Submitted:
08 November 2023
Posted:
09 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
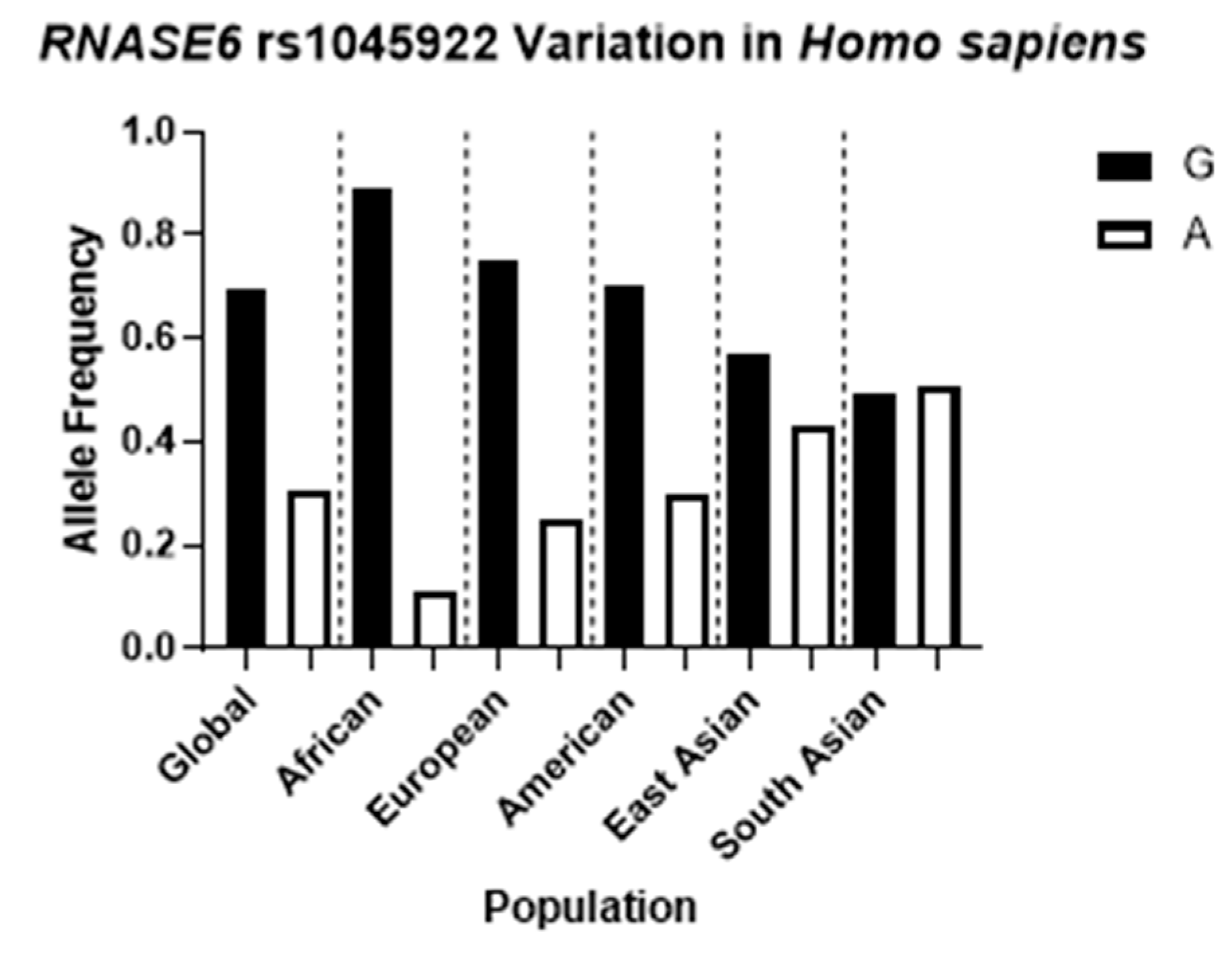
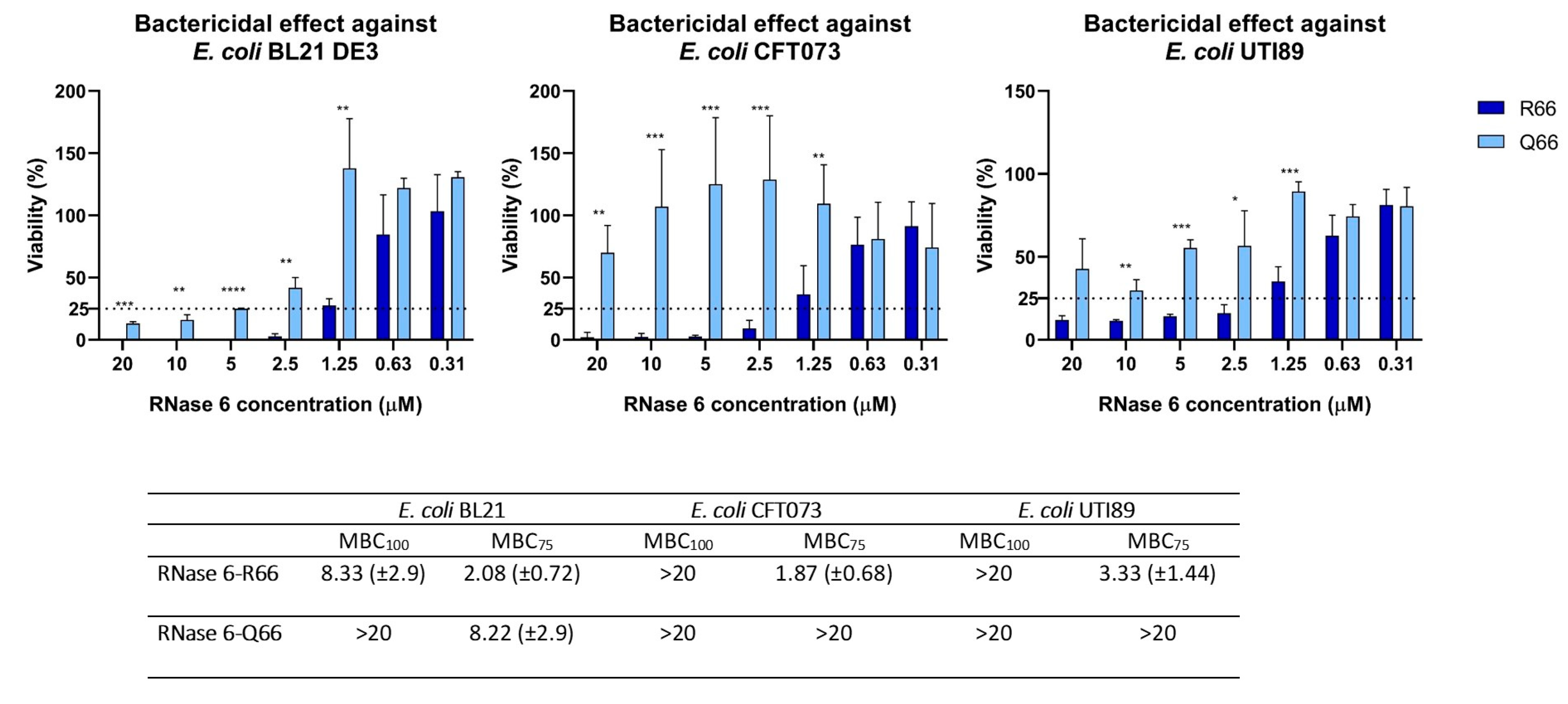
2.1. RNASE6 rs1045922 Is a Common, Non-Synonymous SNP That Alters RNase 6 Antimicrobial Activity
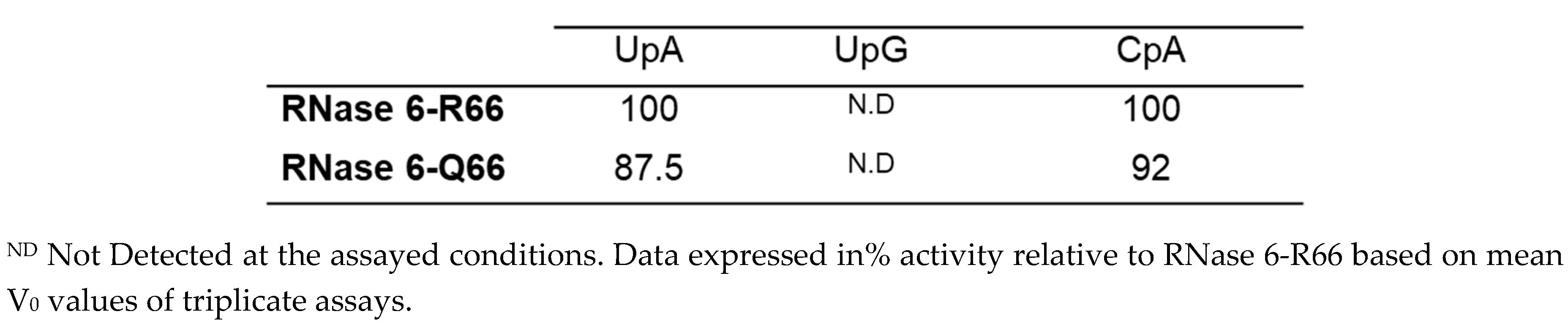
2.2. Both SNP RNase6-R66 and -Q66 Display the Same Catalytic Activity
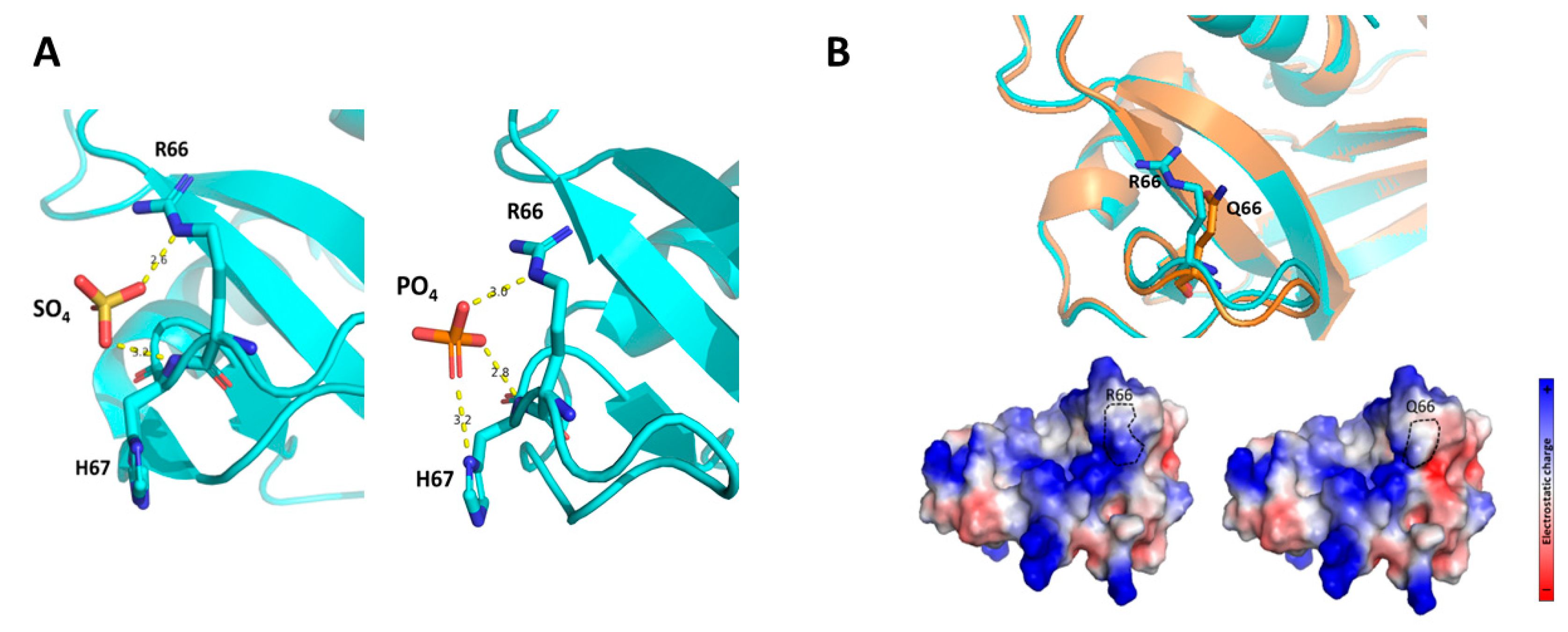
2.3. The RNase6 R66Q Substitution Significantly Reduces the Positive Electrostatic Charge at the Protein Surface
3. Discussion
4. Materials and Methods
Materials
Expression of RNase6-R66 and -Q66 Variants
Minimum Bactericidal Concentration (MBC) Determination
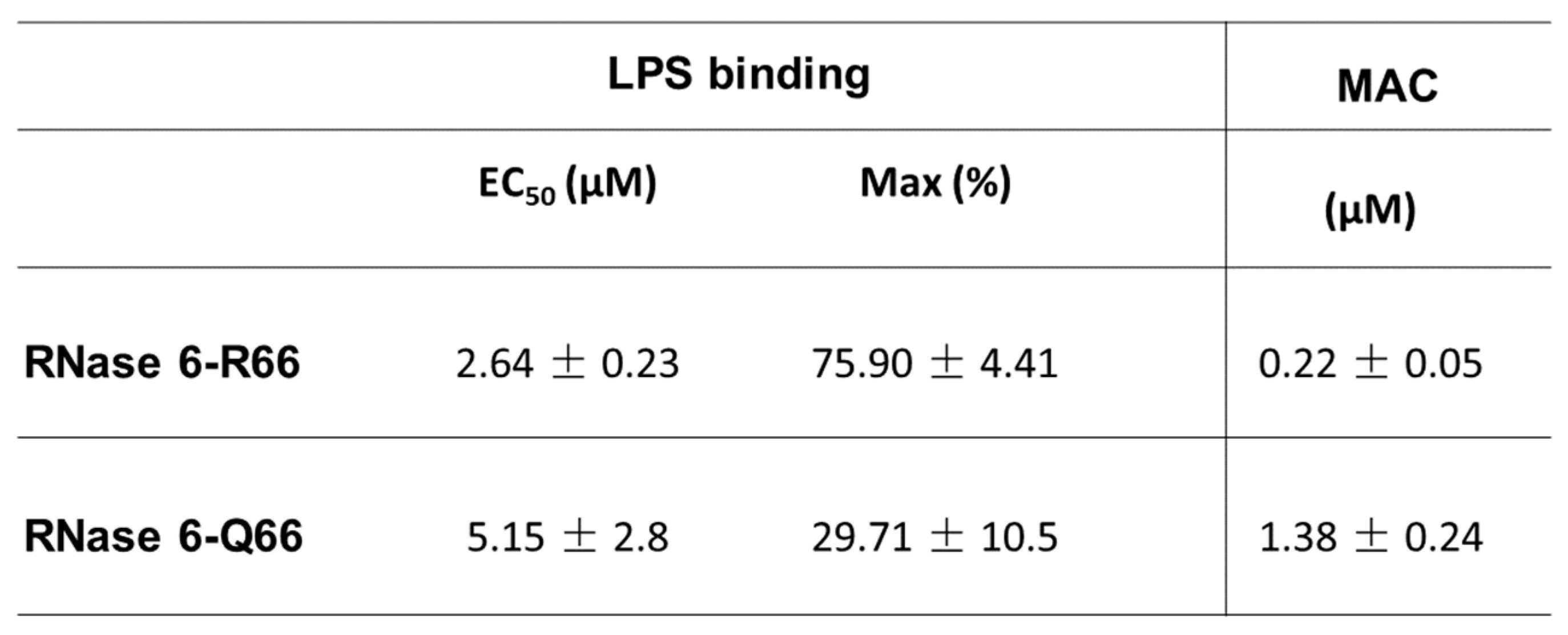
Lipopolysaccharide Binding Assay
Bacterial Agglutination Assay
Spectrophotometric Kinetic Assay
3D Structure Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Medina M, Castillo-Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urol 2019, 11, 1756287219832172. [Google Scholar]
- Foxman, B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Johnson JR, Russo TA. Acute Pyelonephritis in Adults. N Engl J Med 2018, 378, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Becknell B, Schober M, Korbel L, Spencer JD. The diagnosis, evaluation and treatment of acute and recurrent pediatric urinary tract infections. Expert Rev Anti Infect Ther 2015, 13, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Korbel L, Howell M, Spencer JD. The clinical diagnosis and management of urinary tract infections in children and adolescents. Paediatr Int Child Health 2017, 37, 273–279. [Google Scholar] [CrossRef]
- Paul, R. State of the Globe: Rising Antimicrobial Resistance of Pathogens in Urinary Tract Infection. J Glob Infect Dis 2018, 10, 117–118. [Google Scholar] [CrossRef]
- Abbott IJ, Peel TN, Cairns KA, Stewardson AJ. Antibiotic management of urinary tract infections in the post-antibiotic era: a narrative review highlighting diagnostic and antimicrobial stewardship. Clin Microbiol Infect 2023, 29, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Kuhn HW, Hreha TN, Hunstad DA. Immune defenses in the urinary tract. Trends Immunol 2023, 44, 701–711. [Google Scholar] [CrossRef]
- Ching C, Schwartz L, Spencer JD, Becknell B. Innate immunity and urinary tract infection. Pediatr Nephrol 2020, 35, 1183–1192. [Google Scholar] [CrossRef]
- Lacerda Mariano L, Ingersoll MA. The immune response to infection in the bladder. Nat Rev Urol 2020, 17, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Becknell B, Schwaderer A, Hains DS, Spencer JD. Amplifying renal immunity: the role of antimicrobial peptides in pyelonephritis. Nat Rev Nephrol 2015, 11, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ali AS, Townes CL, Hall J, Pickard RS. Maintaining a sterile urinary tract: the role of antimicrobial peptides. J Urol 2009, 182, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [CrossRef]
- Becknell B, Ching C, Spencer JD. The Responses of the Ribonuclease A Superfamily to Urinary Tract Infection. Front Immunol 2019, 10, 2786. [Google Scholar] [CrossRef] [PubMed]
- Becknell B, Eichler TE, Beceiro S, Li B, Easterling RS, Carpenter AR, James CL, McHugh KM, Hains DS, Partida-Sanchez S, Spencer JD. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int 2015, 87, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Wang G, Mishra B, Lau K, Lushnikova T, Golla R, Wang X. Antimicrobial peptides in 2014. Pharmaceuticals (Basel) 2015, 8, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med 2006, 12, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Spencer JD, Schwaderer AL, Wang H, Bartz J, Kline J, Eichler T, DeSouza KR, Sims-Lucas S, Baker P, Hains DS. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int 2013, 83, 615–625. [Google Scholar] [CrossRef]
- Forster CS, Johnson K, Patel V, Wax R, Rodig N, Barasch J, Bachur R, Lee RS. Urinary NGAL deficiency in recurrent urinary tract infections. Pediatr Nephrol 2017, 32, 1077–1080. [Google Scholar] [CrossRef]
- Eichler T, Bender K, Murtha MJ, Schwartz L, Metheny J, Solden L, Jaggers RM, Bailey MT, Gupta S, Mosquera C, Ching C, La Perle K, Li B, Becknell B, Spencer JD. Ribonuclease 7 Shields the Kidney and Bladder from Invasive Uropathogenic Escherichia coli Infection. J Am Soc Nephrol 2019, 30, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Bender K, Schwartz LL, Cohen A, Vasquez CM, Murtha MJ, Eichler T, Thomas JP, Jackson A, Spencer JD. Expression and function of human ribonuclease 4 in the kidney and urinary tract. Am J Physiol Renal Physiol 2021, 320, F972–F83. [Google Scholar] [CrossRef] [PubMed]
- Hains DS, Polley S, Liang D, Saxena V, Arregui S, Ketz J, Barr-Beare E, Rawson A, Spencer JD, Cohen A, Hansen PL, Tuttolomondo M, Casella C, Ditzel HJ, Cohen D, Hollox EJ, Schwaderer AL. Deleted in malignant brain tumor 1 genetic variation confers urinary tract infection risk in children and mice. Clin Transl Med 2021, 11, e477. [Google Scholar] [CrossRef] [PubMed]
- Schwaderer AL, Wang H, Kim S, Kline JM, Liang D, Brophy PD, McHugh KM, Tseng GC, Saxena V, Barr-Beare E, Pierce KR, Shaikh N, Manak JR, Cohen DM, Becknell B, Spencer JD, Baker PB, Yu CY, Hains DS. Polymorphisms in alpha-Defensin-Encoding DEFA1A3 Associate with Urinary Tract Infection Risk in Children with Vesicoureteral Reflux. J Am Soc Nephrol 2016, 27, 3175–3186. [Google Scholar] [CrossRef] [PubMed]
- Pierce KR, Eichler T, Mosquera Vasquez C, Schwaderer AL, Simoni A, Creacy S, Hains DS, Spencer JD. Ribonuclease 7 polymorphism rs1263872 reduces antimicrobial activity and associates with pediatric urinary tract infections. Ribonuclease 7 polymorphism rs1263872 reduces antimicrobial activity and associates with pediatric urinary tract infections. J Clin Invest 2021, 131. [Google Scholar]
- Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res 1996, 24, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Lu L, Li J, Moussaoui M, Boix E. Immune Modulation by Human Secreted RNases at the Extracellular Space. Front Immunol 2018, 9, 1012. [Google Scholar] [CrossRef] [PubMed]
- Pulido D, Arranz-Trullen J, Prats-Ejarque G, Velazquez D, Torrent M, Moussaoui M, Boix E. Insights into the Antimicrobial Mechanism of Action of Human RNase6: Structural Determinants for Bacterial Cell Agglutination and Membrane Permeation. Int J Mol Sci 2016, 17, 552. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rosado, JD. Human Ribonuclease 6 has a Protective Role During Experimental Urinary Tract Infection. 2023, 15, 865–875. 2023; 15, 865–875.
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 2001, 69, 4572–4579. [Google Scholar] [CrossRef]
- Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 1990, 58, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Prats-Ejarque G, Arranz-Trullen J, Blanco JA, Pulido D, Nogues MV, Moussaoui M, Boix E. The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement. Biochem J 2016, 473, 1523–1536. [Google Scholar] [CrossRef]
- Prats-Ejarque G, Blanco JA, Salazar VA, Nogues VM, Moussaoui M, Boix E. Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates. Biochim Biophys Acta Gen Subj 2019, 1863, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 2022, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Santiago B, Serrano CJ, Enciso-Moreno JA. Susceptibility to infectious diseases based on antimicrobial peptide production. Infect Immun 2009, 77, 4690–4695. [Google Scholar] [CrossRef] [PubMed]
- Eriksson J, Reimert CM, Kabatereine NB, Kazibwe F, Ireri E, Kadzo H, Eltahir HB, Mohamed AO, Vennervald BJ, Venge P. The 434(G>C) polymorphism within the coding sequence of Eosinophil Cationic Protein (ECP) correlates with the natural course of Schistosoma mansoni infection. Int J Parasitol 2007, 37, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Jonsson UB, Bystrom J, Stalenheim G, Venge P. Polymorphism of the eosinophil cationic protein-gene is related to the expression of allergic symptoms. Clin Exp Allergy 2002, 32, 1092–1095. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin RL, Phukan J, McCormack W, Lynch DS, Greenway M, Cronin S, Saunders J, Slowik A, Tomik B, Andersen PM, Bradley DG, Jakeman P, Hardiman O. Angiogenin levels and ANG genotypes: dysregulation in amyotrophic lateral sclerosis. PLoS One 2010, 5, e15402. [Google Scholar]
- Boix E, Salazar VA, Torrent M, Pulido D, Nogues MV, Moussaoui M. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol Chem 2012, 393, 801–815. [Google Scholar] [CrossRef]
- Pulido D, Garcia-Mayoral MF, Moussaoui M, Velazquez D, Torrent M, Bruix M, Boix E. Structural basis for endotoxin neutralization by the eosinophil cationic protein. FEBS J 2016, 283, 4176–4191. [Google Scholar] [CrossRef]
- Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob Agents Chemother 2012, 56, 2378–2385. [Google Scholar] [CrossRef] [PubMed]
- Narayanan C, Bernard DN, Letourneau M, Gagnon J, Gagne D, Bafna K, Calmettes C, Couture JF, Agarwal PK, Doucet N. Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding. Biochemistry 2020, 59, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Prats-Ejarque G, Lu L, Salazar VA, Moussaoui M, Boix E. Evolutionary Trends in RNA Base Selectivity Within the RNase A Superfamily. Front Pharmacol 2019, 10, 1170. [Google Scholar] [CrossRef] [PubMed]
- Deming MS, Dyer KD, Bankier AT, Piper MB, Dear PH, Rosenberg HF. Ribonuclease k6: chromosomal mapping and divergent rates of evolution within the RNase A gene superfamily. Genome Res 1998, 8, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed]
- Prats-Ejarque G, Lorente H, Villalba C, Anguita R, Lu L, Vazquez-Monteagudo S, Fernandez-Millan P, Boix E. Structure-Based Design of an RNase Chimera for Antimicrobial Therapy. Int J Mol Sci 2021, 23, 95. [Google Scholar] [CrossRef] [PubMed]
- Torrent M, Navarro S, Moussaoui M, Nogues MV, Boix E. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–9. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





