Submitted:
22 November 2023
Posted:
23 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study participants
2.2. Serological Data
2.2. Data Splitting
2.3. Machine Learning Approach
2.4. Bioinformatic analysis
2.5. Statistical Software
3. Results
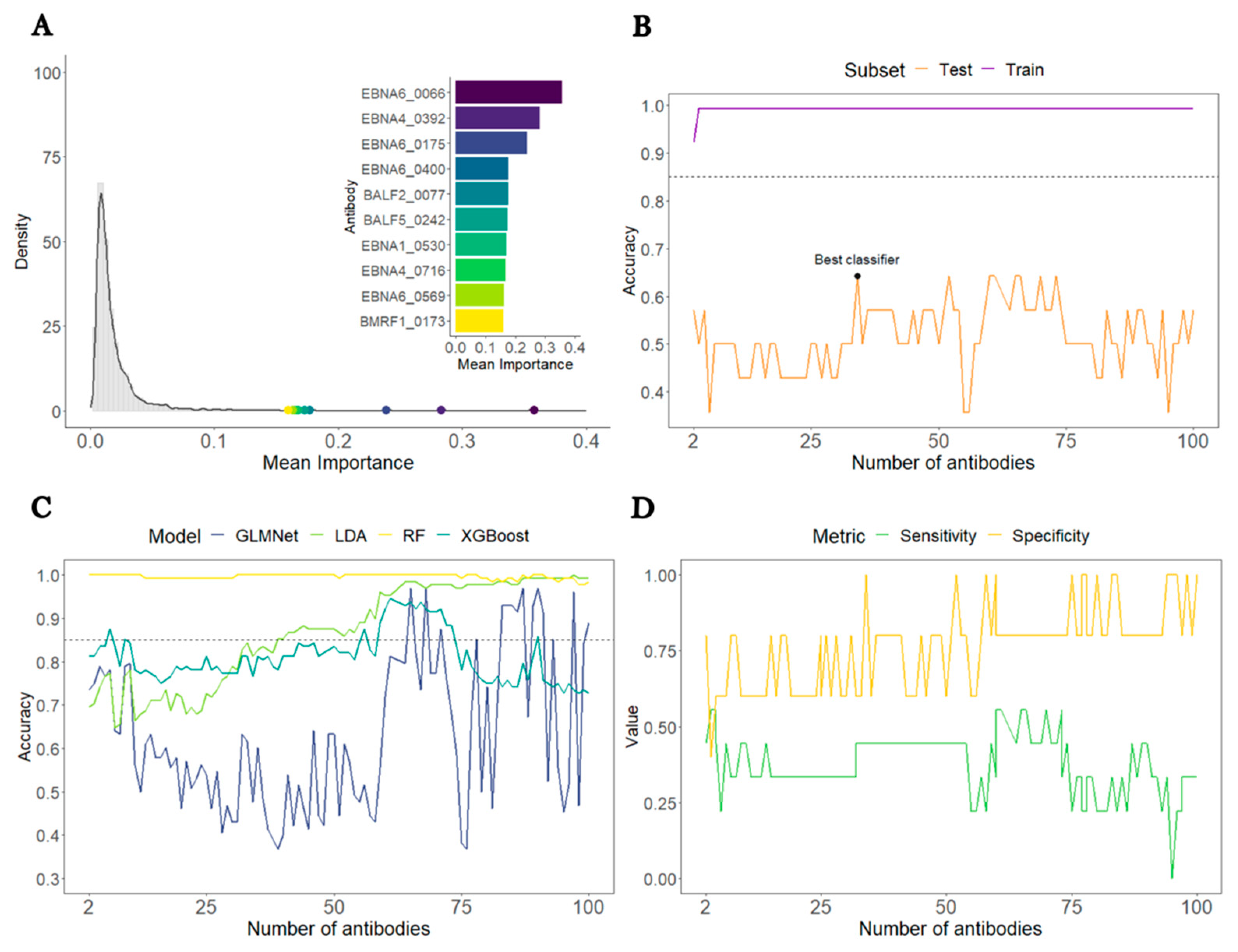
3.1. Analysis of all ME/CFS patients against HCs
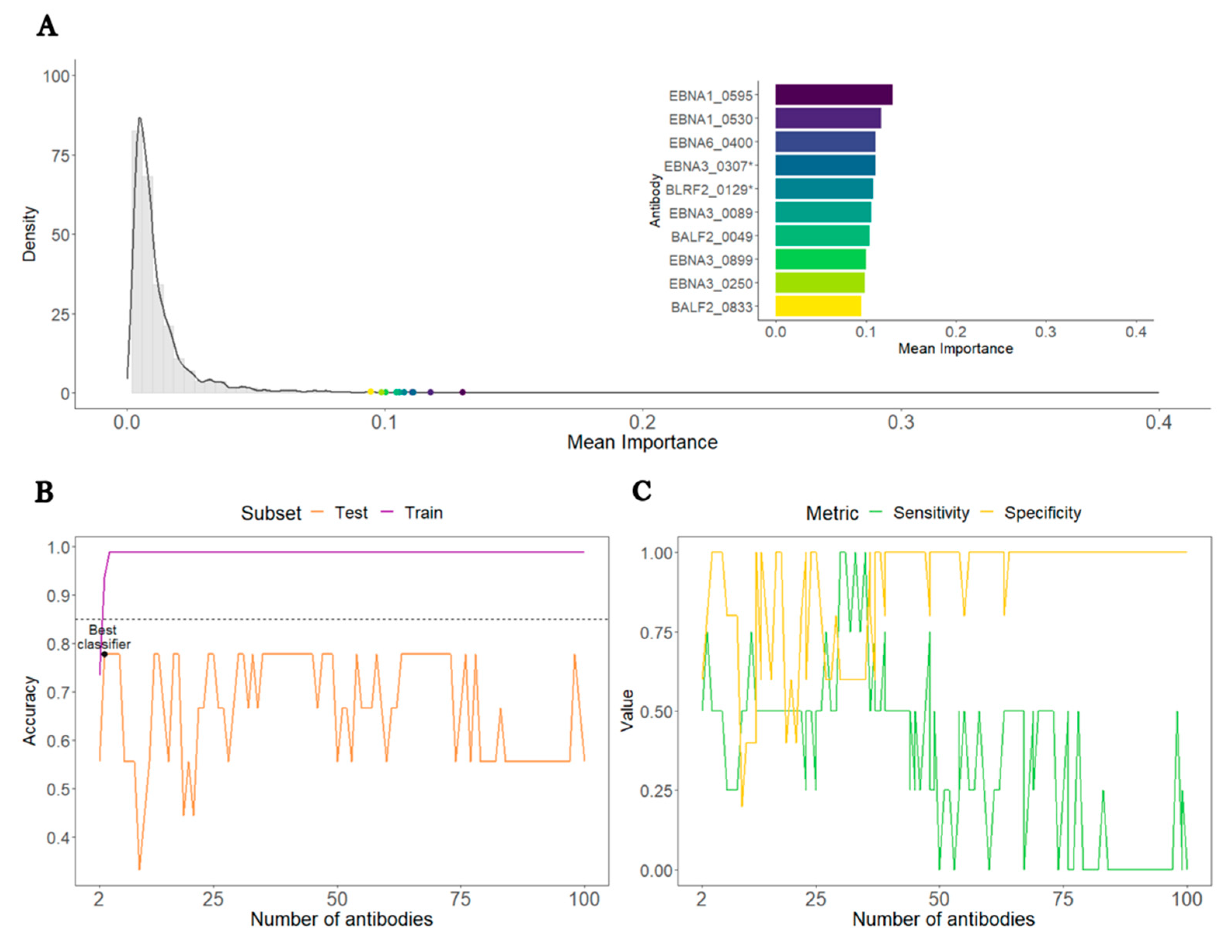
3.2. Analysis of ME/CFS patients with non-infectious or unknown disease trigger against HCs
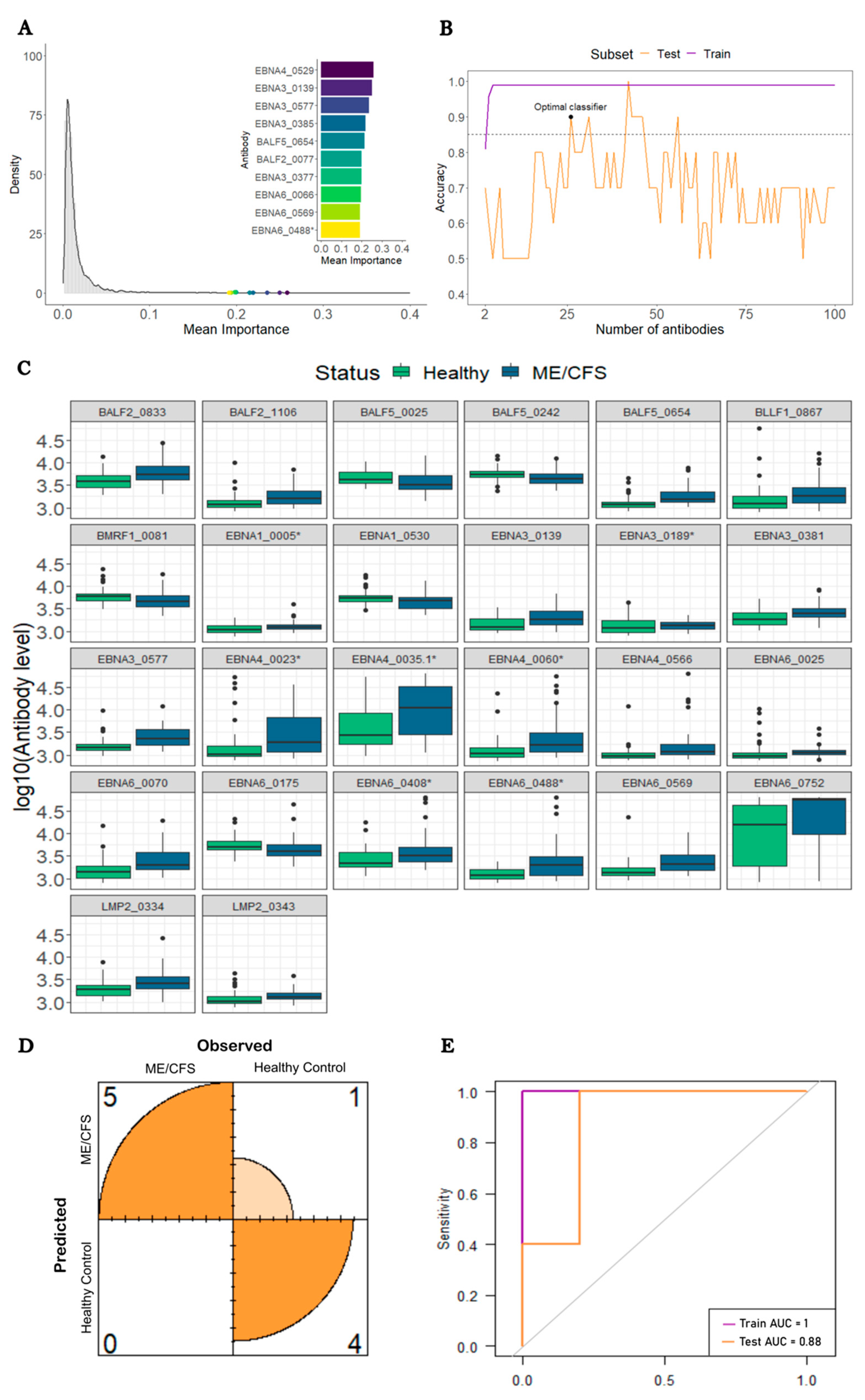
3.3. Analysis of ME/CFS patients with a putative infectious disease trigger against HCs
3.4. Bioinformatic analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rivera MC, Mastronardi C, Silva-Aldana CT, Arcos-Burgos M, Lidbury BA. Myalgic encephalomyelitis/chronic fatigue syndrome: A comprehensive review. Diagnostics 2019;9:91. [CrossRef]
- Fluge Ø, Tronstad KJ, Mella O. Pathomechanisms and possible interventions in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). J Clin Invest 2021;131:e150377. [CrossRef]
- Stanculescu D, Larsson L, Bergquist J. Hypothesis: Mechanisms That Prevent Recovery in Prolonged ICU Patients Also Underlie Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). Front Med 2021;8:628029. [CrossRef]
- Stanculescu D, Sepúlveda N, Lim CL, Bergquist J. Lessons From Heat Stroke for Understanding Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front Neurol 2021;12. [CrossRef]
- Nacul L, O’Boyle S, Palla L, Nacul FE, Mudie K, Kingdon CC, et al. How Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) Progresses: The Natural History of ME/CFS. Front Neurol 2020;11:826. [CrossRef]
- Blomberg J, Gottfries CG, Elfaitouri A, Rizwan M, Rosén A. Infection elicited autoimmunity and Myalgic encephalomyelitis/chronic fatigue syndrome: An explanatory model. Front Immunol 2018;9:229. [CrossRef]
- Sotzny F, Blanco J, Capelli E, Castro-Marrero J, Steiner S, Murovska M, et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome – Evidence for an autoimmune disease. Autoimmun Rev 2018;17:601–9. [CrossRef]
- Morris G, Berk M, Galecki P, Maes M. The emerging role of autoimmunity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/cfs). Mol Neurobiol 2014;49:741–56. [CrossRef]
- Petracek LS, Suskauer SJ, Vickers RF, Patel NR, Violand RL, Swope RL, et al. Adolescent and Young Adult ME/CFS After Confirmed or Probable COVID-19. Front Med 2021;8:668944. [CrossRef]
- Davis HE, Assaf GS, McCorkell L, Wei H, Low RJ, Re’em Y, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine 2021;38. [CrossRef]
- Jason LA, Dorri JA. ME/CFS and Post-Exertional Malaise among Patients with Long COVID. Neurol Int 2022;15:1–11. [CrossRef]
- Vélez-Santamaría R, Fernández-Solana J, Méndez-López F, Domínguez-García M, González-Bernal JJ, Magallón-Botaya R, et al. Functionality, physical activity, fatigue and quality of life in patients with acute COVID-19 and Long COVID infection. Sci Reports 2023 131 2023;13:1–9. [CrossRef]
- Ruiz-Pablos M, Paiva B, Montero-Mateo R, Garcia N, Zabaleta A. Epstein-Barr Virus and the Origin of Myalgic Encephalomyelitis or Chronic Fatigue Syndrome. Front Immunol 2021;12:656797. [CrossRef]
- Ariza, ME. Myalgic encephalomyelitis/chronic fatigue syndrome: The human herpesviruses are back! Biomolecules 2021;11:1–17. [CrossRef]
- Sepúlveda N, Carneiro J, Lacerda E, Nacul L. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome as a Hyper-Regulated Immune System Driven by an Interplay Between Regulatory T Cells and Chronic Human Herpesvirus Infections. Front Immunol 2019;10:2684. [CrossRef]
- Ruiz-Pablos M, Paiva B, Zabaleta A. Epstein–Barr virus-acquired immunodeficiency in myalgic encephalomyelitis—Is it present in long COVID? J Transl Med 2023;21:633. [CrossRef]
- Capone G, Calabrò M, Lucchese G, Fasano C, Girardi B, Polimeno L, et al. Peptide matching between Epstein-Barr virus and human proteins. Pathog Dis 2013;69:205–12. [CrossRef]
- Baboonian C, Venables PJW, Williams DG, Williams RO, Maini RN. Cross reaction of antibodies to a glycine/alanine repeat sequence of Epstein-Barr virus nuclear antigen-i with collagen, cytokeratin, and actin. Ann Rheum Dis 1991;50:772–5. [CrossRef]
- Blomberg J, Rizwan M, Böhlin-Wiener A, Elfaitouri A, Julin P, Zachrisson O, et al. Antibodies to human herpesviruses in myalgic encephalomyelitis/chronic fatigue syndrome patients. Front Immunol 2019;10:1946. [CrossRef]
- Lerner AM, Ariza ME, Williams M, Jason L, Beqaj S, Fitzgerald JT, et al. Antibody to Epstein-Barr virus deoxyuridine triphosphate nucleotidohydrolase and deoxyribonucleotide polymerase in a chronic fatigue syndrome subset. PLoS One 2012;7:e47891. [CrossRef]
- Kerr, JR. Epstein-Barr Virus Induced Gene-2 Upregulation Identifies a Particular Subtype of Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Front Pediatr 2019;7:59. [CrossRef]
- Domingues TD, Grabowska AD, Lee JS, Ameijeiras-Alonso J, Westermeier F, Scheibenbogen C, et al. Herpesviruses Serology Distinguishes Different Subgroups of Patients From the United Kingdom Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Biobank. Front Med 2021;8:686736. [CrossRef]
- Malato J, Graça L, Sepúlveda N. Impact of Misdiagnosis in Case-Control Studies of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Diagnostics (Basel, Switzerland) 2023;13:531. [CrossRef]
- Nacul L, Lacerda EM, Kingdon CC, Curran H, Bowman EW. How have selection bias and disease misclassification undermined the validity of myalgic encephalomyelitis/chronic fatigue syndrome studies? J Health Psychol 2019;24:1765–9. [CrossRef]
- Ariza, ME. Commentary: Antibodies to Human Herpesviruses in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Patients. Front Immunol 2020;11:1945. [CrossRef]
- Loebel M, Eckey M, Sotzny F, Hahn E, Bauer S, Grabowski P, et al. Serological profiling of the EBV immune response in Chronic Fatigue Syndrome using a peptide microarray. PLoS One 2017;12:e0179124. [CrossRef]
- Sepúlveda N, Malato J, Sotzny F, Grabowska AD, Fonseca A, Cordeiro C, et al. Revisiting IgG Antibody Reactivity to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Its Potential Application to Disease Diagnosis. Front Med 2022;9:921101. [CrossRef]
- Roshan V, Stewart JHM, Joseph R, Stewart HM. Optimal ratio for data splitting. Stat Anal Data Min ASA Data Sci J 2022;15:531–8. [CrossRef]
- Van Der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat Appl Genet Mol Biol 2007;6:25. [CrossRef]
- López-Ratón M, Rodríguez-Álvarez MX, Cadarso-Suárez C, Gude-Sampedro F. Optimalcutpoints: An R package for selecting optimal cutpoints in diagnostic tests. J Stat Softw 2014;61:1–36. [CrossRef]
- Valletta JJ, Recker M. Identification of immune signatures predictive of clinical protection from malaria. PLOS Comput Biol 2017;13:e1005812. [CrossRef]
- Wilson RC, Shenhav A, Straccia M, Cohen JD. The Eighty Five Percent Rule for optimal learning. Nat Commun 2019;10. [CrossRef]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45. [CrossRef]
- Karlin S, Altschul SF. Applications and statistics for multiple high-scoring segments in molecular sequences. Proc Natl Acad Sci U S A 1993;90:5873–7. [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J Stat Softw 2008;28:1–26. [CrossRef]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:1–8. [CrossRef]
- Wright MN, Ziegler A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J Stat Softw 2017;77:1–17. [CrossRef]
- Polley E, LeDell E, Kennedy C, van der Laan M. SuperLearner: Super Learner Prediction 2021.
- Tengvall K, Huang J, Hellström C, Kammer P, Biström M, Ayoglu B, et al. Molecular mimicry between Anoctamin 2 and Epstein-Barr virus nuclear antigen 1 associates with multiple sclerosis risk. Proc Natl Acad Sci U S A 2019;116:16955–60. [CrossRef]
- Sepúlveda, N. Impact of genetic variation on the molecular mimicry between Anoctamin-2 and Epstein-Barr virus nuclear antigen 1 in Multiple Sclerosis. Immunol Lett 2021;238:29–31. [CrossRef]
- O’Neal AJ, Glass KA, Emig CJ, Vitug AA, Henry SJ, Shungu DC, et al. Survey of Anti-Pathogen Antibody Levels in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Proteomes 2022, Vol 10, Page 21 2022;10:21. [CrossRef]
- Bjornevik K, Cortese M, Healy BC, Kuhle J, Mina MJ, Leng Y, et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science (80- ) 2022;375:296–301. [CrossRef]
- Jason L, Ohanian D, Brown A, Sunnquist M, McManimen S, Klebek L, et al. Differentiating Multiple Sclerosis from Myalgic Encephalomyelitis and Chronic Fatigue Syndrome. Insights Biomed 2017;2:11. [CrossRef]
- Ohanian D, Brown A, Sunnquist M, Furst J, Nicholson L, Klebek L, et al. Identifying Key Symptoms Differentiating Myalgic Encephalomyelitis and Chronic Fatigue Syndrome from Multiple Sclerosis. Neurol 2016;4:41.
- Domingues TD, Malato J, Grabowska AD, Lee JS, Ameijeiras-Alonso J, Biecek P, et al. Association analysis between symptomology and herpesvirus IgG antibody concentrations in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and multiple sclerosis. Heliyon 2023;9. [CrossRef]
- Fluge Ø, Rekeland IG, Lien K, Thürmer H, Borchgrevink PC, Schäfer C, et al. B-Lymphocyte Depletion in Patients With Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Randomized, Double-Blind, Placebo-Controlled Trial. Ann Intern Med 2019;170:585–93. [CrossRef]
- Bastiaan Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc B Biol Sci 2013;368. [CrossRef]
- Dehingia B, Milewska M, Janowski M, Pe Z Kowska A. CTCF shapes chromatin structure and gene expression in health and disease. EMBO Rep 2022;23:e55146. [CrossRef]
- DiSpirito JR, Zemmour D, Ramanan D, Cho J, Zilionis R, Klein AM, et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol 2018;3:5861. [CrossRef]
- Chandrasekaran V, Oparina N, Garcia-Bonete MJ, Wasén C, Erlandsson MC, Malmhäll-Bah E, et al. Cohesin-Mediated Chromatin Interactions and Autoimmunity. Front Immunol 2022;13:840002. [CrossRef]
- Kerr, JR. Gene profiling of patients with chronic fatigue syndrome/myalgic encephalomyelitis. Curr Rheumatol Rep 2008;10:482–91. [CrossRef]
- Kaushik N, Fear D, Richards SCM, McDermott CR, Nuwaysir EF, Kellam P, et al. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol 2005;58:826–32. [CrossRef]
- Sweetman E, Ryan M, Edgar C, Mackay A, Vallings R, Tate W. Changes in the transcriptome of circulating immune cells of a New Zealand cohort with myalgic encephalomyelitis/chronic fatigue syndrome. Int J Immunopathol Pharmacol 2019;33:1–8. [CrossRef]
- Almenar-Pérez E, Ovejero T, Sánchez-Fito T, Espejo JA, Nathanson L, Oltra E. Epigenetic Components of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Uncover Potential Transposable Element Activation. Clin Ther 2019;41:675–98. [CrossRef]
- De Vega WC, Herrera S, Vernon SD, McGowan PO. Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). BMC Med Genomics 2017;10:11. [CrossRef]
- Lorusso L, Mikhaylova S V., Capelli E, Ferrari D, Ngonga GK, Ricevuti G. Immunological aspects of chronic fatigue syndrome. Autoimmun Rev 2009;8:287–91. [CrossRef]
- Majdalawieh AF, Massri M, Ro HS. AEBP1 is a Novel Oncogene: Mechanisms of Action and Signaling Pathways. J Oncol 2020;2020. [CrossRef]
- Angwin C, Ghali N, van Dijk FS. Case report: Two individuals with AEBP1-related classical-like EDS: Further clinical characterisation and description of novel AEBP1 variants. Front Genet 2023;14. [CrossRef]
- Ritelli M, Cinquina V, Venturini M, Pezzaioli L, Formenti AM, Chiarelli N, et al. Expanding the Clinical and Mutational Spectrum of Recessive AEBP1-Related Classical-Like Ehlers-Danlos Syndrome. Genes (Basel) 2019;10. [CrossRef]
- Castori M, Celletti C, Camerota F, Grammatico P. Chronic fatigue syndrome is commonly diagnosed in patients with Ehlers-Danlos syndrome hypermobility type/joint hypermobility syndrome. Clin Exp Rheumatol 2011;29:597–8.
- Hakim A, De Wandele I, O’Callaghan C, Pocinki A, Rowe P. Chronic fatigue in Ehlers-Danlos syndrome-Hypermobile type. Am J Med Genet C Semin Med Genet 2017;175:175–80. [CrossRef]
- Nacul L, Authier FJ, Scheibenbogen C, Lorusso L, Helland IB, Martin JA, et al. European Network on Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (EUROMENE): Expert Consensus on the Diagnosis, Service Provision, and Care of People with ME/CFS in Europe. Medicina (Kaunas) 2021;57:510. [CrossRef]
- Schlauch KA, Khaiboullina SF, De Meirleir KL, Rawat S, Petereit J, Rizvanov AA, et al. Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome. Transl Psychiatry 2016;6:e730. [CrossRef]
- Dibble JJ, McGrath SJ, Ponting CP. Genetic risk factors of ME/CFS: A critical review. Hum Mol Genet 2020;29:R118–25. [CrossRef]
- Herrera S, de Vega WC, Ashbrook D, Vernon SD, McGowan PO. Genome-epigenome interactions associated with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Epigenetics 2018;13:1174–90. [CrossRef]
- Hajdarevic R, Lande A, Mehlsen J, Rydland A, Sosa DD, Strand EB, et al. Genetic association study in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) identifies several potential risk loci. Brain Behav Immun 2022;102:362–9. [CrossRef]
- Das S, Taylor K, Kozubek J, Sardell J, Gardner S. Genetic risk factors for ME/CFS identified using combinatorial analysis. J Transl Med 2022;20. [CrossRef]
- Haffke M, Freitag H, Rudolf G, Seifert M, Doehner W, Scherbakov N, et al. Endothelial dysfunction and altered endothelial biomarkers in patients with post-COVID-19 syndrome and chronic fatigue syndrome (ME/CFS). J Transl Med 2022;20. [CrossRef]
- Scherbakov N, Szklarski M, Hartwig J, Sotzny F, Lorenz S, Meyer A, et al. Peripheral endothelial dysfunction in myalgic encephalomyelitis/chronic fatigue syndrome. ESC Hear Fail 2020;7:1064–71. [CrossRef]
- Blauensteiner J, Bertinat R, León LE, Riederer M, Sepúlveda N, Westermeier F. Altered endothelial dysfunction-related miRs in plasma from ME/CFS patients. Sci Rep 2021;11:10604. [CrossRef]
- Wang J, Jelcic I, Mühlenbruch L, Haunerdinger V, Toussaint NC, Zhao Y, et al. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 2020;183:1264-1281.e20. [CrossRef]
- Pradeu T, Cooper EL. The danger theory: 20 years later. Front Immunol 2012;3. [CrossRef]
- Gallucci S, Matzinger P. Danger signals: SOS to the immune system. Curr Opin Immunol 2001;13:114–9. [CrossRef]
- Cohen IR, Young DB. Autoimmunity, microbial immunity and the immunological homunculus. Immunol Today 1991;12:105–10. [CrossRef]
- Quintana FJ, Mimran A, Carmi P, Mor F, Cohen IR. HSP60 as a target of anti-ergotypic regulatory T cells. PLoS One 2008;3. [CrossRef]
- Jammes Y, Steinberg JG, Delliaux S. Chronic fatigue syndrome: acute infection and history of physical activity affect resting levels and response to exercise of plasma oxidant/antioxidant status and heat shock proteins. J Intern Med 2012;272:74–84. [CrossRef]
- Thambirajah AA, Sleigh K, Stiver HG, Chow AW. Differential heat shock protein responses to strenuous standardized exercise in chronic fatigue syndrome patients and matched healthy controls. Clin Investig Med 2008;31. [CrossRef]
- Clayton, EW. Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: An IOM Report on Redefining an Illness. JAMA 2015;313:1101–2. [CrossRef]
- Elfaitouri A, Herrmann B, Bölin-Wiener A, Wang Y, Gottfries CG, Zachrisson O, et al. Epitopes of microbial and human heat shock protein 60 and their recognition in myalgic encephalomyelitis. PLoS One 2013;8. [CrossRef]
- Delneste Y, Herbault N, Galea B, Magistrelli G, Bazin I, Bonnefoy J-Y, et al. Vasoactive Intestinal Peptide Synergizes with TNF-α in Inducing Human Dendritic Cell Maturation. J Immunol 1999;163:3071–5. [CrossRef]
- Gomariz RP, Juarranz Y, Abad C, Arranz A, Leceta J, Martinez C. VIP-PACAP system in immunity: new insights for multitarget therapy. Ann N Y Acad Sci 2006;1070:51–74. [CrossRef]
- Staines, DR. Is chronic fatigue syndrome an autoimmune disorder of endogenous neuropeptides, exogenous infection and molecular mimicry? Med Hypotheses 2004;62:646–52. [CrossRef]
- Brenu EW, van Driel ML, Staines DR, Ashton KJ, Ramos SB, Keane J, et al. Immunological abnormalities as potential biomarkers in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J Transl Med 2011;9. [CrossRef]
- Gonzalez-Rey E, Delgado M. Vasoactive intestinal peptide and regulatory T-cell induction: a new mechanism and therapeutic potential for immune homeostasis. Trends Mol Med 2007;13:241–51. [CrossRef]
- Blundell S, Ray KK, Buckland M, White PD. Chronic fatigue syndrome and circulating cytokines: A systematic review. Brain Behav Immun 2015;50:186–95. [CrossRef]
- Carlo-Stella N, Badulli C, De Silvestri A, Bazzichi L, Martinetti M, Lorusso L, et al. A first study of cytokine genomic polymorphisms in CFS: Positive association of TNF-857 and IFNγ874 rare alleles. Clin Exp Rheumatol 2006;24:179–82.
- Steiner S, Becker SC, Hartwig J, Sotzny F, Lorenz S, Bauer S, et al. Autoimmunity-Related Risk Variants in PTPN22 and CTLA4 Are Associated With ME/CFS With Infectious Onset. Front Immunol 2020;11:578. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




