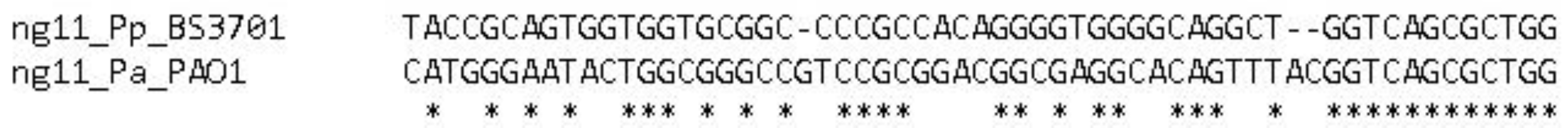1. Introduction
Comparative sequence analysis allows to predict cis- and trans-regulators that are common for the taxa of the genus, family, order and even phylum levels (for example, [
1]). Thus, data on such a well-studied regulon as NtrC-regulon [
2] has been supplemented. Transcription factor NtrC regulates nitrogen metabolism in cells of
Escherichia coli[
3],
Klebsiella pneumonlae [
4],
Salmonella typhimurium [
5],
Rhizobium meliloti [
6],
Stutzerimonas stutzeri [
7],
Pseudomonas aeruginosa [
8],
Pseudomonas fluorescens [
9] and
Pseudomonas putida [
10]. The decrease in nitrogen content causes the NtrC protein phosphorylation by the NtrB kinase, which leads to the activation of sigma54-dependent genes, for example, the glnA glutamine synthetase gene [
11]. The NtrC-regulon of
Stutzerimonas stutzeri A1501 (formerly
Pseudomonas stutzeri) includes small RNAs NFIs and NfiR, which are involved in regulating the expression of nitrogen fixation genes [
12,
13,
14]. |The
Pseudomonas aeruginosa PAO1 regulon includes small RNA NrsZ, which is involved in stimulating the translation of rhlA rhamnosyltransferase gene and in the manifestation of swarming motility [
2]. However, not only gene expression is stimulated in microorganisms of the genus
Pseudomonas under nitrogen deficiency. According to transcriptomic analysis conducted by Hervás and co-authors in 2008, the mRNAs quantity of carbon catabolism genes decreased in
P. putida KT2440 cells [
15]. Pozdnyakova-Filatova and co-authors observed a decrease in the intensity of carbon metabolism in the
P. putida BS3701 strain under nitrogen deficiency in their article in 2020 [
16]. A mechanism for reducing gene expression under a lack of macronutrients has been described for pseudomonas. Thus, PrrF ncRNAs are synthesized in
Pseudomonas aeruginosa PAO1 with iron deficiency, which leads to a quantity decrease of iron-cofactored superoxide dismutase sodB, heme-cofactored katalase katA, succinatedehydrogenase sdh and TF antR gene transcripts [
17,
18]. There are no data on Pseudomonas spp small RNAs capable of stimulating the degradation of target mRNAs under nitrogen deficiency in the literature.
We have used the following selection criteria to identify new small RNAs of
P. putida BS3701 that may be involved in inhibiting the expression of carbon catabolism genes: 1) the presence of the interest gene of the NtrC binding site and a sigma54-dependent promoter in the upstream area, 2) the conservative location of the interest gene in the
P. putida and
P. aeruginosa genomes, 3) increase in the product quantity of the interest gene in a cell under nitrogen deficiency on two different carbon sources (succinate and naphthalene), 4) the product detection of the interest gene in a fraction enriched with small RNAs. The expression of two new small RNAs has increased twice or more times under nitrogen deficiency on both succinate and naphthalene: expression of ng171 and ng379. ng379 contains narK RNA motif [
1].
2. Results
2.1. Bioinformatic search for NtrC- and sigma54-dependent ncRNAs
400 small RNA genes were predicted in the
Pseudomonas putida BS3701 genome (RefSeq NZ_CP059052.1): the prokka program predicted 12 genes by homology (version 1.14.6,
Table S1), and the PresRAT program [
19] predicted 388 genes de novo (
Table S1). The NtrC binding site and a sigma54-dependent promoter were predicted in the upstream area of 7 genes (nrsZ, rsmZ, ng11, ng127, ng171, ng279, ng379). The genes have a conservative arrangement for putida and aeruginosa species.
2.1.1. Prokka prediction. nrsZ
Nicolas Winner and co-authors found non-coding NrsZ RNA in
Pseudomonas aeruginosa PAO1 in 2014 [
2]. The RNA has a length of 227 nucleotides in
Pseudomonas aeruginosa PAO1, and its orthologue
in Pseudomonas putida BS3701 has a length of 179 nucleotides. The downstream of operon ntrBC is located, which encodes the transcriptional regulator NtrC and its kinase NtrB. The NtrC binding site (cgcaccctattggtgcg) is predicted using the SigmoID program at a distance of 5658 bp upstream the assumed +1 point of the nrsZ gene. The iPro54-PseKNC program has predicted a sigma 54-dependent promoter upstream the nrsZ gene (tggtttattcaccggtgagtaaatgtgtccgcggaaataccttgcaagctccatcgcttgCtcgcctggctgaacccgcta, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 1.
Alignment of the nrsZ gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 1.
Alignment of the nrsZ gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
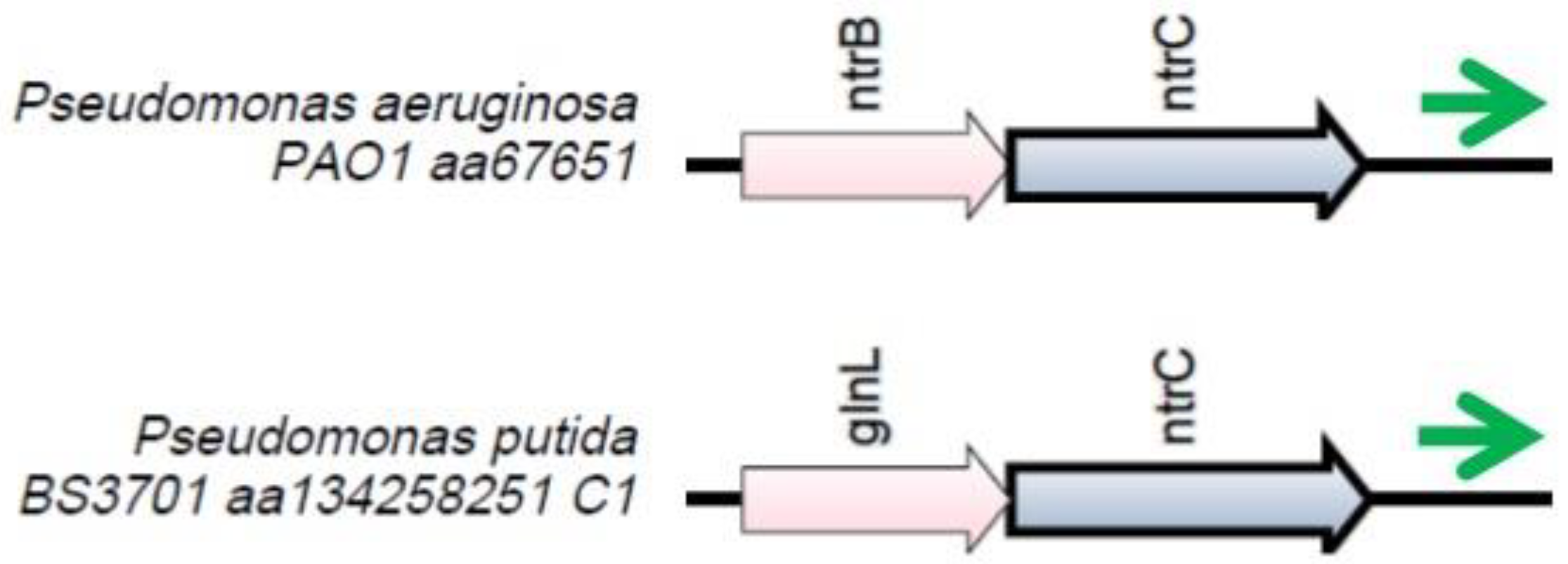
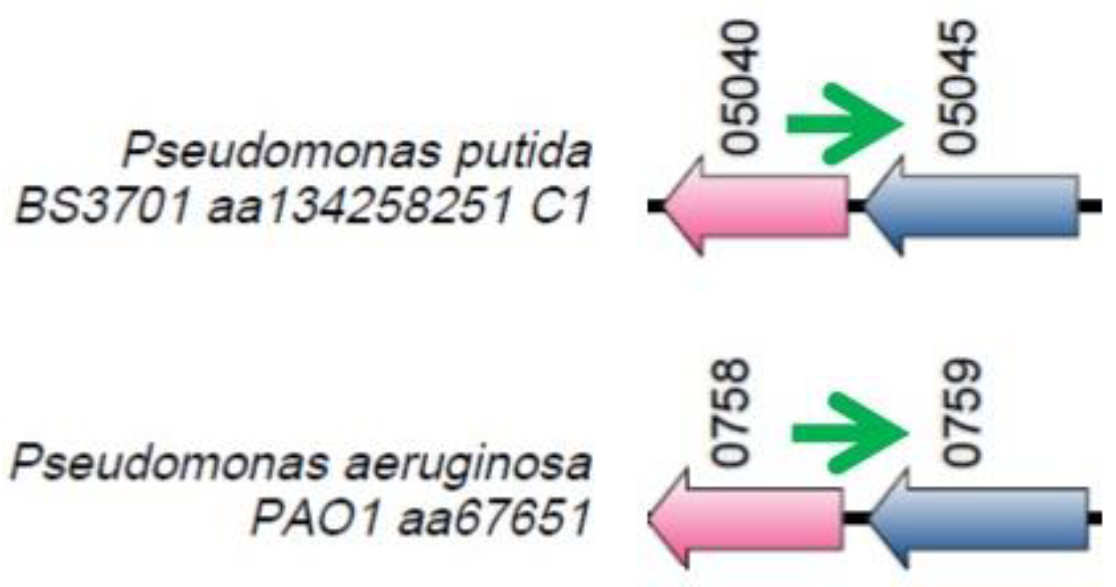
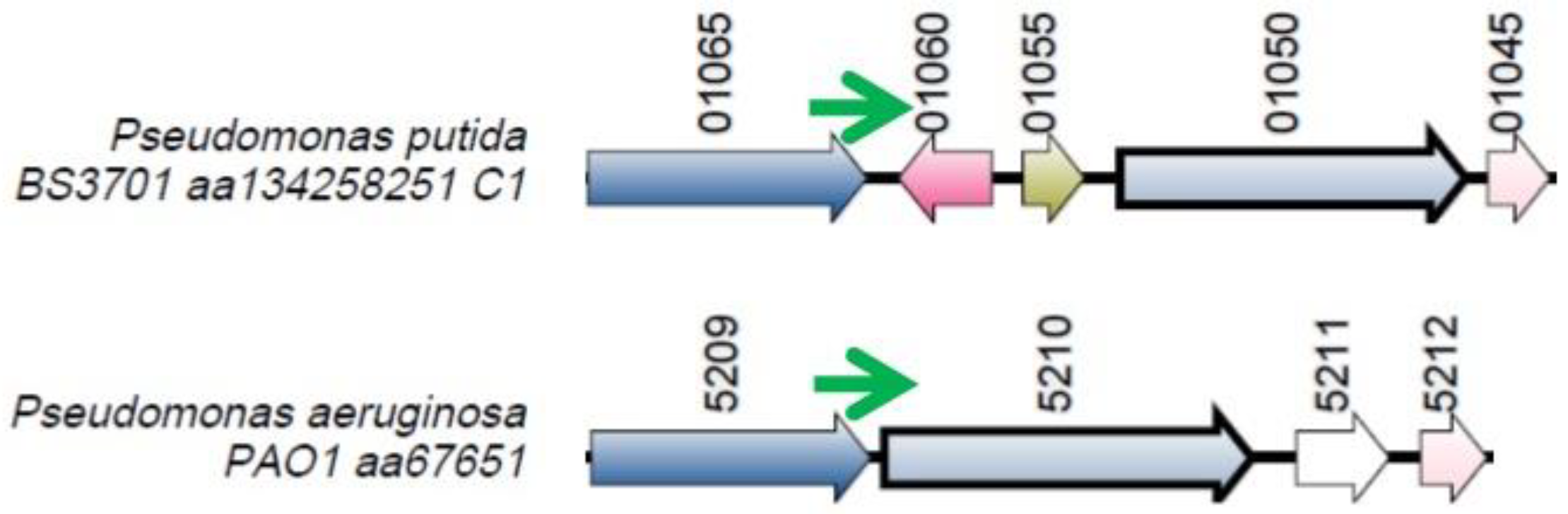
Figure 2.
The locus where the nrsZ gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 2.
The locus where the nrsZ gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
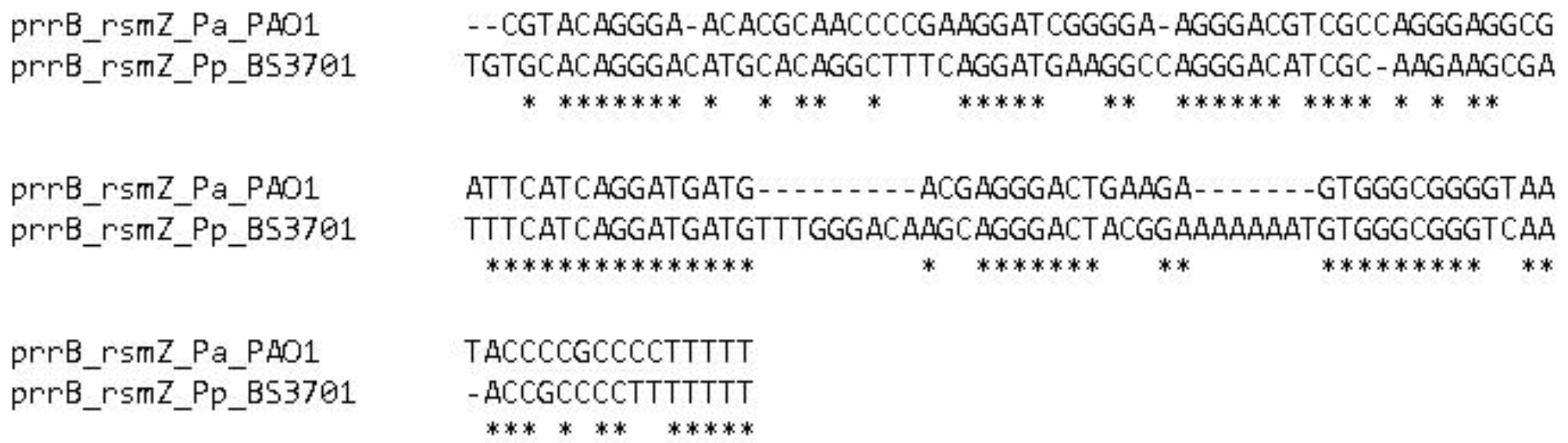
2.1.2. Prokka prediction. rsmZ
Christophe Dubuis and co-authors detected non-coding RsmZ RNA in
Pseudomonas fluorescens CHA0 [
20] in 2006 and they identified it in
Pseudomonas aeruginosa PAO1 [
21] in the same year. The RsmZ orthologue has a length of 134 nucleotides in the
P. putida BS3701 strain, and its gene is located downstream towards the rpoS gene, which encodes the sigma S RNAP subunit. The NtrC binding site (cgcaccacggtggtgtc) is predicted at a distance of 1983 bp upstream the assumed +1 point of the rsmZ gene. A sigma 54-dependent promoter has been predicted upstream the rsmZ gene (tggtttattcaccggtgagtaaatgtgtccgcggaaataccttgcaagctccatcgcttgCtcgcctggctgaacccgcta, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 3.
Alignment of rsmZ gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 3.
Alignment of rsmZ gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
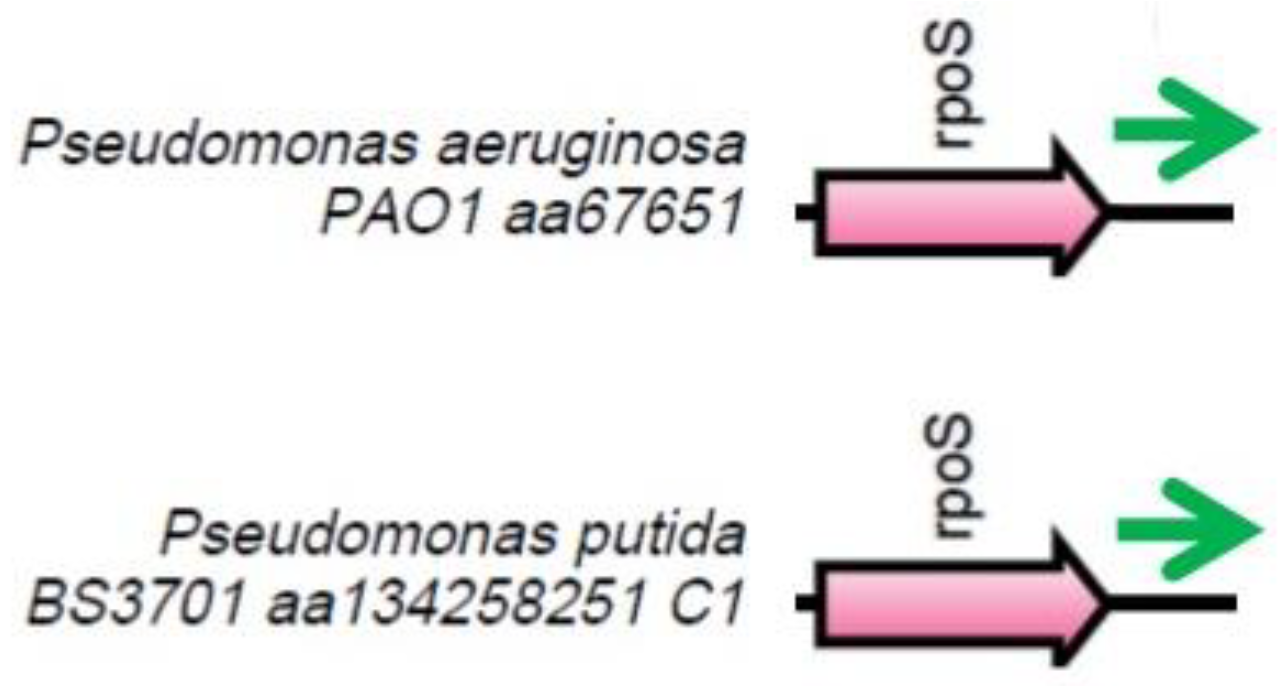
Figure 4.
The locus where the rsmZ gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 4.
The locus where the rsmZ gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
2.1.3. PresRAT prediction. ng11
ng11 small RNA is predicted for the first time. ncRNA has a length of 56 nucleotides, and its gene is located between the genes that encode HDOD domain-containing protein and folate-binding protein YgfZ. The NtrC binding site (tgcaccttgctggtgct) is predicted at a distance of 3076 bp upstream the assumed +1 point of the gene. A sigma54-dependent promoter is predicted upstream the gene (gcagggttggcaggaccaggtcatccttttcgatggcgtcgagcaattgtgcttgaaccaActcggccagcttgttcatgg, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 5.
Alignment of the ng11 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 5.
Alignment of the ng11 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 6.
The locus where the ng11 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 6.
The locus where the ng11 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
2.1.4. PresRAT prediction. ng127
ng127 small RNA gene is predicted for the first time. ncRNA has a length of 141 nucleotides, and its gene is located between the genes that encode porin and nitric oxide-sensing protein NosP [
25]. The NtrC binding site (ggcgccataacgatgcc) is predicted at a distance of 3316 bp upstream the assumed +1 point of the gene. A sigma54-dependent promoter is predicted upstream the gene (ctgagcggccaaccgtgcgtcggtagcctgcgacatggcactgaccacaccgtcgttatgAgcctgctgcatcgtggtctc, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 7.
Alignment of the ng127 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 7.
Alignment of the ng127 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 8.
The locus where the ng127 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 8.
The locus where the ng127 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
2.1.5. PresRAT prediction. ng171
ng171 small RNA gene is predicted for the first time. ncRNA has a length of 59 nucleotides, and its gene is located between the genes that encode metal ABC transporter ATPase and aldo/keto reductase. The NntrC binding site (ggcaccgtgaaggtgct) is predicted at a distance of 1117 bp upstream the assumed +1 point of the gene. A sigma54-dependent promoter is predicted upstream the gene (taaatcaagccttcgggcgtggcctcgacatgcagtggcagtgtcttgaagttgctcgggTtcttgcggatcagcgttcgc, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 9.
Alignment of the ng171 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 9.
Alignment of the ng171 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 10.
The locus where the ng171 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 10.
The locus where the ng171 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
2.1.6. PresRAT prediction. ng279
ng279 small RNA gene is predicted for the first time. ncRNA has a length of 186 nucleotides, and its gene is located downstream the gene that encodes inorganic triphosphatase. The NtrC binding site (tgcgccgcgatggcgac) is predicted at a distance of 2199 bp upstream the assumed +1 point of the gene. A sigma54-dependent promoter is predicted upstream the gene (ctggcaccgggccgaggtaattgcacagaaactgcgtaacctggtgtgtgtgggtgaggtGagcctgaccaacttgcagcg, the predicted iPro54-PseKNC +1 point is capitalized).
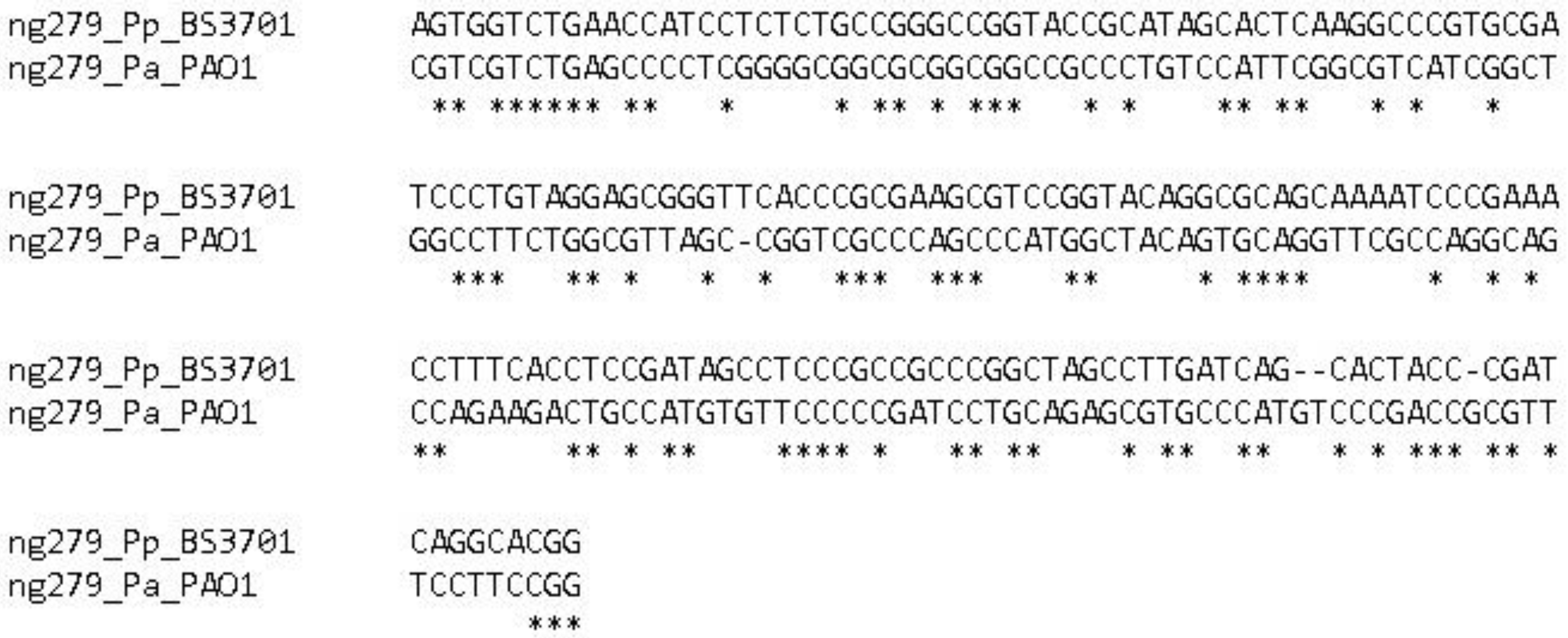
Figure 11.
Alignment of the ng279 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 11.
Alignment of the ng279 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
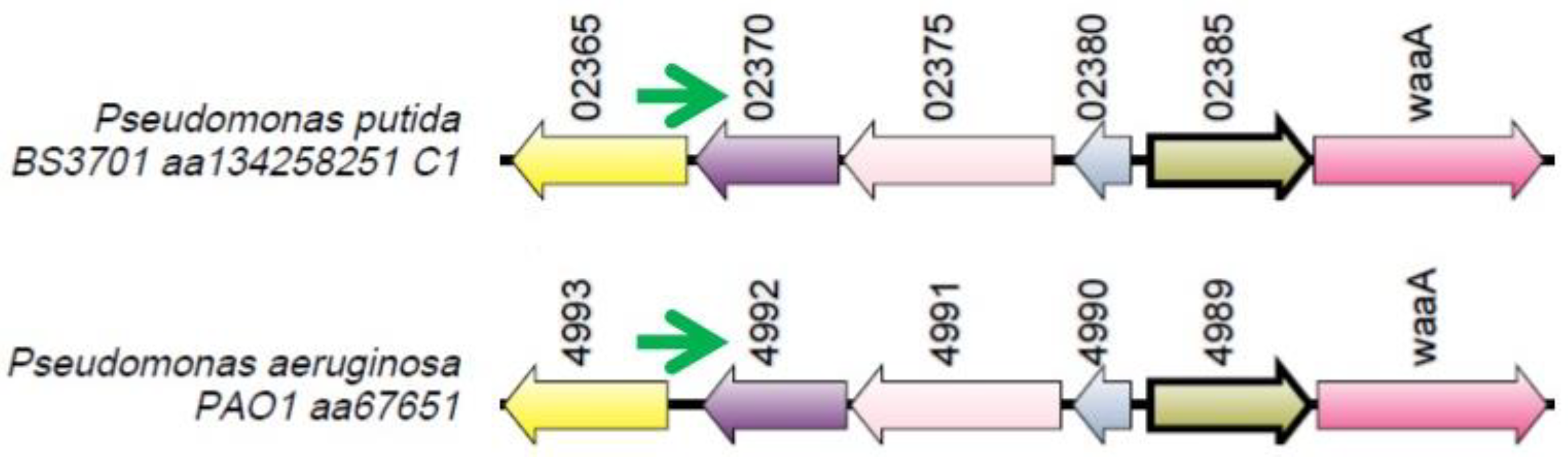
Figure 12.
The locus where the ng279 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
Figure 12.
The locus where the ng279 gene (green arrow) is located. Orthologous genes are indicated by arrows that have the same color.
2.1.7. PresRAT prediction. ng379
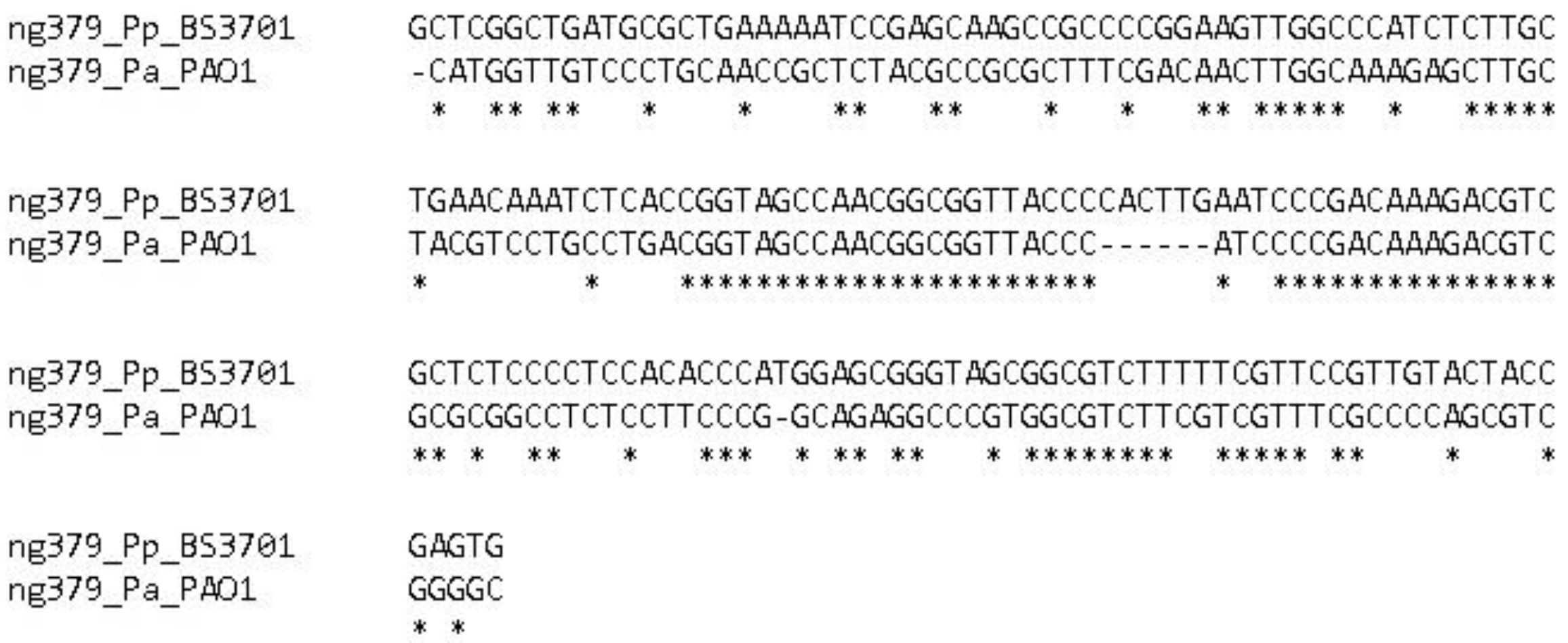
ng379 small RNA gene is predicted for the first time. There is narK-motif in its sequence, predicted bioinformatically in 2017, as a conservative cis-regulatory element in beta and gamma proteobacteria [
1]. ng379 has a length of 185 nucleotides, and its gene is located upstream the nasA gene that encodes the nitrate transporter. The NtrC binding site (tgcaccagcatggcgca) has been predicted at a distance of 1382 bp upstream the assumed +1 point of the gene. A sigma54-dependent promoter is predicted upstream the gene (caccctgatgcgtcgccaggccatgagccggcagcagaagctgatccaggtggccgaacaGatcattgccatgcatgagat, the predicted iPro54-PseKNC +1 point is capitalized).
Figure 13.
Alignment of the ng379 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
Figure 13.
Alignment of the ng379 gene in Pseudomonas aeruginosa PAO1 and Pseudomonas putida BS3701.
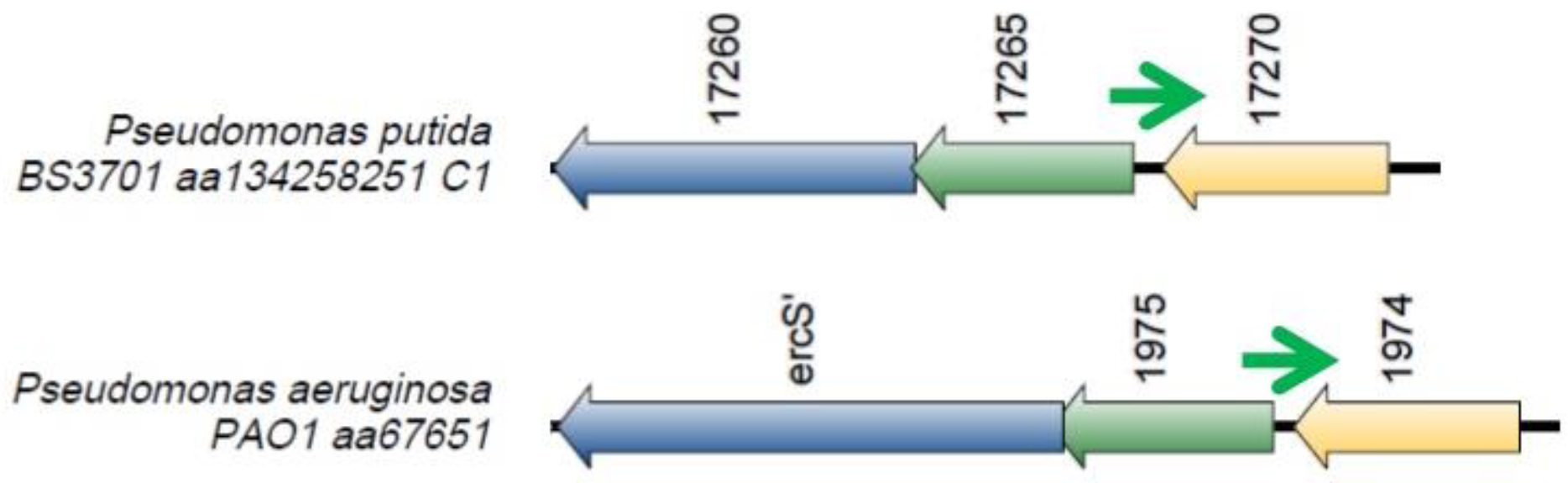
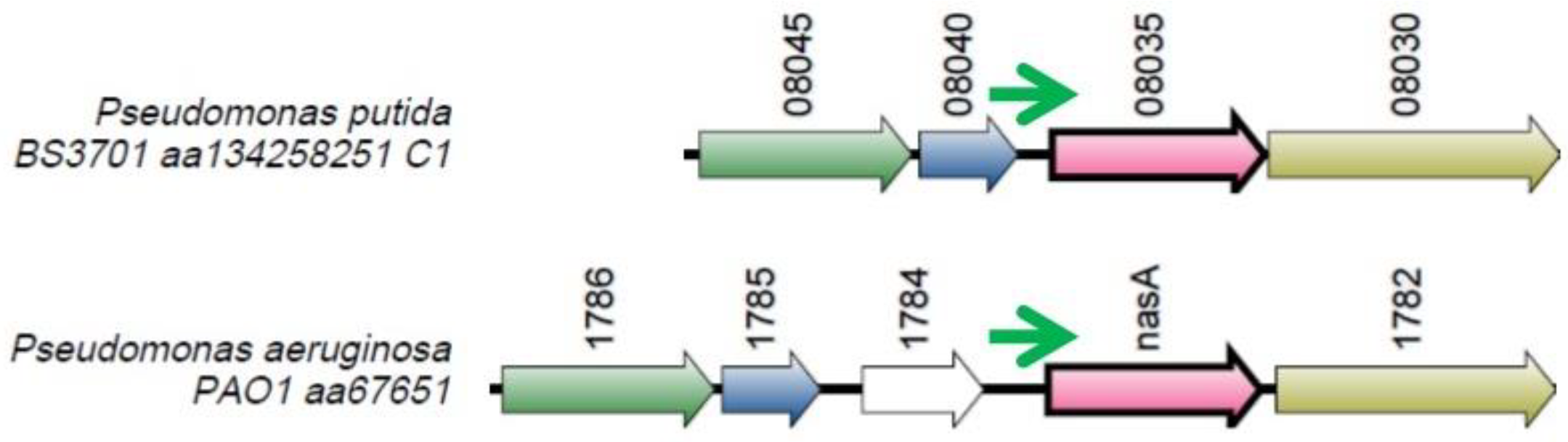
Figure 14.
The locus where the ng379 gene is located. Orthologous genes are indicated by arrows that have the same color.
Figure 14.
The locus where the ng379 gene is located. Orthologous genes are indicated by arrows that have the same color.
2.2. Modeling of nitrogen deficiency in Pseudomonas putida BS3701 cells
Nitrogen-limited conditions are often modeled by replacing ammonium with another nitrogen source [
2,
10,
15,
22]. This activates regulatory networks associated not only with the adaptation of the cell to a deficiency of ammonium ions, but also with adaptation to another nitrogen source. We have reduced the concentration of ammonium ions in the M9 mineral medium without another nitrogen source to observe changes related only to adaptation to the deficiency of them. The ammonium concentration in the normal M9 mineral medium is 18.6 mM. We have compared the growth pattern of
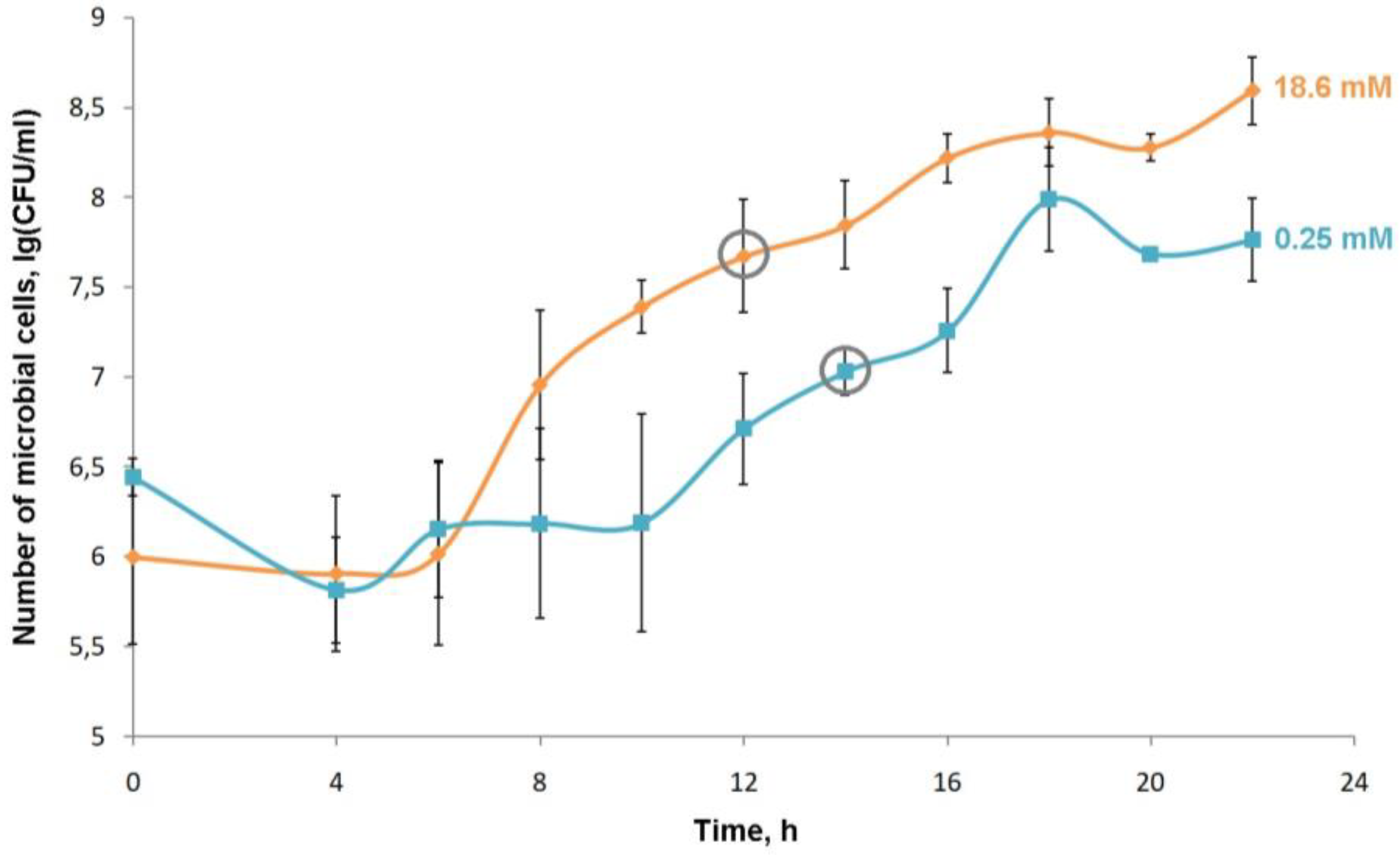
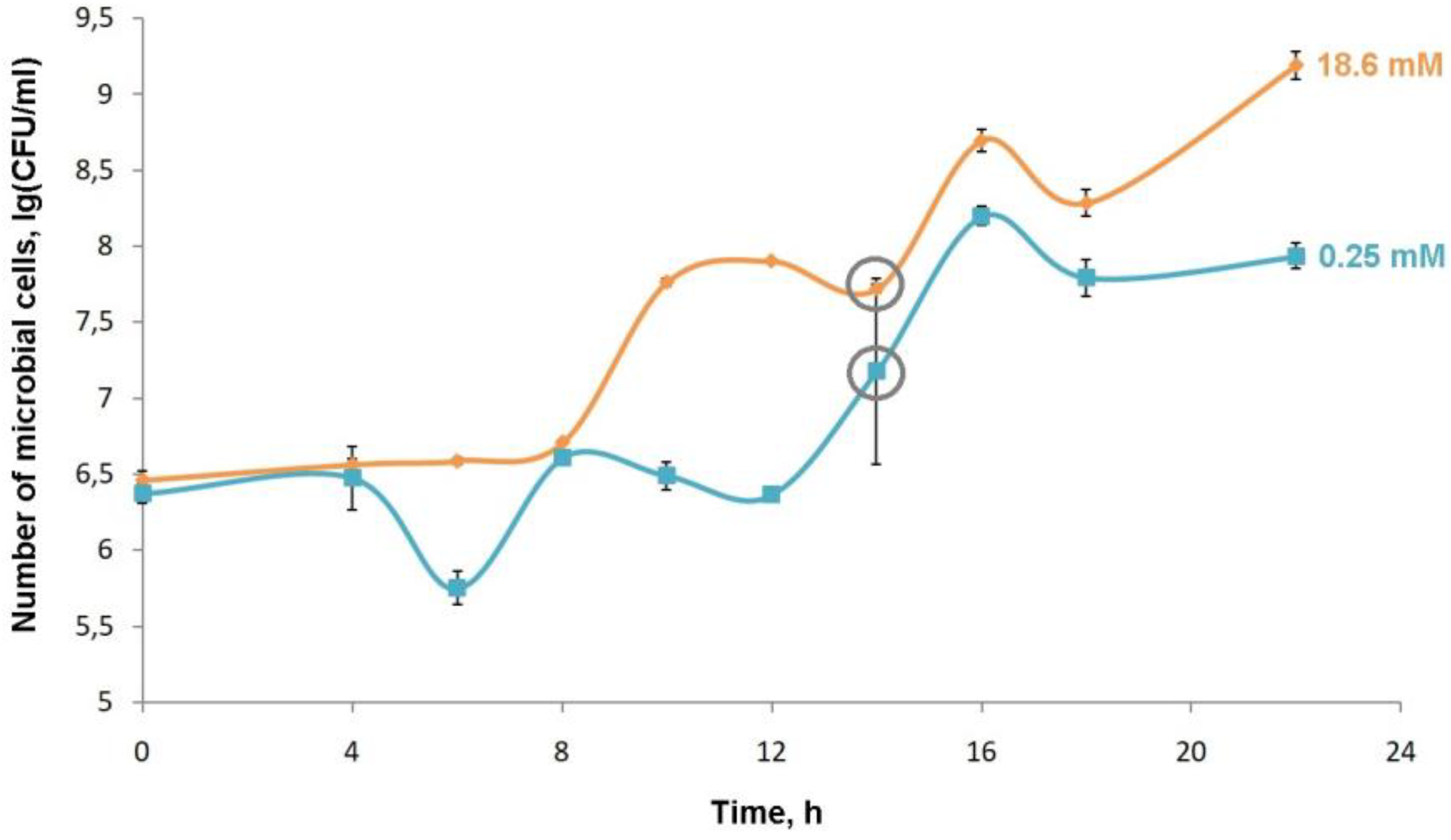
P.putida BS3701 both on a standard medium and on media with a concentration of ammonium ions 5, 2.5, 1, 0.25, 0.1, 0.05, 0.01 Mm. The pattern of the growth curve is comparable to that on the normal M9 medium at the concentration of ammonium ions in the culture medium of 5, 2.5, 1 and 0.25 mM. However, less biomass is accumulated in the stationary phase of growth at the concentration of ammonium ions of 0.25mM. We have observed a significant elongation of the lag phase and a decrease in the growth rate at concentrations of 0.1, 0.05 and 0.01 mM (data are not given). A modified M9 medium with an ammonium ion concentration of 0.25 mM is chosen as the nitrogen-limited conditions (
Figure 15 and
Figure 16).
Figure 15.
Growth curves of P.putida BS3701 grown on M9 medium with different concentrations of NH4Cl (0.25 and 18.6 mM) and the addition of succinate as the only carbon source. A grey circle marks the collection points of biomass to measure the amount of RNA in the cell.
Figure 15.
Growth curves of P.putida BS3701 grown on M9 medium with different concentrations of NH4Cl (0.25 and 18.6 mM) and the addition of succinate as the only carbon source. A grey circle marks the collection points of biomass to measure the amount of RNA in the cell.
Figure 16.
Growth curves of P.putida BS3701 grown on M9 medium with different concentrations of NH4Cl (0.25 and 18.6 mM) and the addition of naphthalene as the only carbon source. A grey circle marks the collection points of biomass to measure the amount of RNA in the cell.
Figure 16.
Growth curves of P.putida BS3701 grown on M9 medium with different concentrations of NH4Cl (0.25 and 18.6 mM) and the addition of naphthalene as the only carbon source. A grey circle marks the collection points of biomass to measure the amount of RNA in the cell.
BS3701 strain cells were grown on M9 medium with the addition of succinate as the only carbon source until the middle of the exponential growth phase, total RNA was isolated and the activity of the glnA glutamine synthetase gene was measured using RT-qPCR and specific primers. It was previously shown that an increase in the expression of the glutamine synthetase gene under conditions of nitrogen deficiency is characteristic of both
E.coli [
22] and
P.putida [
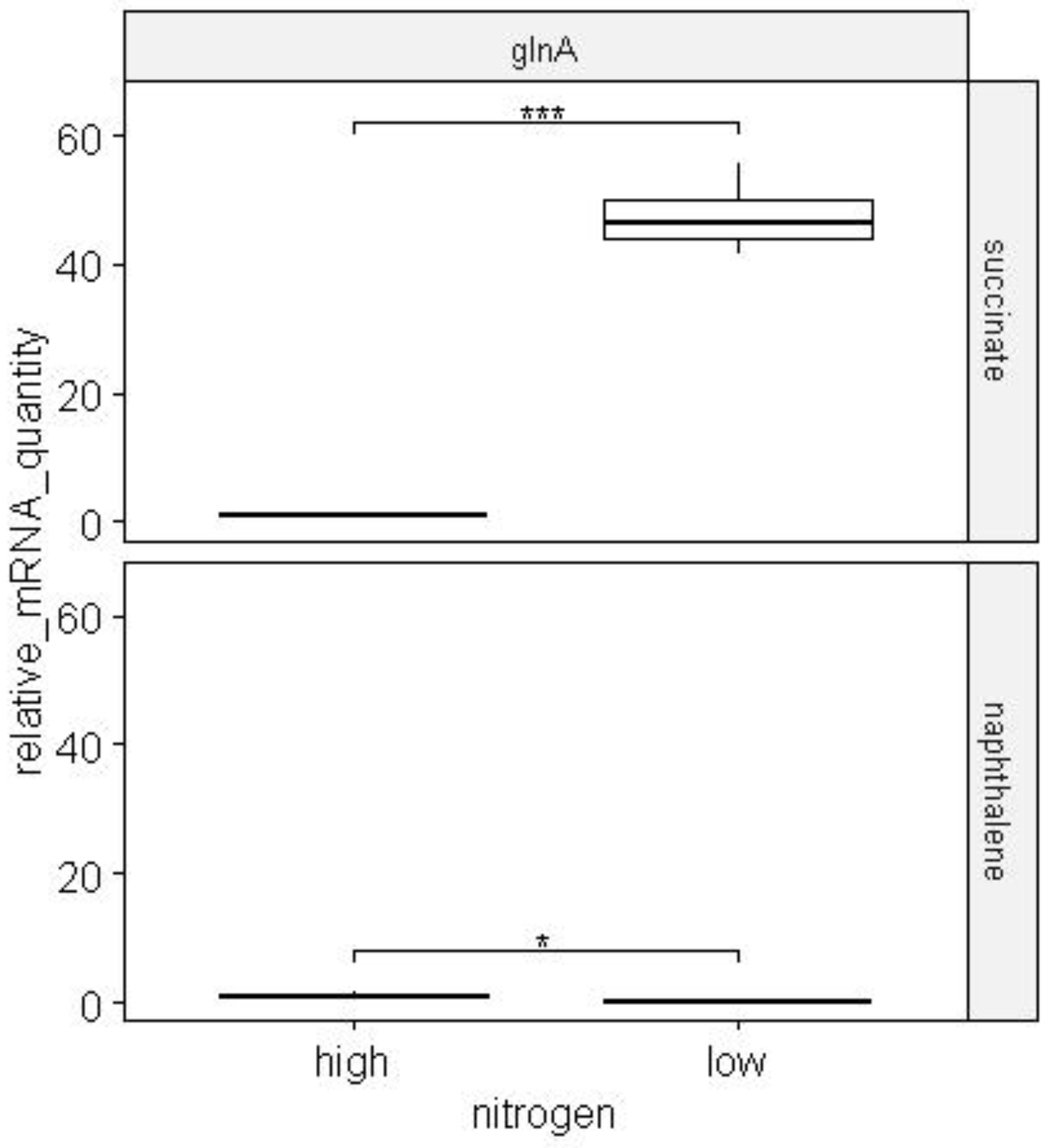
15]. The amount of mRNA of the glutamine synthetase gene of BS3701 strain increased by ~45 times with a decrease in the concentration of ammonium ions in the culture medium (
Figure 17). We also evaluated the activity of the glutamine synthetase gene when cultivating the BS3701 strain on an alternative carbon source, naphthalene (
Figure 17).
Figure 17.
The change in the amount of glutamine synthetase glnA mRNA during the cultivation of BS3701 strain cells in nitrogen-excess and nitrogen-limited conditions. The experiments were performed in 4 biological repeats. * - p-value < 0.05, *** - p-value < 0.001.
Figure 17.
The change in the amount of glutamine synthetase glnA mRNA during the cultivation of BS3701 strain cells in nitrogen-excess and nitrogen-limited conditions. The experiments were performed in 4 biological repeats. * - p-value < 0.05, *** - p-value < 0.001.
glnA expression decreased 5-fold when the strain grew on naphthalene in nitrogen-limited conditions, while glnA expression increased significantly when growing on succinate (
Figure 17). It may indicate a different functioning of the NtrC-regulon when the cell uses polyaromatic hydrocarbons.
The regulating mechanisms of PAH transformation genes, which lead to a decrease in the catabolism efficiency in conditions of nitrogen deficiency, have not been studied to date. We have used both cells grown on succinate, for which the pattern of changes in the mRNA quantity of the glutamine synthetase gene is comparable with the literature data, and cells grown on naphthalene, discover new small RNAs.
2.3. Evaluation of differential expression and the ncRNA presence in the fraction enriched with small RNAs
Specific primers were developed for the nrsZ, rsmZ, ng127, ng171, ng279 and ng379 genes (but not for ng11), which made it possible to estimate the change in the ncRNA quantity in the cell.
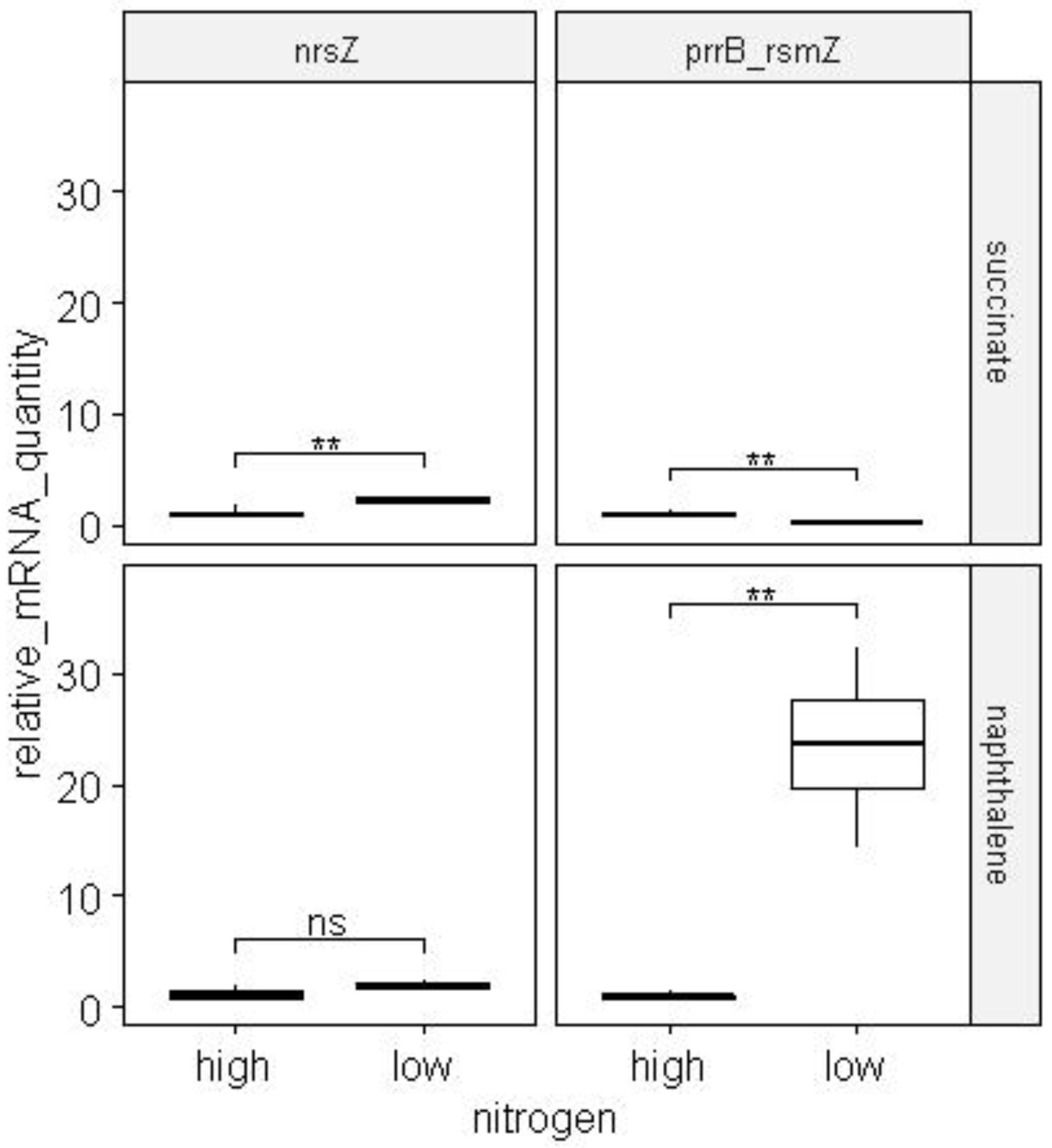
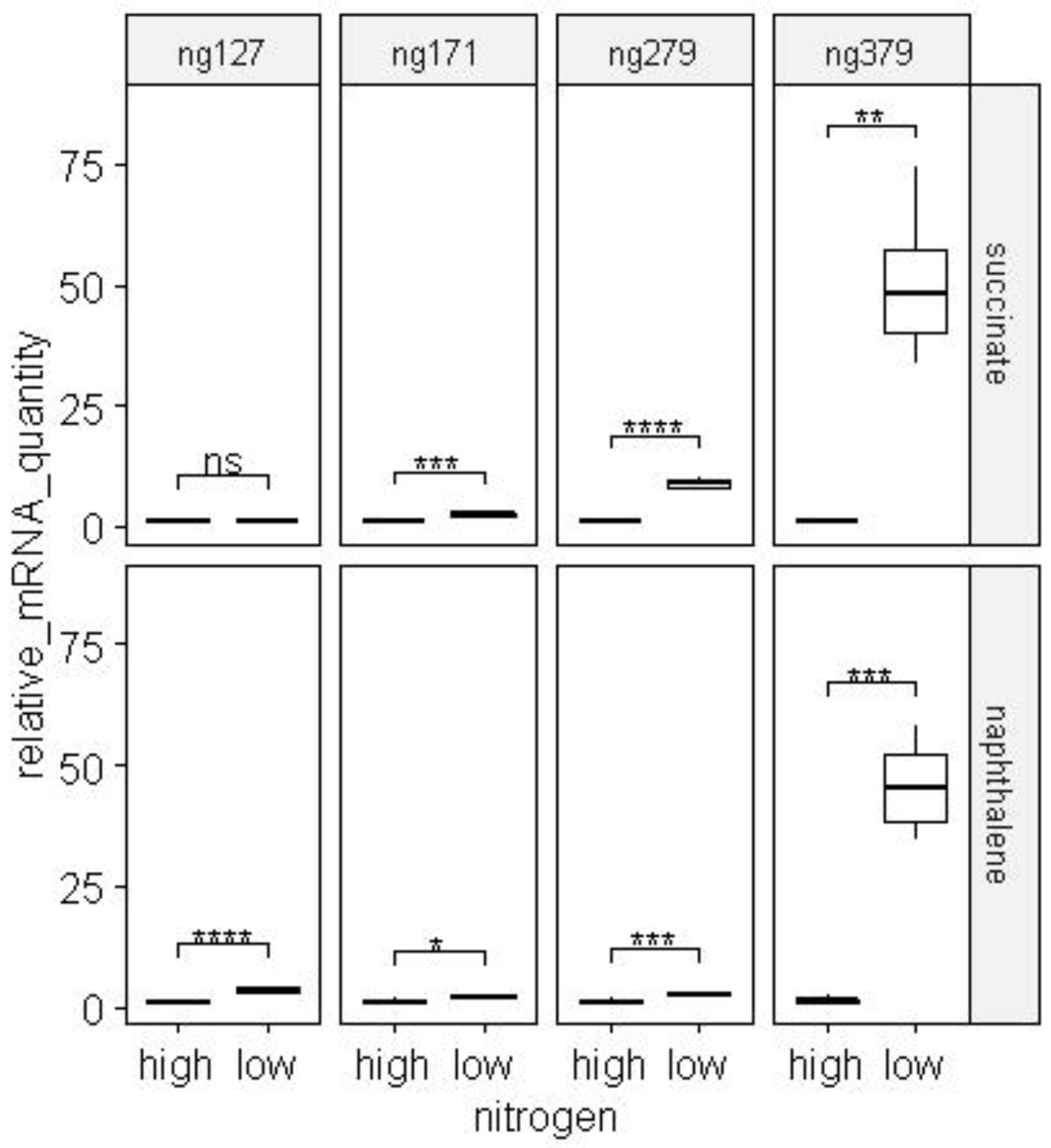
When cells were cultured on succinate in nitrogen-limited conditions, the number of products of the ng379 genes increased 50 times, ng279 - 8.5 times, ng171 - 2 times, nrsZ - 2 times (
Figure 18 and
Figure 19).
When cells were cultured on naphthalene in nitrogen-limited conditions, the number of gene products increased: ng379 - 40 times, ng279 - 8.5 times, ng127 - 3 times, and ng171 - 2 times (
Figure 18 and
Figure 19).
Figure 18.
The change in the mRNAs quantity of ncRNA genes predicted by the Prokka homology program during the cultivation of BS3701 strain cells in nitrogen-excess and nitrogen-limited conditions. The experiments were performed in 4 biological repeats. ** - p-value below 0.01.
Figure 18.
The change in the mRNAs quantity of ncRNA genes predicted by the Prokka homology program during the cultivation of BS3701 strain cells in nitrogen-excess and nitrogen-limited conditions. The experiments were performed in 4 biological repeats. ** - p-value below 0.01.
Figure 19.
The change in the mRNAs quantity of ncRNA genes predicted by the PresRAT program during the cultivation of BS3701 strain cells in nitrogen-excess (orange fill) and nitrogen-limited conditions (blue fill). The experiments were performed in 4 biological repeats. * - p-value below 0.05, ** - p-value below 0.01, ***- p-value below 0.001, ****- p-value below 0.0001.
Figure 19.
The change in the mRNAs quantity of ncRNA genes predicted by the PresRAT program during the cultivation of BS3701 strain cells in nitrogen-excess (orange fill) and nitrogen-limited conditions (blue fill). The experiments were performed in 4 biological repeats. * - p-value below 0.05, ** - p-value below 0.01, ***- p-value below 0.001, ****- p-value below 0.0001.
RNA preparations enriched with small RNAs were isolated from BS3701 strain cells grown in nitrogen-limited conditions and in the presence of succinate. To determine the limit size of the RNA detected in the resulting enriched fraction, we identified the presence not only of the studied ncRNAs, but also of the mRNA genes of the nucleoid mvaT10 proteins (gene size 378bp) and dps (gene size 474bp), as well as the gene encoding the sigma54 RNAP subunit RpoN (gene size 1470bp). The resulting fraction is guaranteed to have no RNAs of ~ 474b in size (
Table 1).
Small RNAs NrsZ, RsmZ, ng379 and ng171 were identified in the enriched fraction (
Table 1). We observed a decrease in the absolute amount of ncRNA when receiving the enriched fraction. The absence of ng127 may be due not to the fact that the predicted ncRNA gene is actually part of the upstream mRNA gene, but because the ncRNA itself is poorly represented in the cell.
3. Discussion
In 2020, we showed [
16] that the residual content of the only carbon source at the end of the exponential growth phase increased and the mRNA quantity of some genes involved in naphthalene and salicylate catabolism decreased when
P. putida BS3701 cells were cultured under conditions of nitrogen deficiency. Data on regulators capable of stimulating the degradation of target mRNA in pseudomonads under conditions of nitrogen deficiency are not available in the literature. In 2022, a small GlnZ RNA was detected in Enterobacteriaceae, which was generated from the 3'UTR of the GS-encoding glnA mRNA and which inhibited the expression of the sucA gene, E1o component of 2-oxoglutarate dehydrogenase at the post-transcriptional level [
23]. However, the expression of glnA
P. putida BS3701, an orthologue of glnA Enterobacteriaceae, was not increased under conditions of nitrogen deficiency when growing on naphthalene. We suggest a different mechanism for inhibiting the expression of catabolic genes. Moreover, we observed no change in the expression of the NtrC-dependent small RNA NrsZ gene, growing on naphthalene, its orthologue was characterized in
Pseudomonas aeruginosa as stimulating swarming motility and the synthesis of rhamnolipids. All this may indicate the presence of additional regulatory factors that affect the functioning of the NtrC-regulon during cell growth on different carbon sources.
In this study, we identified 2 new small RNAs whose expression increased in conditions of nitrogen deficiency by 2 or more times on both succinate and naphthalene ng171 and ng379.
Weinberg and co-authors was predicted narK RNA motif as a conservative motif of proteobacteria [
1] using bioinformatic tools in 2017. The authors have suggested that the motif is part of a cis-regulatory factor, but there are still no experimental data confirming its existence. Small RNA ng379 contains narK RNA motif
The ng379 is located at loci associated with nitrogen metabolism. The ng379 small RNA gene is located upstream the gene whose orthologue encodes the NasA nitrate transporter [
24] and is co-directed with it.
Currently, small RNAs capable of regulating carbon/nitrogen homeostasis in conditions of nitrogen deficiency are unknown for pseudomonads. The small RNAs, that we have selected, are promising objects for studying the adaptation process of pseudomonads to the lack of macronutrients
Authors should discuss the results and how they can be interpreted from the perspective of previous studies and of the working hypotheses. The findings and their implications should be discussed in the broadest context possible. Future research directions may also be highlighted.
4. Materials and Methods
4.1. NCBI GenBank accession number
Pseudomonas putida BS3701 genome have been deposited in GenBank under accession no. NZ_CP059052.1, Pseudomonas putida KT2440 genome - NC_002947.4
4.2. Search for small RNA genes in the P.putida BS3701 genome
Small RNA genes whose homologues were previously described for other microorganisms were predicted using the prokka server (version 1.14.6). Using the PresRAT server, small RNA genes were predicted de novo.
4.3. Search for the sigma-54 promoter, NtrC binding site and small RNA genomic context analysis
The SigmoID v2 program [
28] and aligned prospective NtrC binding sites of Pseudomonas putida BS3701 strain were used to search for the binding site of the transcriptional activator NtrC (
Table S2).
The Syntax server [
29] was used to assess the conservativeness of loci containing the genes of sigma54- and NtrC-dependent small RNAs
4.4. Cultivation environments and conditions
The strain Pseudomonas putida BS3701 was used in the work. The cells were cultured at a temperature of 28 °C on M9 mineral medium (8.5g/l Na2NPO4, 3g/l KH2PO4, 0.7 g/l NaCl, 0.5g/l MgSO4, 0.01g/l CaCl2, 1g/l NH4Cl) with the addition of sodium succinate (1 g/l) or naphthalene (2 g/l) as the only carbon sources.
A daily culture grown on M9 medium with ammonium chloride at a concentration of 1g/l and the addition of succinate (1 g/l) was used as an inoculum. The resulting suspension of cells was precipitated for 10 minutes at 4 ° C at 8000 rpm, the biomass precipitate was resuspended in M9 medium containing an appropriate amount of ammonium chloride. The final concentration of cells after resuspending was ~ 3×108 CFU/ml. 1 ml of inoculate per 100 ml of medium was added. The experiments were carried out in 750 ml rocking flasks containing 100 ml of mineral medium and substrate.
4.5. RT-qPCR
Specific primers have been developed using the Primer-BLAST tool (
www.ncbi.nlm.nih.gov/tools/primerblast) (
Table 2). A set containing SYBR Green (catalog number R-402, Syntol, Russia) was used for qPCR. Amplification program: (1) 95 °C 3 min, (2) 95 °C 10 sec, (3) 60 °C 20 sec, (4) 72 ° C 5 sec, stages (2)-(4) 40 cycles. The efficiency of amplification was determined for each pair of primers by the method of serial DNA dilutions. The specificity of the reaction was evaluated by electrophoresis in agarose gel.
10 ml of 5% alcohol solution of phenol was added to 100 ml of cell culture before collecting biomass. Trizol reagent (Sigma-Aldrich, USA) was used to isolate total RNA. A kit for isolating total RNA and microRNA from the Lira reagent (cells, tissues) (LRU-100-50) (Biolabmix, Russia) was used to extract the fraction enriched with small RNAs. The RevertAid RT Reverse Transcription Kit (Thermo, USA) was used in accordance with the manufacturer's protocol to carry out the reverse transcription reaction. 100 ng of total RNA and random hexamer primers were taken into the reaction. Genomic DNA (gDNA) contamination was quantified (1/E^(Cp«-RT»-Cp«+RT»)) and did not exceed 3%. A reference gene was selected among gyrB, rpoN, mvaT10, hupA, ihfA, dps to validate the RT-qPCR method. The hupA gene (SD 0.38; CV 1.65%) was the most stable (according to the BestKeeper program). The relative number of transcripts was calculated according to the Pfaffl formula, taking into account the efficiency of amplification.
All experiments were carried out in 4 biological repeats. The data were processed using RStudio Desktop 2023.09.0+463. The normality of the distribution was evaluated using the Shapiro–Wilk test, and an unpaid t-test was used to evaluate the differential expression of genes.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: Small RNA analysis; Table S2: Probable NtrC-dependent genes。.
Author Contributions
Conceptualization, IP and MZ; methodology, IP, EI, KP, AR, and AF; software, IP; validation IP, formal analysis, IP; investigation, IP, EI, KP, AR, and AF; resources, IP, KP and MZ; data curation, IP; writing—original draft preparation, IP and AF ; writing—review and editing, IP and MZ; visualization, IP; supervision, IP; project administration, IP; funding acquisition, MZ.
Funding
This research was funded by Russian Science Foundation, grant № 22-24-01138, https://rscf.ru/project/22-24-01138/.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Weinberg, Z.; Lünse, C.E; Corbino, K.A.; Ames, T.D.; Nelson, J.W.; Roth, A.; Perkins, K.R.; Sherlock, M.E.; Breaker, R.R. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017, 45, 10811–10823. [Google Scholar] [CrossRef] [PubMed]
- Wenner, N.; Maes, A.; Cotado-Sampayo, M.; Lapouge, K. NrsZ: a novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ Microbiol. 2014, 16, 1053–1068. [Google Scholar] [CrossRef]
- Brown, D.R.; Barton, G.; Pan, Z.; Buck, M.; Wigneshweraraj, S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun. 2014, 5, 4115:1–4115:8. [Google Scholar] [CrossRef]
- Austin, S.; Kundrot, C.; Dixon, R. Influence of a mutation in the putative nucleotide binding site of the nitrogen regulatory protein NTRC on its positive control function. Nucleic Acids Res. 1991, 19, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Ferro-Luzzi Ames, G.; Nikaido, K. Nitrogen regulation in Salmonella typhimurium. Identification of anntrC protein-binding site and definition of a consensus binding sequence. EMBO J 1985, 4, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Patriarca, E.J.; Riccio, A.; Colonna-Romano, S.; Defez, R.; Iaccarino, M. DNA binding activity of NtrC from Rhizobium grown on different nitrogen sources. FEBS Lett. 1994, 354, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Q.; Yan, Y.; Ke, X.; Han, Y.; Wu, S.; Lv, F.; Shao, Y.; Jiang, S.; Lin, M.; Zhang, Y.; Zhan, Y. Master regulator NtrC controls the utilization of alternative nitrogen sources in Pseudomonas stutzeri A1501. World J MicrobiolBiotechnol 2021, 37, 177:1–177:12. [Google Scholar] [CrossRef] [PubMed]
- Alford, M.A.; Baghela, A.; Yeung, A.T.Y.; Pletzer, D.; Hancock, R.E.W. NtrBC Regulates Invasiveness and Virulence of Pseudomonas aeruginosa During High-Density Infection. Front Microbiol 2020, 11, 773:1–773:17. [Google Scholar] [CrossRef] [PubMed]
- Naren, N.; Zhang, X.X. Role of a local transcription factor in governing cellular carbon/nitrogen homeostasis in Pseudomonas fluorescens. Nucleic Acids Res. 2021, 49, 3204–3216. [Google Scholar] [CrossRef]
- Hervás, A.B.; Canosa, I.; Little, R.; Dixon, R.; Santero, E. NtrC-dependent regulatory network for nitrogen assimilation in Pseudomonas putida. J Bacteriol. 2009, 191, 6123–6135. [Google Scholar] [CrossRef]
- van Heeswijk, W.C.; Westerhoff, H.V.; Boogerd, F.C. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol Mol Biol Rev. 2013, 77, 628–695. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Yan, Y.; Deng, Z.; Chen, M.; Lu, W.; Lu, C.; Shang, L.; Yang, Z.; Zhang, W.; Wang, W.; Li, Y.; Ke, Q.; Lu, J.; Xu, Y.; Zhang, L.; Xie, Z.; Cheng, Q.; Elmerich, C.; Lin, M. The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501. Proc Natl Acad Sci U S A. 2016, 113, E4348–E4356. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Deng, Z.; Yan, Y.; Zhang, H.; Lu, C.; Yang, Z.; Shang, L.; Huang, Y.; Lv, F.; Liu, Y.; Liu, Y.; Wang, S.; Chen, S.; Zhang, X.X.; Cheng, Q.; Lin, M. NfiR, a New Regulatory Noncoding RNA (ncRNA), Is Required in Concert with the NfiS ncRNA for Optimal Expression of Nitrogenase Genes in Pseudomonas stutzeri A1501. Appl Environ Microbiol. 2019, 85, e00762–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhan, Y.; Yan, Y.; Liu, Y.; Hu, G.; Wang, S.; Yang, H.; Qiu, X.; Liu, Y.; Li, J.; Lu, W.; Elmerich, C.; Lin, M. The Pseudomonas stutzeri-Specific Regulatory Noncoding RNA NfiS Targets katB mRNA Encoding a Catalase Essential for Optimal Oxidative Resistance and Nitrogenase Activity. J Bacteriol. 2019, 201, e00334–19. [Google Scholar] [CrossRef] [PubMed]
- Hervás, A.B.; Canosa, I.; Santero, E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008, 190, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Pozdnyakova-Filatova, I.; Petrikov, K.; Vetrova, A.; Frolova, A.; Streletskii, R.; Zakharova, M. The Naphthalene Catabolic Genes of Pseudomonas putida BS3701: Additional Regulatory Control. Front Microbiol. 2020, 11, 1217:1–1217:12. [Google Scholar] [CrossRef]
- Reinhart, A.A.; Nguyen, A.T.; Brewer, L.K.; Bevere, J.; Jones, J.W.; Kane, M.A.; Damron, F.H.; Barbier, M.; Oglesby-Sherrouse, A.G. The Pseudomonas aeruginosa PrrF Small RNAs Regulate Iron Homeostasis during Acute Murine Lung Infection. Infect Immun. 2017, 85, e00764–16. [Google Scholar] [CrossRef]
- Djapgne, L.; Panja, S.; Brewer, L.K.; Gans, J.H.; Kane, M.A.; Woodson, S.A.; Oglesby-Sherrouse, A.G. The Pseudomonas aeruginosa PrrF1 and PrrF2 Small Regulatory RNAs Promote 2-Alkyl-4-Quinolone Production through Redundant Regulation of the antR mRNA. J Bacteriol. 2018, 200, e00704–17. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Chakraborty, A.; Chakrabarti, S. PresRAT: a server for identification of bacterial small-RNA sequences and their targets with probable binding region. RNA Biol. 2021, 18, 1152–1159. [Google Scholar] [CrossRef]
- Dubuis, C.; Rolli, J.; Lutz, M.; Défago, G.; Haas, D. Thiamine-auxotrophic mutants of Pseudomonas fluorescens CHA0 are defective in cell-cell signaling and biocontrol factor expression. Appl Environ Microbiol. 2006, 72, 2606–2613. [Google Scholar] [CrossRef]
- Livny, J.; Brencic, A.; Lory, S.; Waldor, M.K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006, 34, 3484–3493. [Google Scholar] [CrossRef] [PubMed]
- Reitzer, L.J.; Magasanik, B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci U S A. 1985, 82, 1979–1983. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Morita, T.; Kobayashi, A.; Berger, A.; Takahashi, H.; Gotoh, Y.; Hayashi, T.; Tanaka, K. Glutamine synthetase mRNA releases sRNA from its 3'UTR to regulate carbon/nitrogen metabolic balance in Enterobacteriaceae. Elife. 2022, 11, e82411:1–e82411-24. [Google Scholar] [CrossRef] [PubMed]
- Sias, S.R.; Ingraham, J.L. Isolation and analysis of mutants of Pseudomonas aeruginosa unable to assimilate nitrate. Arch Microbiol. 1979, 122, 263–270. [Google Scholar] [CrossRef]
- Hossain, S.; Boon, E.M. Discovery of a Novel Nitric Oxide Binding Protein and Nitric-Oxide-Responsive Signaling Pathway in Pseudomonas aeruginosa. ACS Infect Dis. 2017, 3, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, A.A.; Vetrova, A.A.; Filonov, A.E.; Boronin, A.M. Phenanthren biodegradation and interaction of Pseudomonas putida BS3701 and Burkholderia sp.BS3702 in plant rhizosphere. Mikrobiologiia. 2009, 78, 484–490. [Google Scholar]
- Lin, H.; Deng, E.Z.; Ding, H.; Chen, W.; Chou, K.C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res. 2014, 42, 12961–12972. [Google Scholar] [CrossRef] [PubMed]
- Nikolaichik, Y.; Damienikan, A.U. SigmoID: a user-friendly tool for improving bacterial genome annotation through analysis of transcription control signals. PeerJ. 2016, 4, e2056:1–e2056:21. [Google Scholar] [CrossRef]
- Oberto, J. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics. 2013, 14, 4:1–4:10. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).