1. Introduction
Male infertility is estimated to be responsible for 50-60% of infertile couples, as a primary or contributing cause (1). Male infertility is defined as either primary, in which male patients have never fathered a child, or secondary, in which a male patient had achieved pregnancy in the past but is unable to achieve a new one (2).
Secondary infertility is believed to constitute a large proportion of male infertility cases and causes a considerable amount stress and economic burden on coupler. Multiple factors have been implicated in secondary male infertility, which include varicocele, infection, smoking, obesity, exposure to high temperature and hazardous materials, and oxidative stress (3).
Oxidative stress occurs when the production of reactive oxygen species (ROS) exceeds ability of the cell to scavenge the ROS. Oxidative stress negatively impacts sperm cells causes DNA damage, mitochondrial dysfunction, and apoptosis, all of which are related to male infertility (4).
Sperm cells are highly vulnerable to ROS production as they contain high levels of polyunsaturated fatty acids in both the membrane and cytoplasm and have a low ability to scavenge ROS compared to other cell types. As a result, sperm cells are dependent on the seminal plasma for oxidative stress protection, which is crucial for maintaining proper sperm function and its fertility potential (5).
The seminal plasma is a complex composition rich in proteins as well as other molecules such as sugars and lipids. This protein rich composition makes it a promising biological fluid for biomarker discovery and a target for proteomics and bioinformatic analysis(6). Multiple proteomic studies have revealed that the protein components of the seminal plasma play a pivotal role in maintaining proper fertility potential and that differential seminal plasma protein expression is associated with oxidative stress and infertility (7).
In this study we aimed to elucidate proteomic modifications occurring within the seminal plasma of individuals afflicted by concomitant oxidative stress and secondary infertility. Our underlying hypothesis posits that oxidative stress increases susceptibility toward secondary infertility. Results of our study could impact clinical care and suggest ways to treat secondary infertility by targeting oxidative stress, which would help to overcome infertility in the future.
2. Materials and Methods
2.1. Ethical Statement
Samples used for proteomic analysis originated from patients and donors after informed consent, pursuant of IRB protocols #11–451 and# 14-235, as previously described (8,9).
2.2. Sample and Data Collection
Semen samples were obtained from patients diagnosed with secondary male infertility, characterized by the inability to achieve conception following one year of unprotected intercourse. Additionally, samples were drawn from fertile donors who have successfully fathered a child within the preceding two years and exhibited elevated semen oxidative markers (Reactive Oxygen Species—ROS >93RLU/sec and Total Antioxidant Capacity—TAC < 1790 MTE). The control group comprised healthy fertile donors who have fathered a child within the past two years and with normal semen characteristics and exhibited no signs of oxidative stress in their semen (Reactive Oxygen Species—ROS <93RLU/sec and Total Antioxidant Capacity—TAC > 1790 MTE). Infertility patients were excluded from the study if they presented with varicocele, azoospermia, severe oligospermia, or when there was an identifiable female contributory factor.
2.3. Semen Analysis and Protein Extraction
Semen specimens were collected and processed in alignment with the World Health Organization (WHO) 2010 fifth edition guideline (World Health Organization., 2010). A chemiluminescence assay was employed to determine ROS levels, while sperm DNA fragmentation (SDF) was ascertained using the terminal deoxynucleotidyl transferase-mediated fluorescein end labeling (TUNEL) assay (citations). Following centrifugation of the samples at 13,000g for 20 minutes, seminal plasma was separated and subsequently preserved at −80 °C for proteomic evaluation.
2.4. Protein Identification and Global Proteomics Analysis
For the proteomic analysis, seminal plasma samples were equilibrated to room temperature and subsequently centrifuged at 3,000g for 30 minutes at 4 °C to remove residual sperm and other biological contaminants. Subsequently, samples were treated with a protease inhibitor cocktail to inhibit proteolysis. After protein quantification, five pooled samples from each group were selected for further evaluation. Seminal plasma concentrations were normalized to ensure equal protein concentrations for each specimen. Pooling of samples was employed to minimize biological variation (11,12). Subsequently, specimens were combined with SDS-PAGE buffer and subjected to 1D-PAGE, conducted in triplicate runs to mitigate technical variation, resulting in a final volume of 30 μL (13).
The extracts were concentrated using a Speedvac and resuspended in 1% acetic acid to reach a 30 μL volume for LC-MS. Finnigan LTQ-Orbitrap Elite hybrid mass spectrometer system was utilized with a Dionex 15 cm × 75 μm Acclaim Pepmap C18 column. Extracts (5 μL) were injected, and peptides eluted using an acetonitrile/0.1% formic acid gradient at 0.25 μL/min, introduced directly into the mass spectrometer. The ion source operated at 2.0 kV, and digestions were analysed using the instrument’s data-dependent multitask capability, acquiring both full scan spectra for peptide weights and MS/MS for amino acid sequences.
2.5. Database Searching and Protein Identification
Proteins were identified using Proteome Discoverer version 1.4.1.288, Mascot (Matrix Science, London, UK; version 2.3.02), Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version 1.4.0.288), and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1)). For validation, Scaffold (version 4.0.6.1, Proteome Software Inc., Portland, OR) was employed. Peptide identification utilized a confidence threshold of 95.0% (14), while protein identification adhered to stringent criteria with a 99.0% probability threshold, ensuring a false detection rate (FDR) below 1.0% and requiring at least two identified peptides per protein. The Protein Prophet algorithm (15) was used to assess protein probabilities. Furthermore, gene ontology (GO) terms from the National Centre for Biotechnology Information (NCBI), retrieved on October 21, 2013, facilitated protein identification.
2.6. Quantitative Proteomics
In the proteomic analysis, protein quantities were determined using spectral counts (SpCs), which represent the total mass spectra matching peptides for specific proteins. To normalize these counts, the NSAF approach was used(16), which adjusts for variations in sample replicates and considers that longer proteins typically have more peptide identifications (17). Differentially expressed proteins (DEPs) were identified using criteria such as significance tests and fold change thresholds based on the average SpC. Given the higher error rate for low-abundance proteins, different constraints were applied to SpC levels, categorizing proteins as High, Medium, Low, or Very Low based on their spectral counts. Each category had specific significance and fold change constraints to ensure accurate protein quantification and genuine biological change detection. These criteria were validated using control experiments with the NSAF method.
2.7. Bioinformatics Analysis
For bioinformatic analysis, multiple software applications were employed. Initially, we utilized Go Term Finder and UniProt for protein annotation. Subsequently, we incorporated complimentary software such as DAVID (Database for Annotation, Visualization, and Integrated Discovery), STRING (Search Tool for the Retrieval of Interacting Genes/Proteins). The DAVID software facilitated the analysis of differentially expressed proteins in terms of biological processes, cellular components, and molecular functions. The STRING software was instrumental in examining protein-protein interaction networks and conducting functional enrichment analysis. After these preliminary analyses, we employed the IPA software (Ingenuity Pathway Analysis) from Ingenuity® Systems. This software enabled a comprehensive analysis of the functional pathways of the differentially expressed proteins, encompassing top canonical pathways, disease and biological pathways, causal networks, and upstream regulators.
2.8. Statistical Analysis
Statistical analyses were performed using MedCalc Statistical Software (version 17.8; MedCalc Software, Ostend, Belgium). For semen analysis, various tests were employed based on variable types, including Analysis of Variance (ANOVA), two-sample T-tests, and the Wilcoxon rank-sum test. A p-value less than 0.05 was considered statistically significant. Quantitative protein expression comparisons between the two groups (fertile donors with high oxidative stress and secondary infertile patients) were also undertaken using two-sample T-test.
3. Results
3.1. Semen Analysis
Table 1 presents the characteristics of the semen samples, including the percentage of sperm DNA fragmentation SDF and the concentration of reactive oxygen species ROS for intracellular oxidative stress assessment.
3.2. Proteomic Profiling
In the pooled samples from both groups, distinct proteomic analyses were conducted.
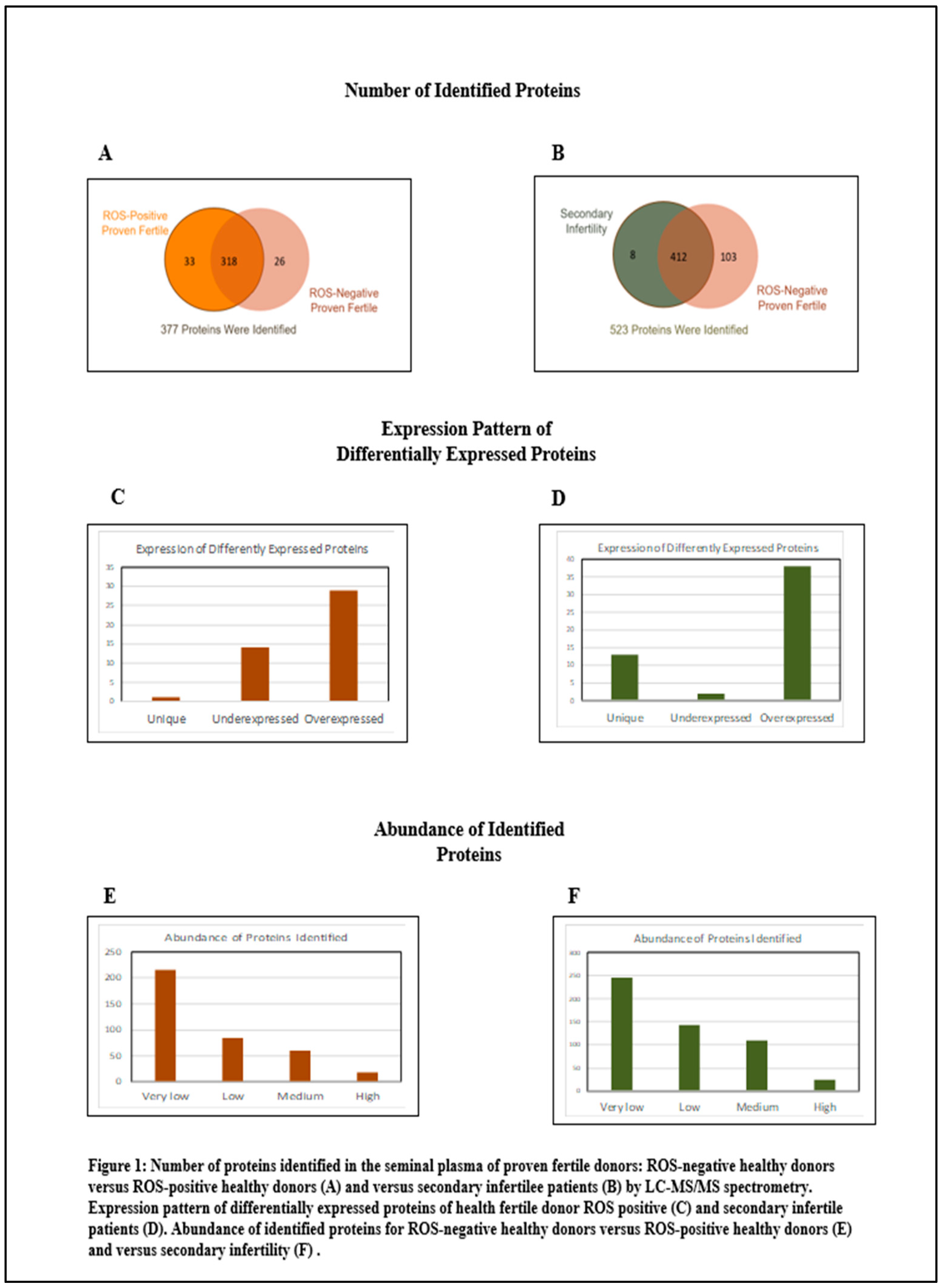
Figure 1 summarizes the distribution of proteins. Within the cohort of fertile donors with high oxidative stress in the semen, 377 proteins were identified. Of these, 44 proteins exhibited differential expression with the control group: 29 were overexpressed and 15 were underexpressed. In terms of protein abundance, 18 proteins were classified as highly abundant, 60 as moderately abundant, 84 as low in abundance, and 216 as very low in abundance. In contrast, for the group of patients with secondary infertility and high oxidative stress in the semen, analysis revealed 523 proteins. Among these, 53 proteins were differentially compared to the control group, with 51 being overexpressed and 2 underexpressed. The abundance categorization for this group was as follows: 25 proteins were highly abundant, 110 were of moderate abundance, 143 had low abundance, and 245 were very low in abundance.
Supplementary Table S1.
3.3. Bioinformatic Analysis Results
Table 2 and
Table 4 present the raw analysis conducted using DAVID software for both the group of fertile donors ROS positive and the group of secondary infertile patients. We had a further analysis and restructured the data to have a deep overview of the involved proteins.
Table 3 and
Table 5 summarize the proteins by their expression trend.
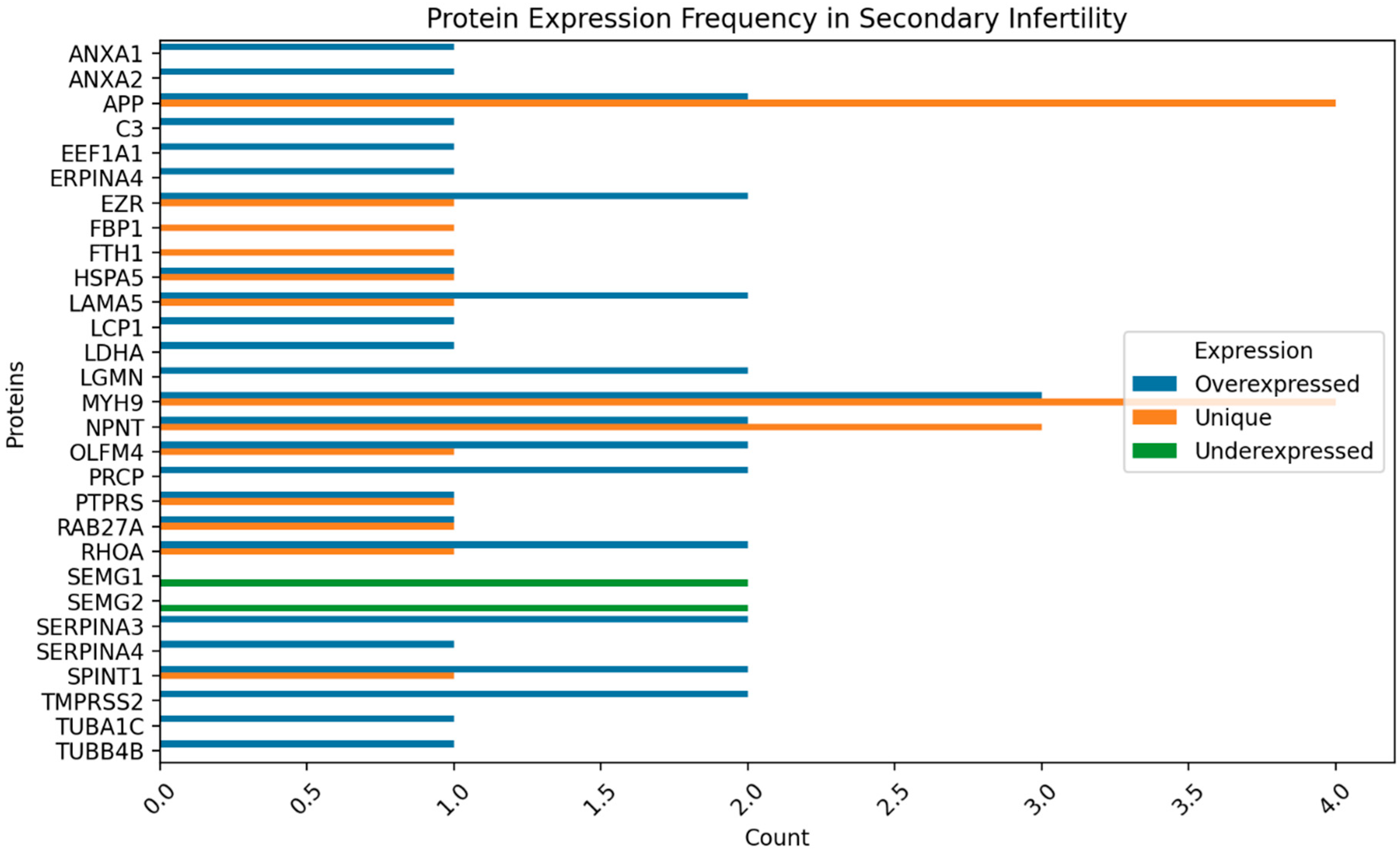
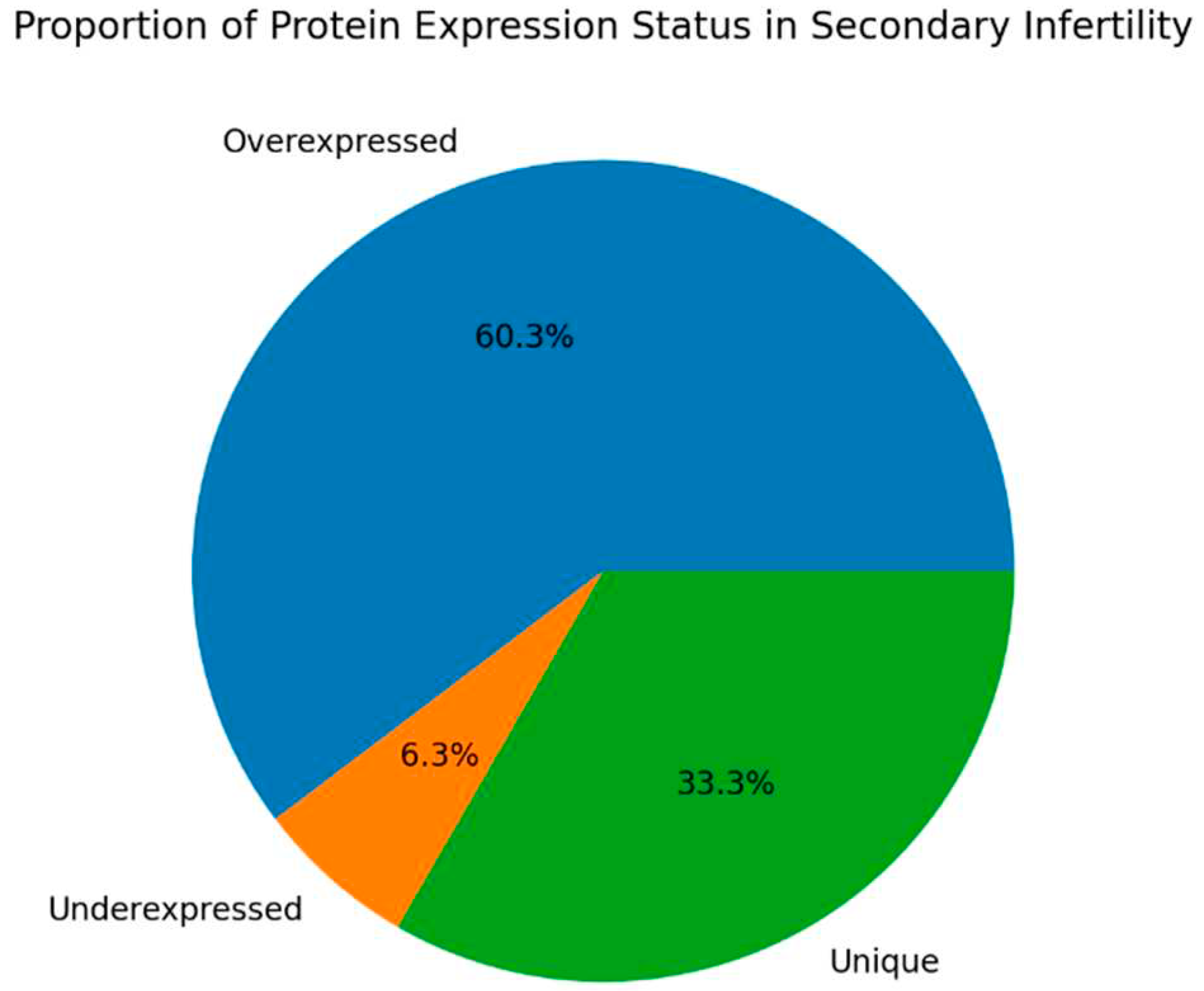
Figure 2,
Figure 3,
Figure 4 and
Figure 5 show the protein expression frequency and proportion of protein expression status of each group.
For fertile donors experiencing high oxidative stress, the overexpressed proteins are associated with antioxidant activity and immune response. Conversely, the underexpressed proteins pertain to protein folding, stabilization, binding, and energy metabolism.
For patients with secondary infertility overexpressed proteins are associated with inflammatory and immune response, energy metabolism and cell stability. Underexpressed proteins are related to sperm cell motility and capacitation. Unique proteins are related to protein binding, cell adhesion and extracellular matrix organization.
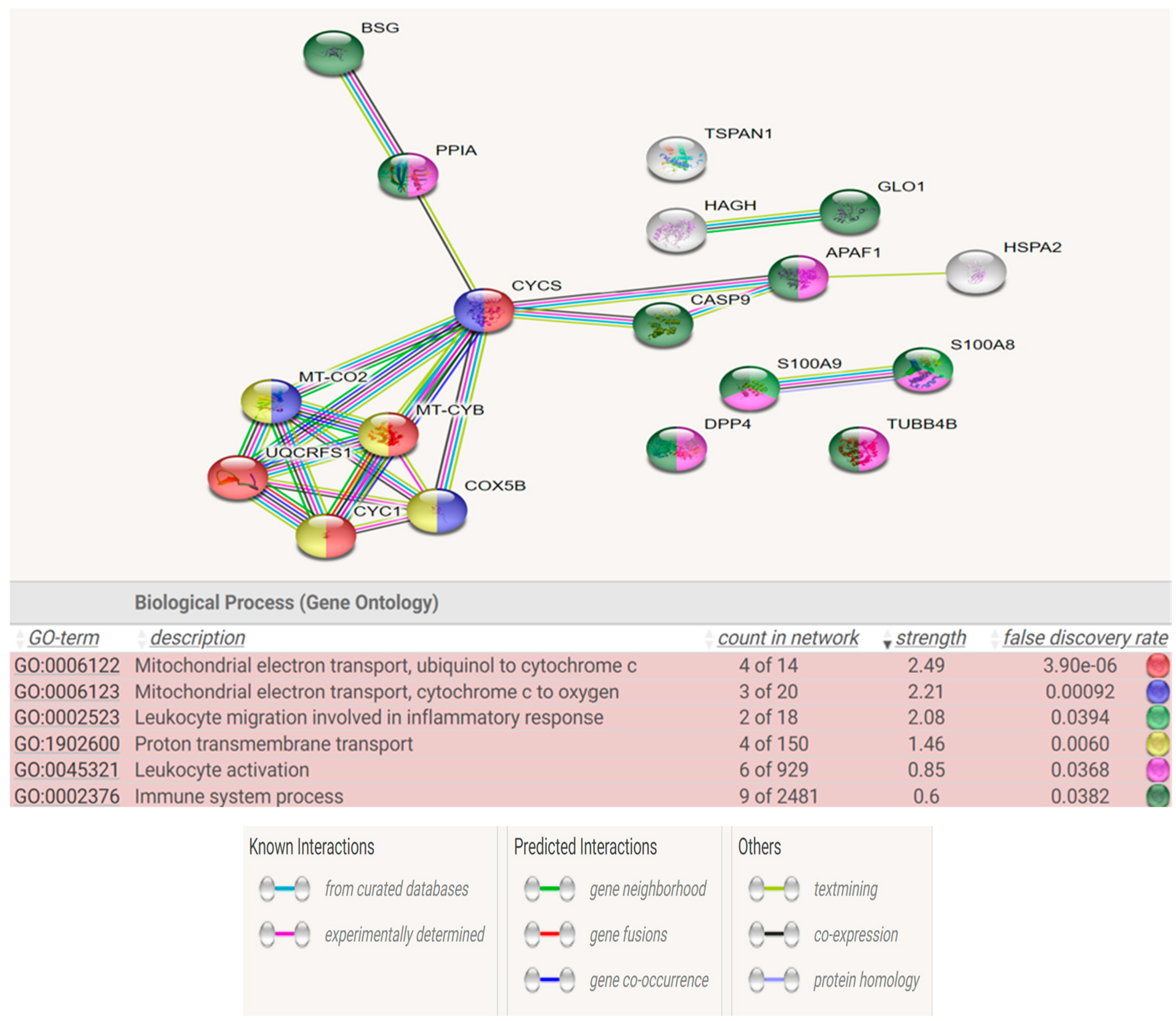
Figure 6 and
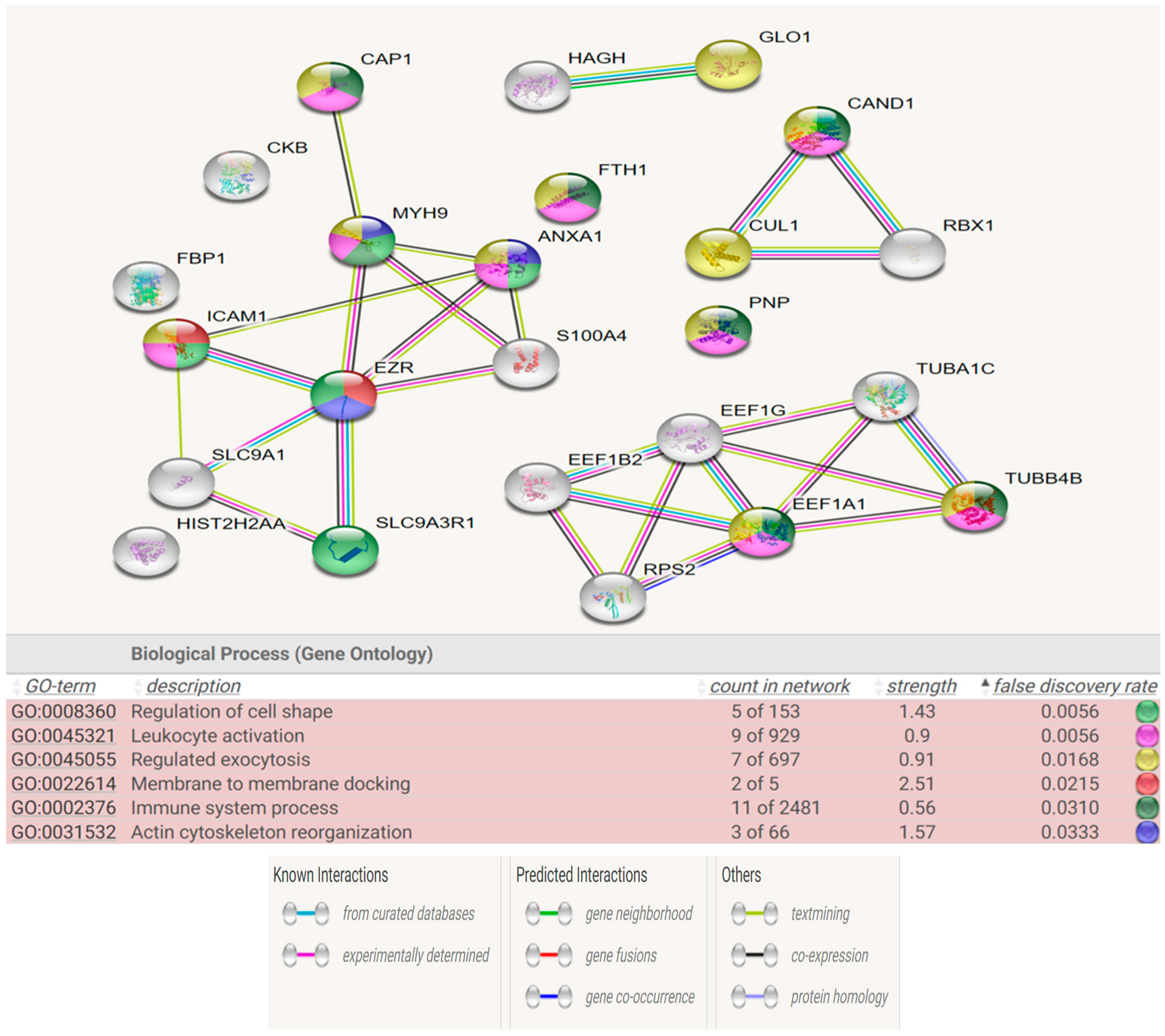
Figure 7 show the STRING analysis for each the group of fertile donors ROS positive and Secondary infertile patients. The analysis shows the interactions between the mapped proteins (known, predicted or other interactions) and the functional enrichments of the biological process.
Figure 8 and
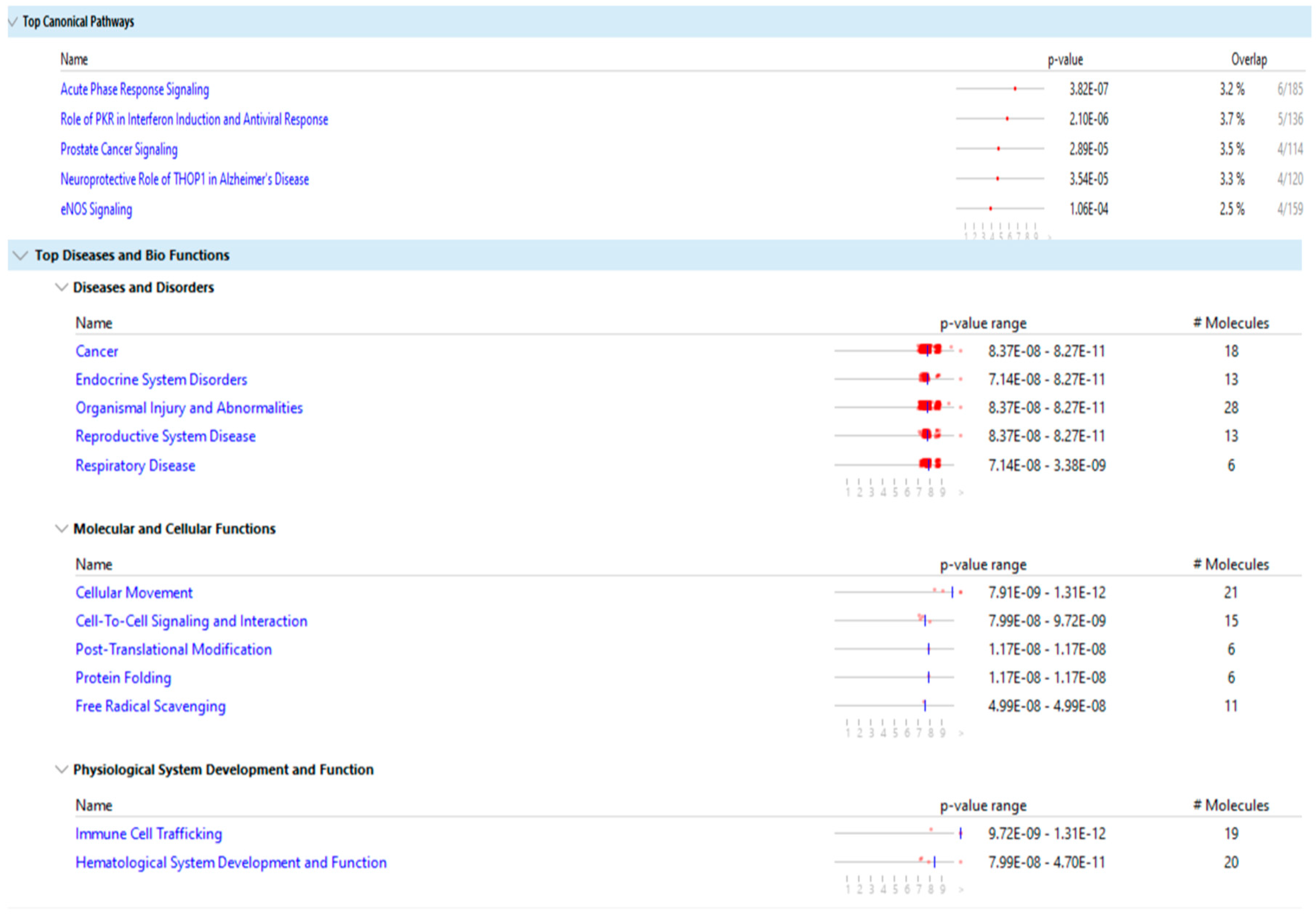
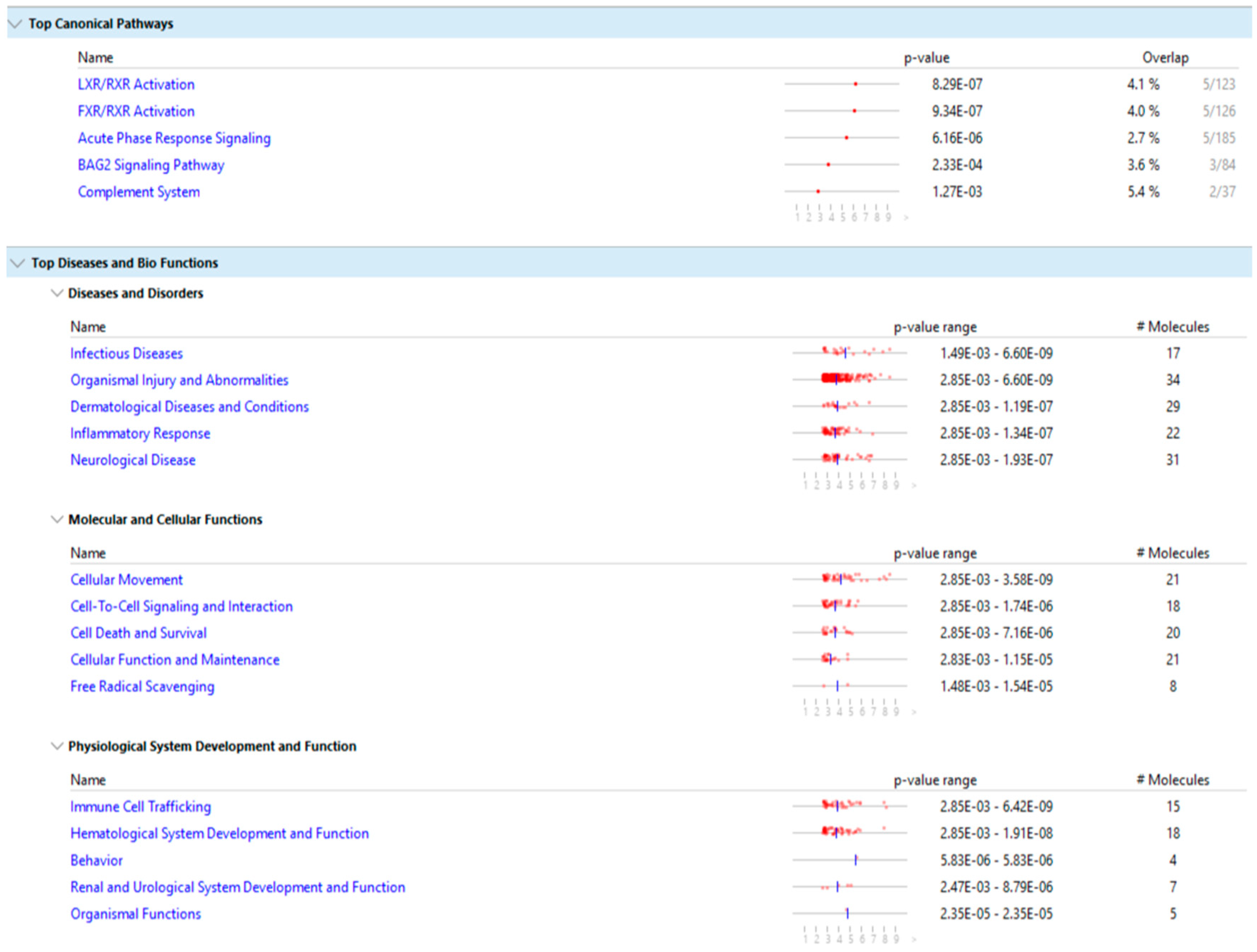
Figure 9 show the main result of the IPA analysis for each group of fertile donors with ROS positive and patients with secondary infertility.
4. Discussion
Seminal plasma is vital for the development of functional sperm cells. It predominantly consists of proteins secreted by the accessory glands of the male reproductive system. These proteins are responsible for the various aspects of male fertility, from ensuring sperm health and motility to facilitating successful ovum fertilization, interaction with the female reproductive system and the conveyance of genetic material.
Moreover, seminal plasma proteins also play a pivotal role in providing antioxidant protection to sperm cells. This ensures their viability, motility, and DNA integrity. As oxidative stress is a known contributing factor to male infertility, these proteins are crucial for maintaining male fertility.
In our current study, we aimed to explore whether high oxidative stress is a risk factor for secondary infertility and to validate our hypothesis, we compared the proteomic profiles of fertile donors with high oxidative stress to those of patients with secondary infertility. We used different software analysis that encompassed shared biological processes, molecular functions, predominant canonical pathways, as well as associated diseases and biological functions.
Our results show that the differentially expressed proteins of the two groups share multiple biological pathways and molecular functions such as: acute phase response signaling, organismal injury and abnormalities, cellular movement, cell to cell signaling and interaction, hematological system development and function, immune cell trafficking and free radical scavenging.
The molecular function of free radicals scavenging is activated in the group of fertile donors with ROS positive, as expected, and is also activated in the group of secondary infertile patients, which suggests that oxidative stress may be a cause of this pathology. Additionally, other shared pathways of acute phase response signaling, organismal injury and abnormalities, cellular movement and immune cell trafficking are also a result of oxidative stress, which further suggests that oxidative stress is a cause of secondary infertility.
The multiple share pathological pathways between the secondary infertile group and the fertile ROS positive group is indicative that oxidative stress is a risk factor for secondary infertility. As such, patients with high oxidative stress in semen could be advised to act and treat the oxidative stress pathology before reaching the point of infertility or potentially to correct infertility. For instance, treatment with antioxidants to reverse the infertility may be fruitful. However, more research in this area is needed.
Beside the issue of infertility, the DEPs of the group of fertile donors with ROS positive were associated with cancer, endocrine system disorders, organismal injury and abnormalities, reproductive system disease and respiratory diseases. As such, treatment of oxidative stress may have benefits beyond treating infertility.
We had a comprehensive analysis of proteins within each biological pathway and process to pinpoint potential markers for future infertility and associated comorbidities. Notably, the C3 protein was consistently present in all previously specified biological pathways and molecular functions,especially the free radical scavenging pathway, exhibiting a positive fold change in expression, indicating its overexpression.The C3 protein, is glycoprotein and an integral component of the complement immune system. It is encoded by the C3 gene situated on chromosome 19(18). Elevated C3 protein levels typically suggest inflammation(19). While increased levels of C3 have been observed in the seminal plasma of infertile patients and in abnormal semen samples (20,21), to our knowledge, this is the first documentation of its presence in the seminal plasma of fertile males exhibiting oxidative stress.
Another protein identified in the free radical scavenging pathway is SERPINA3. This protein is a serine protease inhibitor and belongs to the Serpins family (22). It is implicated in processes like inflammation and oxidative stress (23). Furthermore, SERPINA3 is associated with various pathologies, including chronic obstructive pulmonary disease, Parkinson’s disease, Alzheimer’s disease, and cancer (24). Past proteomic research has linked SERPINA3 with male infertility following spinal cord injuries and in patients with varicocele(25,26). However, our study is the first to demonstrate a connection between this protein and secondary male infertility, likely stemming from oxidative stress and inflammation.
These two pivotal proteins, C3 and SERPINA3, may act as biological biomarkers for potential infertility and a spectrum of other diseases and pathologies in fertile patients experiencing oxidative stress, providing avenues for early diagnosis and pre-emptive treatment options.
5. Conclusions
Our study robustly establishes oxidative stress as a critical factor in secondary infertility. The discovery of key proteins such as C3 and SERPINA3 as overexpressed biomarkers reinforces the link between oxidative stress and inflammatory responses in male fertility and are a potential biomarker for future diagnosis and treatment. These findings not only pave the way for antioxidant-based therapeutic interventions to address fertility issues and mitigate associated health conditions, but also highlight the broader impact of oxidative stress on male health, underscoring the need for early detection and preventive measures against its potentially multifaceted consequences.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization RSK and SL; methodology, RSK and MKPS.; software, RSK.; validation, SL, MKPS, SM and MAW.; formal analysis, RSK.; investigation, RSK and SM.; resources, RSK.; data curation, RSK.; writing—original draft preparation, RSK.; writing—review and editing, SL,MKPS,SK,MAW,and SV; visualization, NA.; supervision, SL and SV.; project administration, SL and SV.; funding acquisition, NA. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Cleveland Clinic Foundation (IRB protocols #11–451 and# 14-235).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The lists of differentially expressed proteins are available as the supplementary materials of this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, et al. Male infertility. Lancet [Internet]. 2021 Jan 23 [cited 2023 Jul 19];397(10271):319–33. Available from: https://pubmed.ncbi.nlm.nih.gov/33308486/.
- Cannarella R, Condorelli RA, Mongioì LM, La Vignera S, Calogero AE. Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility. Int J Mol Sci [Internet]. 2020 Mar 1 [cited 2023 Jul 19];21(5). Available from: https://pubmed.ncbi.nlm.nih.gov/32138324/. https://doi.org/10.3390/ijms21051728. [CrossRef]
- Punab M, Poolamets O, Paju P, Vihljajev V, Pomm K, Ladva R, et al. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod [Internet]. 2017 Jan 1 [cited 2023 Jul 19];32(1):18. Available from: /pmc/articles/PMC5165077/. https://doi.org/10.1093/humrep/dew284. [CrossRef]
- Bisht S, Faiq M, Tolahunase M, Dada R. Oxidative stress and male infertility. Nat Rev Urol. 2017 Aug 1;14(8):470–85. https://doi.org/10.1038/nrurol.2017.69. [CrossRef]
- Aitken RJ, Smith TB, Jobling MS, Baker MA, De Iuliis GN. Oxidative stress and male reproductive health. Asian J Androl [Internet]. 2014 Jan 1 [cited 2023 Oct 29];16(1):31–8. Available from: https://pubmed.ncbi.nlm.nih.gov/24369131/. https://doi.org/10.4103/1008-682x.122203. [CrossRef]
- Druart X, Rickard JP, Tsikis G, de Graaf SP. Seminal plasma proteins as markers of sperm fertility. Theriogenology. 2019 Oct 1;137:30–5.
- Samanta L, Parida R, Dias TR, Agarwal A. The enigmatic seminal plasma: A proteomics insight from ejaculation to fertilization. Reproductive Biology and Endocrinology. 2018 Apr 28;16(1). doi:10.1186/s12958-018-0358-6. [CrossRef]
- Martins AD, Panner Selvam MK, Agarwal A, Alves MG, Baskaran S. Alterations in seminal plasma proteomic profile in men with primary and secondary infertility. Sci Rep. 2020 Dec 1;10(1). doi: 10.1038/s41598-020-64434-1. [CrossRef]
- Dias TR, Samanta L, Agarwal A, Pushparaj PN, Selvam MKP, Sharma R. Proteomic signatures reveal differences in stress response, antioxidant defense and proteasomal activity in fertile men with high seminal ROS levels. Int J Mol Sci. 2019 Jan 1;20(1). https://doi.org/10.3390/ijms20010203. [CrossRef]
- World Health Organization. WHO laboratory manual for the examination and processing of human semen. World Health Organization; 2010. 271 p.
- Bogle OA, Kumar K, Attardo-Parrinello C, Lewis SEM, Estanyol JM, Ballescà JL, et al. Identification of protein changes in human spermatozoa throughout the cryopreservation process. Andrology [Internet]. 2017 Jan 1 [cited 2023 Sep 24];5(1):10–22. Available from: https://pubmed.ncbi.nlm.nih.gov/27860400/. https://doi.org/10.1111/andr.12279. [CrossRef]
- Ayaz A, Agarwal A, Sharma R, Arafa M, Elbardisi H, Cui Z. Impact of precise modulation of reactive oxygen species levels on spermatozoa proteins in infertile men. Clin Proteomics [Internet]. 2015 Feb 9 [cited 2023 Sep 24];12(1). Available from: https://pubmed.ncbi.nlm.nih.gov/25972767/. https://doi.org/10.1186/1559-0275-12-4. [CrossRef]
- Molloy MP, Brzezinski EE, Hang J, McDowell MT, VanBogelen RA. Overcoming technical variation and biological variation in quantitative proteomics. Proteomics [Internet]. 2003 Oct [cited 2023 Sep 24];3(10):1912–9. Available from: https://pubmed.ncbi.nlm.nih.gov/14625853/. https://doi.org/10.1002/pmic.200300534. [CrossRef]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom [Internet]. 1994 [cited 2023 Sep 26];5(11):976–89. Available from: https://pubmed.ncbi.nlm.nih.gov/24226387/. doi:10.1016/1044-0305(94)80016-2. [CrossRef]
- Serang O, Noble W. A review of statistical methods for protein identification using tandem mass spectrometry. Stat Interface. 2012;5(1):3–20. https://doi.org/10.4310/sii.2012.v5.n1.a2. [CrossRef]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem [Internet]. 2010 Mar 15 [cited 2023 Sep 17];82(6):2272–81. Available from: https://pubmed.ncbi.nlm.nih.gov/20166708/. https://doi.org/10.1021/ac9023999. [CrossRef]
- Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem [Internet]. 2005 Oct 1 [cited 2023 Sep 17];77(19):6218–24. Available from: https://pubmed.ncbi.nlm.nih.gov/16194081/. https://doi.org/10.1021/ac050846r. [CrossRef]
- Delanghe JR, Speeckaert R, Speeckaert MM. Complement C3 and its polymorphism: Biological and clinical consequences. Pathology. 2014;46(1):1–10. doi: 10.1097/PAT.0000000000000042. [CrossRef]
- Geisbrecht B V., Lambris JD, Gros P. Complement component C3: A structural perspective and potential therapeutic implications. Semin Immunol. 2022 Jan 1;59. https://doi.org/10.1016/j.smim.2022.101627. [CrossRef]
- Sullivan H, Quinlivan WLG. Immunoglobulins in the semen of men with azoospermia, oligospermia, or self-agglutination of spermatozoa. Fertil Steril [Internet]. 1980 [cited 2023 Oct 26];34(5):465–8. Available from: https://pubmed.ncbi.nlm.nih.gov/7439412/. https://doi.org/10.1016/s0015-0282(16)45139-3. [CrossRef]
- Ulèová-Gallová Z, Krauz V, Mohamed AM, Rokyta Z. Immunity to spermatozoa and male fertility. Andrologia. 1999;31(5):318–9.
- Sánchez-Navarro A, Murillo-de-Ozores AR, Pérez-Villalva R, Linares N, Carbajal-Contreras H, Flores ME, et al. Transient response of serpinA3 during cellular stress. FASEB Journal. 2022 Mar 1;36(3). https://doi.org/10.1096/fj.202101912r. [CrossRef]
- de Mezer M, Rogaliński J, Przewoźny S, Chojnicki M, Niepolski L, Sobieska M, et al. SERPINA3: Stimulator or Inhibitor of Pathological Changes. Biomedicines. 2023 Jan 1;11(1). https://doi.org/10.3390/biomedicines11010156. [CrossRef]
- Li B, Lei Z, Wu Y, Li B, Zhai M, Zhong Y, et al. The Association and Pathogenesis of SERPINA3 in Coronary Artery Disease. Front Cardiovasc Med. 2021 Dec 8;8. https://doi.org/10.3389/fcvm.2021.756889. [CrossRef]
- Silva BF Da, Meng C, Helm D, Pachl F, Schiller J, Ibrahimo E, et al. Towards understanding male infertility after spinal cord injury using quantitative proteomics. Molecular and Cellular Proteomics. 2016 Apr 1;15(4):1424–34. doi: 10.1074/mcp.M115.052175. [CrossRef]
- Agarwal A, Sharma R, Durairajanayagam D, Ayaz A, Cui Z, Willard B, et al. Major protein alterations in spermatozoa from infertile men with unilateral varicocele. Reproductive Biology and Endocrinology. 2015 Feb 22;13(1). https://doi.org/10.1186/s12958-015-0007-2. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).