of recent work in RGB based camera.
| Author |
Contribution |
Plant diseases |
Method |
Limitation /Future scope |
| 1.Salvador Gutierrez et al. Deep learning for the differentiation of downy mildew and spider mite in grapevine under field conditions. |
Finding spider mites & downy mildew on grape vines differentiating their signs. |
1.downy mildew 2.spider mite |
1.Convolutional Neural Networks (CNNs)
2.Data augmentation.
Evaluation matrix:
1.F1 score
2. Recall
3.Accuracy |
Viticulture and agriculture enable quick in-field disease & insect identification Limitation: leaf images taken in actual field settings. |
| 2. Miaomiao Ji et al. Automatic detection and severity analysis of grape black measles disease based on deep learning and fuzzy logic. |
Grades of healthy, mild, medium & severe grape leaves are used to classify them. |
1.Black measles |
1.DeepLabV3+
using ResNet50-
2. Fuzzy logic
Inference system.
Evaluation matrix:
1. mAS
2. mIOU
3.Confusion matrix
|
Deep learning in semantic segmentation. Limitation: model classified medium samples as mild samples. |
| 3. Peng Wang et al. Improved Lightweight Attention Network-Based Method for Recognising Fine-Grained Grape Leaf Diseases. |
Use picture fine-grained features to quickly & affordably identify illnesses in orchard grape leaves. |
1. Orchard
|
1.ECA-SNet lightweight model.
2.Channel interaction
2. ShuffleNet-v2.
Evaluation matrix:
1.F1-score
2. MAC |
Data collection for orchard grape disease in real time. |
2. Multispectral camera
In the paper
I. Fernandez et al. [
4] proposed powdery mildew disease detection model for cucumber plants. Here in most commercialized planthouse was pre-owned to detect plant diseases where images are accumulated using a MicaSense RedEdge camera for close-range non-georeferenced multispectral imagery which was staged on a cart at 1.5m above top of the plant. Planthouse ceiling ranged from 3m to 6m and expanse between plants assorted from 40 to 45 cm with corresponding temperature between 20
0 and 23
0C with companion humidity between 60-70%. Alike environmental state favours for disease spread.
After collecting 20 images through MicaSense RedEdge camera which was staged on a wheeled cart so that fluctuations in the camera position down the greenhouse passage becomes convenient. A computer at a distance placed to control the RedEdge camera facing horizontal field view of 47.20 and vertical FOV of 35.4o captures images of pixel size 0.10 cm free from artificial light. For camera calibration was achieved by owning a MicaSense reflectance white panel with an area of 225 cm2, which was mounted on the top of canopy 1.5 m in distinction to camera.
Entire data were processed using MATLAB R2020b (MathWorks, inc., Natick, MA, USA) workspace. Functions and constants associated with MATLAB R2020b were got from www.mathworks.com. Where images are transformed from uint16 to uint8 file form with the im2uint8 function. The model arranges identical points for the two fixed and in motion images using Speeded-Up Robust Features (SURF) to figure out blob-like formations (SURF features). Motion images are modified into fixed images through geometric transformation techniques. To work with a little training dataset a Gaussian Support Vector Machine (SVM) algorithm can be applied to classify healthy and infected pixels from the recorded bands and associated vegetation indices.
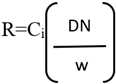
Digital Numbers (DN) of the processed images were transformed using below Equation (2.1.1)
R, means band reflectance.
DN, means digital number of all pixels in the recorded band image.
w, means digital number out of whiteboard panel image calibration.
Ci, means reflectance coefficient (where i = 1 to 5)
To obtain a higher contrast between bright and dark skeletal factors and to minimize regional mean intensity the images were normalized in the middle of 0 and 1. imbinarize function which generates binary image out of the grayscale image was accounted to clear away the background. A dataset with 1000 infected and 1000 healthy pixels was obtained, from which 70% of dataset in given to the SVM algorithm for training and remaining 30% for validation, a confusion matrix and the Sensitivity, F1 score, Precision and total accuracy were evaluated.
For the bands red, green, and blue bands were calculated using corresponding coefficient C1, C2, C3 respectively. The band with highest esteem specifies that specific band had highest radiance. Finally, from results got to know that blue band had the highest illumination out of aligned RGB image, and it is a best band with accuracy of 89%. The worse band was out to be red-edge band with accuracy of 59%. Classification of diseased and healthy leaf areas became convenient when re-examine the images of bands NIR and RE with an unsupervised classification which provides higher reflectance intensity of mock-inoculated than the pathogen-infected class. For the trained model the overall accuracies of RGB and other band reflectance were 93% and 99% respectively.
Esca fungi causing Grapevine trunk disease (GTD) is reported in the paper
Bendel et al. [
2]. During three successive years 2016 to 2018 multispectral imaging was performed for active detection of foliar Esca indications. Disease detection model was amid symptomatic and asymptomatic utilizing two of original data and annotated data. The vineyard incorporated with 120 m length rows originating from east-west direction. Spacing of 1.8 and 1.3 m between interrow and grapevine respectively. The accurate position of every plant was collected using a portable GPS (SPS585, Trimble, Sunnyvale, CA, USA).
Systematized image acquisition is performed using two 300W short-wave spotlights and PTFE spectralon having smaller calibration were used with two different line scanning cameras. In 2016 spectra in the visible and near-infrared range were filed by a line scanning camera HySpex VNIR 1600 and other two years 2017 and 2018 with a HySpex VNIR 1800. Nearest wavelength interpolation strategy was applied for camera-to-camera transformation. Vine based GPS labelling and Manual labelling of Esca symptoms were two perspectives for labelling leaf stuffs.
Data processing was managed by the Fraunhofer IFF HawkSpex Flow software, which is tooled inside Matlab workspace. Two distinct models for VNIR and SWIR image processing were designed. Positive reflectance instance pattern linked with negative instance pattern to obtain a new set for the modelling experiments. The mask was obtained on the VNIR image and forecasted on the SWIR images to produce instance pattern from both wavelength spectrum.
An algorithm is interpreted for each approach which finds ideal parameter of a model. A coded method i.e. coding -1 for control and +1 for infection was adopted which represents visual rating class correlated with data points, then for optimization either gradient descent or closed form mathematical computation are used. Performance criteria for all models was as follows in the given below equations 2.1.2, 2.1.3 and 2.1.4
Evaluation begins with an n-fold cross validation method, where all models were estimated and amended on n-1 folds while evaluated on nth-fold. 10,000 random samples were acquired using weighting profile used as a probability function(pdf). To place characteristic wavelengths dataset is used with Neural Gas algorithm which is vector quantization algorithm. Band of 30 nm full-width-half-maximum (FWHM) is accepted to be ideal for multispectral cameras.
CA, TPR, FPR values of experiment for both models and individual years outcomes overall accuracy with CAs of 73%, 70% and 77% (VNIR) and 73%, 81% and 80% (SWIR). When considered only symptomatic leaves outcome was more appreciable TPRs up to 100% with FPRs 0 to 11% leads to CAs of 88 to 95%. Results show that VNIR seems to deliver the more stable wavelength set, whereas SWIR range is suitable for pre-symptomatic disease detection.
A remedial management of fungal leaf disease caused by fungi
Botrytis cinerea is explained in the paper
Fahrentrapp et al. [
3]. The five bands recorded were green, blue, red, near infrared (NIR, 840 nm) and rededge (RE, 720 nm). Here tomato leaflets were pathogen and mock-inoculated at same time for 5 h inside a greenhouse having open windows. During sunny days the temperature was maintained with 20-26
o and during cloudy days 40
0. Lighting of 80 kW per square meter for 16 h per day was done using artificial light.
Frame by frame experiment supplemented with lighting for image acquisition where images are taken with multispectral camera MicaSense RedEdge assembling 10-40nm wide bands in blue, green, red, near-infrared, and rededge which captures 1280 X 960 pixels, every pixel envelops approx. 0.2 mm2 at a span of 66.6 cm above the pathogen and mock-inoculated leaflets, taken every 80 min. Python v2.7 script was implanted to gather imagery with proper camera and lights.
ImageJ software bundled in the FIJI distribution are used for image processing and time lapse study. Since the multi-lens and multi-sensor configuration of the camera model leads to band misregistration result, this problem is resolved when every spectral band processed band-wise. It consists of five steps i.e., to identify regions of interest, statistical outlier image detection using stack, Outlier image removal and calculation of mean image intensities on frame scale when image is stabilized, Frame co-registration, profiling images, excluding background.
To spot infected and healthy segment of leaflets an unsupervised classification in addition with masking strategy was implemented. To separate the masks of the image into two classes an iterative self-organizing (ISO) classifier was used. The class with the least constant is predicted as an infected portion of the leaflets. Linear regression models were developed to forecast temporal flip in the band intensities.
To originate 95% confidence intervals a parametric bootstrap approach for linear regression constants to report for temporal dependence and the measures regression parameters and model fit: R2(coefficient of determination) and RMSE (root mean squared error) were accounted. -x resampling approach was accounted for substituting original residuals in individual iterations.
Linear regression model depicts NIR and RE are the superb descriptive data sets in the process of extricating pathogen and mock-inoculated leaf Regions of Interest (ROI). Reflectance intensity of the green band was decreasing over the time. The best overall accuracy was accomplished with RGB composite (89%) and with the PMVI-2 image (81.83%). The outcome of this experiment specifies that leaf infection symptoms can be recognized at the earliest at 9 h post infection (hpi) within informative bands NIR and RE.
Table 2.
Tabular column 2.1 summary of recent publication in Multispectral imaging-based technique papers.
Table 2.
Tabular column 2.1 summary of recent publication in Multispectral imaging-based technique papers.
| Author |
Contribution |
Plant diseases |
Method |
Limitation/Future scope |
1.I. Fernandez et al. [4].
Detecting Infected Cucumber Plants with Close Range Multispectral Imagery
|
Powdery mildew caused by Podosphaera xanthii was detected using close-range multispectral imagery over cucumber plants. |
Powdery mildew |
1.Speeded-Up Robust Features (SURF).
2.Gaussian Support Vector Machine algorithm.
Evaluation metrics:
1.confusion matrix
2.Sensitivity
3.Precision
4.F1 score
5.Overall Accuracy
|
Every time we provide a new or large dataset to a detection model, it is always difficult.
More research is needed before the symptoms manifest to put the imagery to the test. |
2.Fahrentrapp et al. [6]
Detection of Gray Mold Leaf Infection Prior to Visual Symptom Appearance Using a Five-Band Multispectral Sensor |
Time-course experiments of detached tomato leaves inoculated with the fungus Bortrytis cinerea and mock infection solution were recorded using a five-lens multispectral imaging.
|
Gray Mold |
1.Unsupervised Schema of classification
2.Masking method
Evaluation metrics:
1.Linear regression
2.Bootstrap approach |
Analyzing the difference in reflectance intensity to determine whether it is due to biotic or abiotic stresses, or a true infection is a limitation. |
3.Bendel et al. [5]
Evaluating the suitability of hyper and multispectral imaging to detect foliar symptoms of the grapevine trunk disease Esca in vineyard |
To assess the feasibility of detecting foliar esca symptoms in the field over three years using ground-based hyperspectral and airborne multispectral imaging. |
Grapevine trunk diseases (GTDs) |
1.Ground-based imaging technique
Evaluation metrics:
1.Gradient descent
2.Cross validation |
Environmental factors influence reflectance. A single-lens multispectral camera can be used to reduce computational steps for accurate band registration. |
3. Thermal imaging
In the paper Dong-Mei Wen et al. [
7] proposed downy mildew disease detection model for cucumber plant. Here on moist gauze in petri dishes, the seeds of cucumber were germinated in a monitored environment for 3 days at 25°C/20°C (day/night temperature) having a relative humidity of 80%±10% and photoperiod of 16hd^-1(RXZ-380D LED lights).in 3:1 mixture of peat and vermiculite germinated seeds were planted. Plants were irrigated & fertilised with tap water every 2-3 days and organic liquid fertilizer (N+P+K≥280g/L, microelement≥2g/L) respectively and utilised for the investigations with the fifth to 6th b genuine leaves were unfurled.
P.cubensis samples were obtained in a plant house effect in Beijing, China from naturally infected cucumber leaves. The infected leaves were turned to the lab and cleaned with distilled water to remove debris such as sporangia and dirt. To stimulate the generation of new sporangia, leaves with downy mildew symptoms were put in a darkened controlled environment room at 20°c and 100% relative humidity for 20 hours. The sporangia were dislodged into distilled water with a gentle brush and filtered three times with a cell strainer(70m). Using a Fuchs-Rosenthal hemocytometer, the concentration was adjusted to 1 106 sporangia mL^1.
Thirty-five plants were chosen for P.cubensis inoculation; all had five true leaves to allow subsequent thermography observations. P.cubensis spore suspension was manually sprayed across the full surface of the leaves with a hand-held sprayer(YZB-A) in a controlled atmosphere of 20°C-25°C.The leaves were covered with a clear polythene bag for at least 2 hours after inoculation to establish a high humidity environment favourable to infection. Plants tests (immunised and non-immunised) were equilibrated in the research center for 1 hr before thermography and the estimation of apparent spectra computerised pictures.
A progression of computerised warm pictures were caught utilising an infrared checking camera(FLIR A615;FLIR Frameworks) with an unearthly responsiveness of 7.5-14 micrometre and a mathematical goal of 0.69 mrad(640x480 pixels).The camera has an extraction ±2°C and a warm responsiveness of 0.05°C.It was put on a stand, and the distance between the camera and the leaves was roughly 0.8m.The warm camera was situated almost opposite to the leaf surface. At the point when capturing the noticeable range pictures, the camera was put on a stand to avoid the leaf tests that was essentially as steady as conceivable with that of the infrared filtering camera.
The FTIR spectrum was obtained by co-adding 32 scans at a 4cm-1 resolution with a VERTEX 70 spectrometer (Bruker Optics) in the 4000 400cm^-1 wavenumber range. Six cucumber plants were inoculated for FTIR spectroscopy measurements. Three infected leaves were randomly excised the morning after inoculation, and for next ten days. Three spectra were acquired and averaged. In the later stages of side effect improvement, tests were extracted from five ailing leaves for ghastly location from (1) the region between the apparently not healthy issues and the healthy tissue, (2) the focal region of effected tissue and (3) the focal region of sound tissue. The range data was provided.
For FTIR spectroscopy estimations, we chose potassium bromide (KBr) to quantify the infrared spectrum as it is normally utilized, has great conveyance and less infrared retention in the infrared locales. To keep away from obstruction in the absorbance range brought about by the water, entire leaf tests and the KBr were dried at 40°C and 100°C,separately,for 48hrs, under infrared light , ground to a fine powder the powder was made from a 1:100 mixture of leaf test and KBr and was ground in an agate mortar to reduce the molecular size to 100 micrometres.
The subsequent powder was squeezed into a plate for transmission infrared spectroscopy. Reference spectra was obtained by gathering a range from an unadulterated KBr plate. The KBr foundation was deducted from the range of each leaf test. We analysed the distinctions in the spectra of the locales between: (1) the unhealthy region and the sound region, (2) the focal piece of the ailing tissues, and (3) the solid tissues. The principle regions got for explicit spectra were likewise determined.
Thermal infrared photos were examined with BM IRV7.4 Thermal Infrared Image Analysis Software after being saved as BIT files (FLIR Systems). The leaves with highest and lowest temperature were measured. The maximum temperature difference (MTD) for the leaf regions was calculated as the highest temperature minus the lowest temperature. Thermal image analysis software was used to monitor the spot temperature on the same leaf from day 1 to day 10. Because apparent leaf symptoms cannot be observed by infrared imaging from one to three days after infection, we located the lesions four days after infection and later located the corresponding position on the leaves to calculate the spot temperatures prior to the occurrence of visible symptoms.
The wave numbers at 2977 cm^-1,1544 cm^-1 and 1050 cm^-1 were sensitive to early infection & the good indicators of the pre-symptomatic stages of P.cubensis infection. The findings of this study suggest that FTIR spectra could be a promising technique for pre-symptomatic detection of downy mildew in cucumber. FTIR has an advantage over traditional methods in that it can detect changes in the plant before visual symptoms caused by the pathogen appear. In comparison to the FTIR method in our study, FTIR-ATR spectroscopy may be more likely to be non-destructive for the early diagnosis of plant diseases. When paired with thermal imaging, FTIR spectroscopy can assist give a foundation for understanding the complicated chemical processes that occur during pathogen invasion for a host.
It is difficult to collect non-destructive samples for early illness detection. One difficulty is that observations of infrared thermography can be impacted by a variety of other elements that affect leaf temperature, including as ambient temperature, humidity, sunshine and wind. Another problem is that, due to technical constraints, thermal imaging at these sizes is now limited to a medium-resolution level. The main issue with utilising FTIR as an infection diagnosis technique is acquiring leaf samples since the samples must be taken from the plant & the sample processing stage is difficult and time-consuming. However, future research must address the limitations of the instruments described in this study.
In the paper
Shamaila Zia-Khan et al. [
8] proposed downy mildew disease detection model for grapevine using infrared imaging. The experiment was place in the University of Hohenheim’s experimental vineyard. The vines under investigation were 33-year-old Pinot Meunier grafted on SO4.The experimental plot was 250m^2 in size & was divided into four rows of 25 plants each, running north-south. Sensors were mounted in four distinct positions around the experimental plot. Each row was photographed infrared such that the camera could capture 1m^2 of area. This region was taped off, & the infrared photos were captured inside this frame.
Every time infrared photos of the indicated region were captured, it was documented. In addition,10 leaves were marked with red tape on the sunny and shaded sides of the canopy, and infrared photos of single leaves were obtained. Irrigation was not used, and single leaf infrared photos were captured. Throughout the trial, no irrigation was used since precipitation was adequate to fulfil crop water requirements. Micro data loggers (Testo 174H, Titisee-Neustadt) were used to measure and record microclimate data in the canopy, such as air temperature and relative humidity.
A resistance grid sensor was used to monitor the moisture of the leaves (PHYTOS 31). When the sensor was dry, it provided roughly 435 leaf moisture counts, however when completely wet, as in rain,the signal jumped to approximately 1100 counts. As a result, numbers greater than 435 were transformed to leaf wetness duration (LWD) via data processing. All of the sensors were installed at a height of 1.3m above the ground, and data was collected at 10-minute intervals. If there was a disagreement in the output of the leaf wetness sensors, a period was considered moist if the mean relative humidity recorded by probes was greater than 85%.
Weather data were collected from a weather station located 75 meters away from the experimental field. The plots were manually monitored every third day of the experiment (DOE) from the beginning to the conclusion to compare the measured illness level. The canopy was checked by virtually evaluating each vine from east to west based on the individual leaves at various heights. Disease level in % was calculated as the percentage of infected leaf area.
Thermal imaging was performed using an infrared camera. Images were only taken on the bright days when the crop’s transpiration rate was high. Each thermal scan covered appropriately 1 m^2.The thermal pictures were analysed using the thermography program IRBIS (InfraTech GnbH), and the emissivity value of 0.95 was chosen. To distinguish between the canopy and the soil surface, an upper temperature threshold was established, and pixels with higher temperatures were rejected by image processing in the given below equations 3.1.1, 3.1.2, 3.1.3
The mean canopy temperature ACT was computed as
Tpixel is the temperature of a pixel in the infrared picture, and n is the number of pixels in the selected polygon.
Within the canopy polygon, the maximum temperature difference (MTD) was computed as follows:
The canopy temperature differential (CTD) was defined as:
where the weather station measured the air temperature Tair.
To assess the influence of downy mildew on ACT, MTD and CTD, all analyses were performed using Origin version 2019.P. viticola’s impact on ACT, MTD and CTD was evaluated using conventional analysis of variance (ANOVA) at a least significant threshold of 0.05.A Pearson regression analysis was done to determine the relationship between illness level and ACT, MTD and CTD.
We differentiated between the sunny and shaded sides of the canopy since there is still the potential of shaded leaves occurring on the sunlit side of the vines within the canopy, and vice versa. Throughout the trial, the MTD of the sunny leaves was greater than that of the shaded leaves, which was very significant (p 0.001). It should be highlighted that looking solely at the darkened side might be deceiving because it does not fluctuate much & only responds when the illness severity is severe. In the CTD, the difference between the shaded & sunny leaves was significant (p 0.001). The CTD of the sun-shaded side was always greater than zero & declined in both sunlit & non-shaded leaves throughout the last days of the measurements.
Our data confirm Fuchs’ observations that there is an increase in temperature variance when stomata close; indeed, there is a definite trend of temperature being higher in sunny leaves than in shaded leaves. As the conclusion of the trial or as the disease level grew, a drop in CTD and an increase in MTD were found, independent of the canopy sides, caused by a loss in photosynthesis activity. At the end of the experiment, the shaded side of the canopy had a minimum temperature of 20°C and the sunny side had a maximum temperature of 38°C.
Two key conclusions may be taken from the data acquired under the field experiment circumstances. To begin, the causative agent of grapevine downy mildew requires temperatures below 20°C to grow, and alternate periods of leaf wetness length and drying, together with the ideal temperature range of 20°C-25°C, will enhance disease severity. Second, because pathogen infestation modifies plant metabolic processes, particularly leaf transpiration rate, infrared imaging techniques can be used to detect the disease’s early start. A 3.2°C increase in canopy temperature was observed in this study well before visible symptoms appeared.
Only the MTD parameter was substantially associated (R^2=0.76) to the illness level, and hence it may be utilized to predict the onset of fungal disease in grapevine. As a result, if and when used, infrared photography helps speed up the plant disease screening process. However, further research is needed to evaluate the impact of light direction on illness development in relation to microclimatic variables.
In the paper
Bar Cohen et al. [
9] proposed downy mildew disease detection model for grapevine using infrared imaging. Experiments on 169 grapevine plants, cultivar ‘Chardonnay,’ were carried out in six campaigns between the end of December 2019 and the end of October 2020. The plants were cultivated in experimental greenhouses at 25/18 degree Celsius (day/night) in plastic pots with a blend of organic soil (Ever Ari Green LTD). Each campaign examined between 15 and 34 plants (depending on availability) and comprised different imaging days (healthy leaves and 1, 2, 4, 5, 6 and 7 days after inoculation). Photographs were not obtained on the third day following inoculation since it fell on a non-working day, making it impossible to capture images to this day. Furthermore, according to literature, symptoms often manifest on thermal imaging on the fourth day following inoculation; hence, the data for the third day were not collected. Plants were watered twice daily with fresh water and organic liquid fertilizer (ICL Fertilizers).
The following stages were featured in each of the six campaigns:
The second unfurled leaf from the apex of each stem was chosen for inoculation and tagged with a colour clip or aluminium foil (2-6 leaves per plant).
Images of healthy leaves were obtained on the first day of the campaign.
Using a hand sprayer, the leaves were inoculated with a 1 104 concentration of P. viticola on the lower surface.
The injected plants were housed in a high humidity room under ideal environmental conditions to allow the virus to infect the host tissue and cause DM to form.
Images of healthy and infected leaves were obtained 1-7 days after inoculation. Photographs were only taken on bright days with a clear sky, which means that if the weather circumstances were unsuitable on a specific day, no images were taken.
Following the last imaging day (day 7), the leaves were put in Petri dishes to assess the extent of disease development, which was visually graded by an expert between 0 and 10 (0- healthy, 10- severe illness).
On October 27, the diurnal response of leaf temperature was observed. Images were taken at seven distinct times (round) during the day, between 7:00 a.m. and 4:30 p.m. Each round lasted around one and a half hours and comprised approximately 87 samples. Six leaves were taken from each plant: three six days after inoculation and three healthy. Each plant received two daily water doses: one before the first cycle and one before the fourth. Imaging was performed outside of the greenhouse to allow for maximum photosynthesis of the plants. The plants were removed from the controlled greenhouse and placed outside for at least one hour before imagine to allow them to acclimate to ambient conditions that differed from those in the greenhouse. To avoid fluctuations in the lighting of the plant surfaces when the sun angle changed, each leaf was put exactly in front of the sun. Meteorological parameters, including air temperature (degree Celsius, solar radiation (W/m2),), relative humidity, wind direction and wind speed (m/s) and were continually recorded.
Two cameras were utilized to capture images of each leaf: a thermal camera (FLIR SC655, FLIR Systems) and an RGB camera (Canon EOS6D, Canon Inc.) for documentation. The infrared camera employs an uncooled microbolometer detector with a resolution of 640 480 pixels, sensitivity in the spectral region of 7.5-1.3m, accuracy of 2 degree Celsius or 2% pf the reading, and thermal sensitivity of 0.05 degree Celsius @+ 30 degree Celsius. The thermal camera captured a half-minute video of each leaf, while the RGB camera captured two photos. For categorisation, one picture from each video was used. The image was chosen by eye, calculating the maximum leaf surface exposed to the camera. There were 1403 entries in the classification dataset (599 healthy leaves and 804 infected leaves). The records contained leaf thermographic measures, meteorological measurements gathered concurrently, computed characteristics from raw data, and manual disease severity rating. To establish the earliest day on which a model can detect the disease, a subset of this dataset was generated that comprised records with real disease severity of 5 or above. This collection contained 1097 records (599 healthy leaves and 498 infected leaves). Outliners were deleted from the new collection, yielding a total of 1012 records (571 healthy and 441 infected leaves).
Edge detection with the ‘Chan-Vese’ active contour method was used to segment leaves. This approach guarantees an impartial contour that may be shrunk or expanded based on picture attributes. The program was created with MATLAB version R2019b (MathWorks Inc) with enhanced functionalities (Shawn Lankton, 2007). The algorithm’s inputs were a grayscale thermal picture and initial mask; the algorithm’s outputs were an image with the leaf contour and a final mask. The original mask position and size were individually selected for each leaf. The mask was subjected to 100iterations of the active contour method with a smoothing term of 0.3. (Lambda).
The mask of the leaf was used to compute the leaf’s characteristics. After being given numbers for ambient temperature, reflected temperature, emissivity, and distance from the item, the thermal camera calculated the absolute temperature of the leaf. Relative values were employed in the analysis by adjusting characteristics expressing temperature by T-Tair. Healthy leaves were averaged 30 degrees Celsius with a standard deviation of 4, while diseased leaves averaged 31.1 degrees Celsius with a standard deviation of 5.22. The temperature difference between healthy and diseased leaves was around 1 degree Celsius. The classification performance was measured using the accuracy, precision, recall, F1 score, and the area under the receiver operating characteristic (ROC) curve, abbreviated as the AUC. The class set had two labels: positive(infected) and negative(healthy) in the given equation 3.1.4 ,3.1.5, 3.1.6, 3.1.7 and 3.1.8
Given a classifier and an instance, there were four possible outcomes:
True positive (TP): the leaf was infected, and it was classified as infected.
False-negative (FN): the leaf was infected, but it was classified as healthy.
True negative (TN): the leaf was healthy, and it was classified as healthy.
False-positive (FP): the leaf was healthy, but it was classified as infected.
Accuracy is defined as the likelihood of properly categorising a test instance:
Precision is also known as positive predictive value, and it is calculated as follows:
Recall is also known as the true positive rate (TPR), and it is calculated as follows:
The F! score is calculated as the harmonic mean of accuracy and recall.
An ROC curve is a graph that displays two parameters that represent the performance of a classification model at all classification thresholds:
Recall (sometimes referred to as TPR);
False Positive Rate(FPR).
The following is how False Positive Rate is defined:
TPR vs FPR is plotted on a ROC curve at various categorisation levels. Lowering the categorisation threshold causes more items to be classified as positive, which means increases both False Positives and True Positives. AUC calculates the area under the full ROC curve in two dimensions.
The findings show that thermograms can identify downy mildew before any obvious signs arise. An SVM model based on a balanced dataset with the following features: MTD, STD, percentile 90, CV and CWSI performed best for distinguishing healthy & sick leaves. The model outperformed all other models evaluated by 10% (81.6% accuracy,77.5% F1 score and 0.874 AUC). The inconsistency of the results (days 4-7 following inoculation) could not be explained. The optimal time inconsistency of day for capturing photos for downy mildew identification was between 10:40 am and 11:30 am, with an accuracy of 80.7%, an F1 score of 80.5% and an AUC of 0. 895.Even if the images are not from the same day, using images from the best hours is likely to improve performance.
Even if the picture capture is performed at the optimal time, fluctuations in light cannot be prevented, resulting in diminished performance. There is a trade-off between employing a vast and diverse database (collected over time) and identifying the illness (the fewer dates, the easier). Thermal imaging can detect illness early and should be improved.
Table 3.
Tabular column 3 summary of recent publication in thermal based papers.
Table 3.
Tabular column 3 summary of recent publication in thermal based papers.
| Author |
Contribution |
Plant diseases |
Method |
Limitation /Future scope |
1.Dong-Mei Wen et al [7].
Downy mildew disease detection model for cucumber plant. |
(1) To see if P.cubensis infection of cucumber can be detected early on using FTIR and thermal imaging techniques, and (2) to select the best discriminating criteria for detecting P.cubensis infection at the pre-symptomatic stage. |
Downy mildew |
1.FTIR spectrum
2.Infrared thermography
Evaluation matrix:
1.F1 score
2. Recall
3.Accuracy |
The main challenge with utilizing FTIR as an infection diagnosis technique is obtaining leaf samples because they must be taken from the plant and the sample processing stage is tedious and time consuming. Future research, however, will need to address the limitations of the instruments described in this study.
|
2. Shamaila Zia-Khan et al [8].
Downy mildew disease detection model for grapevine using infrared imaging. |
The purpose of this study was to see if high-resolution thermal imagery and physiological indices could be used to detect P. viticola infection in grapevine. |
Downy mildew |
1.Remote sensing
2.Thermal imaging |
Further research is needed to evaluate the impact of light direction on illness development in relation to microclimatic variables. |
| 3 Cohen et al. [9] Downy mildew disease detection model for grapevine using infrared imaging. k
|
The particular goals were to:
(i)extract classification features based on temperature and image processing methods.
(ii) create classification models to discriminate between infected and healthy grapevine leaves; and
(iii) find the ideal time of day to gather thermal pictures for DM detection. |
Downy mildew
|
1.Decision tree
2.Logistic regression
3.Navie Bayes
4.Support vector machine (SVM)
5.Ensemble |
Even if the picture capture is performed at the optimal time, fluctuations in light cannot be prevented, resulting in diminished performance. There is a trade-off between employing a vast and diverse database (collected over time) and identifying the illness (the fewer dates, the easier). Thermal imaging can detect illness early and should be improved. |
Conclusion
The study’s overarching goal was to evaluate spectral sensors for the plant diseases detection. However, transferring disease detection models to unknown data continues to be a difficult task. The images are captured spatially and spectrally and are used to calculate vegetation indices. The current research is part of an ongoing project to develop deeper into deep learning architecture boost for plant disease. When there is a wide range of symptoms addition of pathogen takes place because of adaption of pathogen to anti-infection solutions they grow more stronger and with new varieties of gene expression due to the evolution happened in that species, the detection model has a difficult time detecting differences in spectral reflectance, which is why experiments must be noticed and update pre-symptomatic changes every year. To summarise our work, we attempted to demonstrate a tool for identifying leaf microbe effecting at a pre-visual stage rather than relying solely on the naked eye.
Acknowledgments
The author is thankful to the reviewer for their critical comments and suggestions.
References
- Salvador Gutierrez, Ines Hernandez, Sara Ceballos, Ignacio Barrio, Ana M. Diez-Navajas, Javier Tardaguila, Deep learning for the differentiation of downy mildew and spider mite in grapevine under field conditions, Computers and Electronics in Agriculture, Volume 182, 2021. [CrossRef]
- Miaomiao Ji, Zhibin Wu, Automatic detection and severity analysis of grape black measles disease based on deep learning and fuzzy logic, Computers and Electronics in Agriculture, Volume 192, 2022. [CrossRef]
- Wang P, Niu T, Mao Y, Liu B, He D and Gao Q, Fine-Grained Grape Leaf Diseases Recognition Method Based on Improved Lightweight Attention Network. Front. Plant Sci. 2021. [CrossRef]
- Fernandez, C.I.; Leblon B.; Wang J.; Haddadi A.; Wang K. Detecting Infected Cucumber Plants with Close-Range Multispectral Imagery. Remote sens. 2021. [CrossRef]
- Bendel,N., A., Backhaus, A. et al., Evaluating the sutiability of hyper- and multispectral imaging to detect foliar symptoms of grapevine trunk disease Esca in vineyards. Plant Methods16, 142 2020. [CrossRef]
- Fahrentrapp J, Ria F, Geilhausen M and Panassaniti B, Detection of Grrey Mold Leaf Infections Prior to Visual Symptoms Appearance Using a Five-Band Multispectral Sensor. Front Plant Sci. 2019. [CrossRef]
- Wen, DM., Chen, MX., Zhao, L. et al. Use of thermal imaging and Fourier transform infrared spectroscopy for the pre-symptomatic detectin of cucumber downy mildew. Eur J Plant Pathol 155, 405-416 (2019). [CrossRef]
- Zia-Khan, S.; Kleb, M.; Merket, N.; Schock, S.; Muller, J. Application of Infrared Imaging for Early Detection of Downy Mildew (Plasmopara viticola) in Grapevine. Agriculture 2002, 12,617. [CrossRef]
-
9. Cohen, B.; Edan, Y.; Levi, A.; Alchanatis, V. Early Detection of Grapevine (Vitis vinifera) Downy Mildew (Peronospora) and Diurnal Variations Using Thermal Imaging. Sensor 2022, 22, 3585. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).