1. Summary
The distribution of the typical bacterial redox enzyme pyruvate-ferredoxin oxidoreductase (PFOR) in eukaryotes remains interestingly puzzling. Previous studies have demonstrated diverse cellular localizations of PFOR in some amitochondriate anaerobic protozoa such as Trichomonas vaginalis, Giardia duodenalis and Entamoeba histolytica (Dyall et al. 2004, Emelyanov et al. 2011). Interestingly, free-living mitochondriate protist-like photosynthetic algae also express homologs of PFOR. In Chlamydomonas reinhardtii, PFOR has been shown to localize in the chloroplastic stroma whereas that of Euglena gracilis localizes in the mitochondrion (Dubini et al. 2009, Noth et al. 2013, van Lis et al. 2013). However, PFOR is not expressed in photosynthetic plants.
PFOR is of particular pharmacological importance because it catalyzes the reductive bio-activation of nitro-based prodrugs to cytotoxic radical metabolites (Wyllie et al. 2013). Some nitro-based prodrugs may interfere with the primary function of PFOR, resulting in a drastic disruption of protein synthesis, growth, and overall survival of anaerobic and microaerophilic parasites, especially under unfavorable oxidative stress. This is the proposed mechanism of antimicrobial activity exerted by nitazoxanide against PFOR-possessing pathogens such as Cryptosporidium parvum (Hoffman et al. 2007, Scior et al. 2015, Shakya et al. 2018). The absence of PFOR in higher eukaryotes makes nitro-based prodrugs highly specific against invading pathogens without significant cytotoxic damage to host cells (Bendesky et al. 2002, Tweats et al. 2012). In bacteria, PFOR plays a similar role in the activation of nitro-based prodrugs (Narikawa 1986, Çelik et al. 2012, Martínez-Júlvez et al. 2012). Based on the functional similarity between the bacterial and protozoan forms of PFOR, an antiprotozoal nitro-based prodrug that undergoes similar activation mechanism can be used as an anti-infective agent in the treatment of some bacterial infections (Hof 1989, Freeman et al. 1997, Sisson et al. 2002). For example, metronidazole was developed in the 1950s originally as an antiprotozoal agent for the treatment of genito-urinary tract infections caused by the sexually transmitted protozoan parasite T. vaginalis (Bendesky et al. 2002, Kusdian 2014). Its antimicrobial spectrum was subsequently expanded as an antibacterial agent, as used routinely in the eradication therapy against Helicobacter pylori-induced peptic ulcer disease (Martinez et al, 2001). Mutations resulting in a substantial decrease or complete absence of PFOR culminate in inherent resistance of T. vaginalis and H. pylori to metronidazole (Hrdý et al. 2005, Martínez-Júlvez et al. 2012, Bradic et al. 2017, Graves et al. 2020).
Gene organization and genome sequencing techniques performed in previous studies suggest that some protozoa possess genes for encoding metabolic enzymes that trace back to common facultative anaerobic eubacterial ancestors (Hrdý et al. 1995, Rosenthal et al. 1997, Rotte et al. 2001, Nixon et al. 2002, Hug et al. 2010, Müller 2019). By applying comparative bioinformatic approaches, this study aims to analyze the distribution of PFOR in eukaryotes, with special focus on pathogenic protozoa. Furthermore, the study seeks to demonstrate that horizontal gene transfer (HGT) is the most likely mechanism by which some protozoan species acquired these genes from bacteria.
2. Background
Based on the presence or absence of mitochondria, protozoa can be generally classified as either mitochondriate or amitochondriate organisms. The oxygen-sensitive enzyme PFOR, which is responsible for the anaerobic fermentative decarboxylation of pyruvate, has varying localizations in amitochondriate protozoa (Chabrière et al. 1999, Charon et al. 1999). Based on the localization of PFOR, amitochondriate protozoa are classified further into type 1 and type 2 protists (Hrdý et al. 1995, Muller 1998). In type 1 protists such as G. duodenalis, E. histolytica and C. parvum, PFOR is localized in the cytosol (Embley et al. 2003, Ctrnacta et al. 2006). The PFOR in type 2 protists is localized in the hydrogenosome, as observed in the trichomonads (Müller 1993, Muller 1998, Kulda 1999). The hydrogenosome is a double-membrane-bounded organelle (Benchimol et al. 1996, Benchimol 2009), which is postulated to have resulted either from an ancestral mitochondrial transformation with subsequent gene loss or from endosymbiosis of an anaerobic bacterium (Roger et al. 1996, Hrdy et al. 2004, Makiuchi et al. 2014, Lewis et al. 2020). The hydrogenosome is the powerhouse for the fermentative decarboxylation of pyruvate to produce hydrogen, carbon dioxide, and ATP molecules in the absence of molecular oxygen (Lindmark et al. 1973, Hackstein et al. 1999, Boxma et al. 2005). Unlike mitochondria, hydrogenosomes have no genetic material; they contain proteins that are encoded by nuclear genes, which are post-translationally translocated into the organelle (Johnson et al. 1993, Rada et al. 2011). Interestingly, the free-living, mitochondriate, photosynthetic, protist-like algae C. reinhardtii is able to switch from aerobic to anaerobic metabolism by upregulating the synthesis of PFOR, which localizes and functions in the chloroplastic stroma (Noth et al. 2013, van Lis et al. 2013). However, the N-terminus of the PFOR of the facultative anaerobic, photosynthetic protist E. gracilis localizes in the mitochondrion (Rotte et al. 2001, Ctrnacta et al. 2006). Some prokaryotes possess orthologs of these enzymes that function in a similar manner as observed in protozoa. Due to the bizarrely restricted occurrence, localization, and distribution of these enzymes in protozoa, potential bacterial origins have been proposed in previous studies (Hrdý et al. 1995, Horner et al. 1999, Rotte et al. 2001). One plausible mechanism to explain this phenomenon is by HGT. This study seeks to test the hypothesis that some ancestors of protozoa initially acquired these redox enzyme-encoding genes by multiple independent horizontal gene transfer events from bacteria.
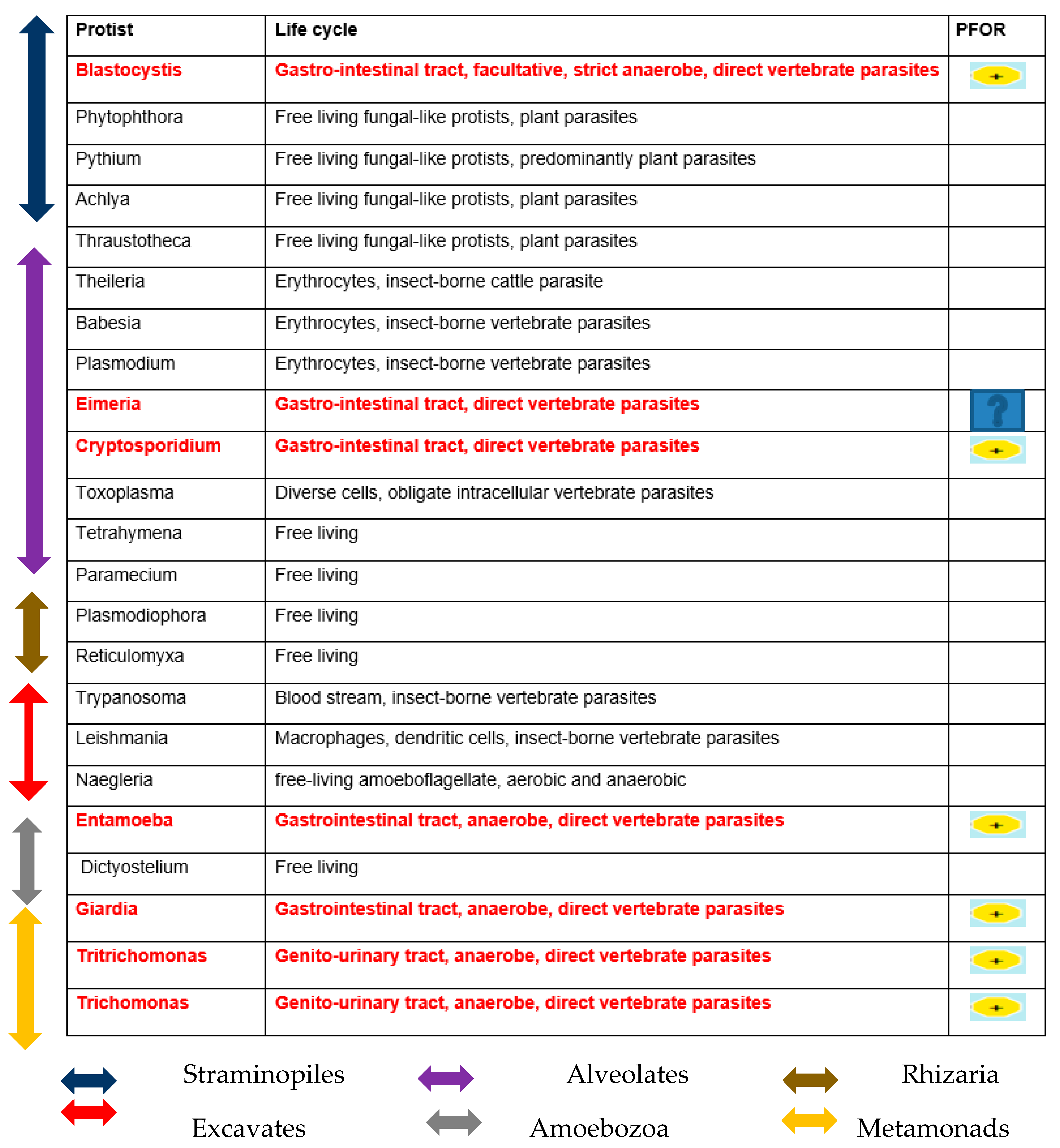
Table 1.
Diversity of the presence and cellular location of PFOR in protists.
Table 1.
Diversity of the presence and cellular location of PFOR in protists.
| Protist |
Localization of PFOR |
Source |
| Entamoeba histolytica |
Cytosol |
(Rodríguez et al. 1998) |
| Giardia duodenalis |
Cytosol |
(Emelyanov et al. 2011) |
| Cryptosporidium parvum |
Cytosol |
(Ctrnacta et al. 2006) |
| Blastocystis hominis |
Hydrogenosome |
(Wawrzyniak et al. 2008) |
| Trichomonas vaginalis |
Hydrogenosome |
(Hrdý et al. 1995) |
| Tritrichomonas foetus |
Hydrogenosome |
(Docampo et al. 1987) |
| Euglena gracilis |
Mitochondrion |
(Rotte et al. 2001) |
| Chlamydomonas reinhardtii |
Chloroplast |
(van Lis et al. 2013) |
Based on the cellular localization of PFOR E. histolytica, G. duodenalis and C. parvum are classified as type-1 protists whereas T. vaginalis, T. foetus and B. hominis are classified as type 2 protists.
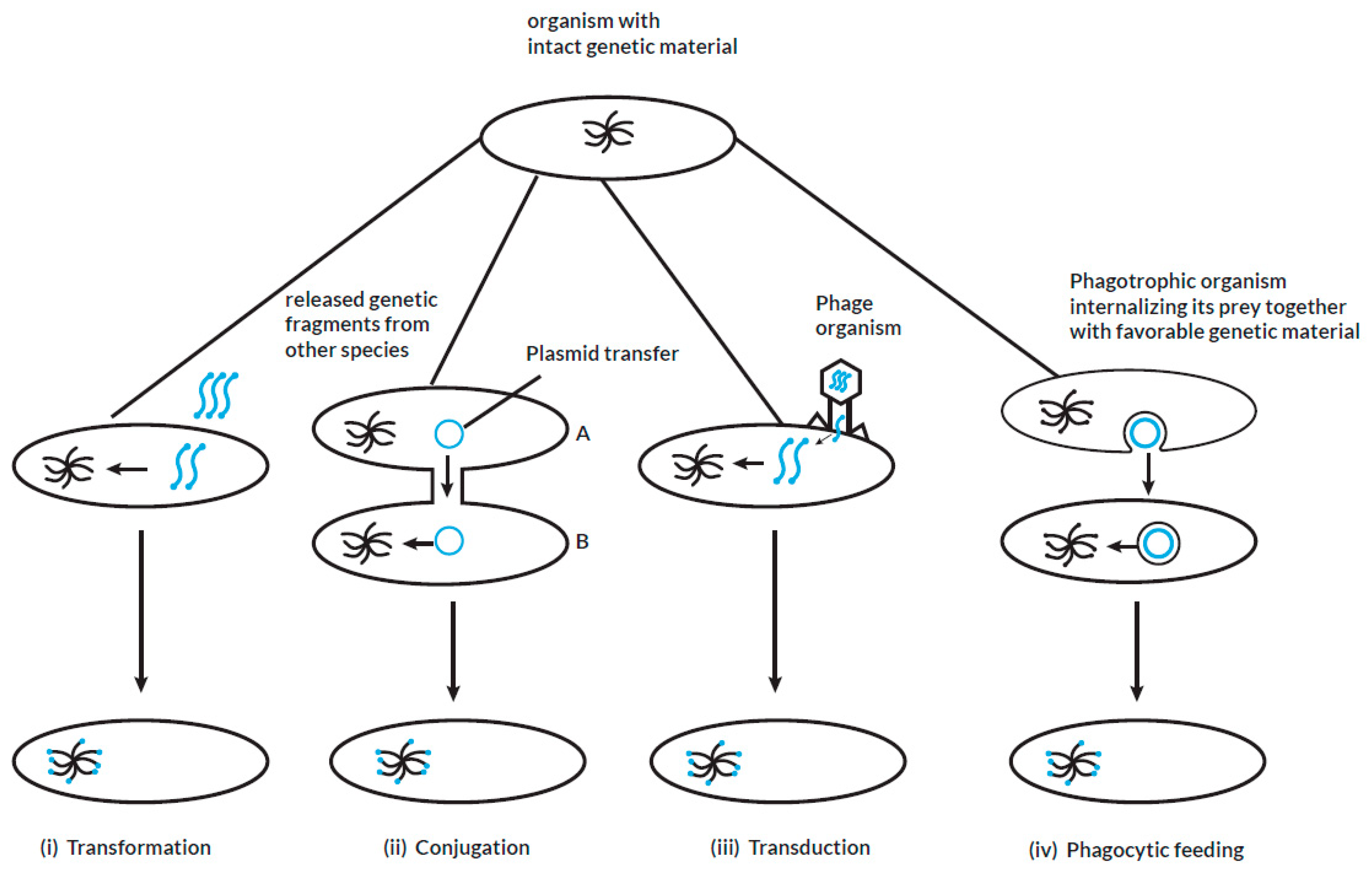
HGT, alternatively referred to as lateral gene transfer (LGT), is a term used in evolutionary genetics to explain the transfer of genetic material or genomic fragments between genetically related or unrelated organisms that do not share an inherent parent-to-offspring reproductive relationship (Soucy et al. 2015, Daubin et al. 2016). HGT is likely to occur through naturally occurring processes such as transformation or transfection, genetic conjugation and transduction: a detailed illustration of the different mechanisms of HGT is shown in
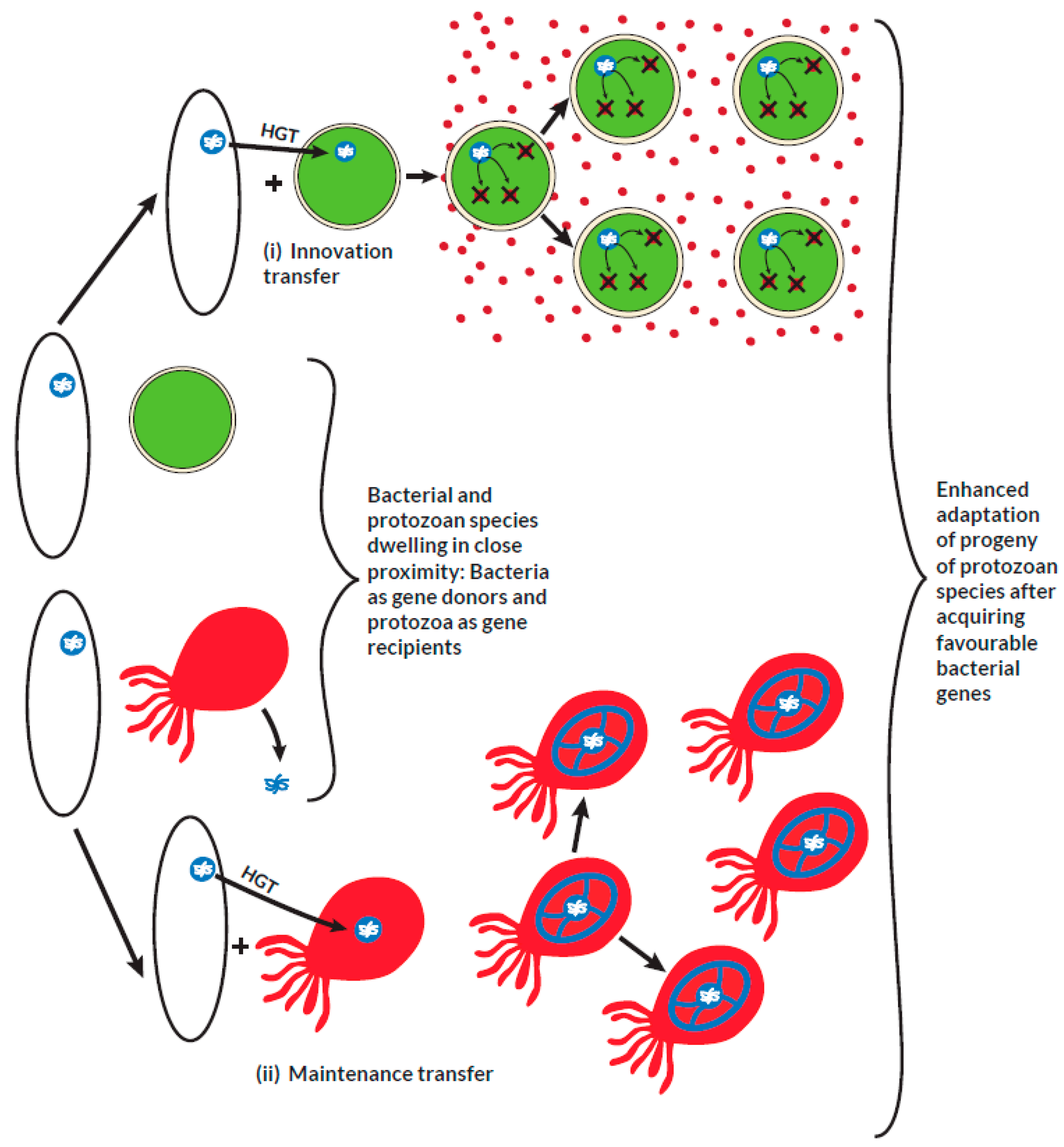
Figure 1. It may result in a substantial change in the genetic composition of the recipient organism or may be subsequently lost, leading to no overall significant genetic change. However, some HGT events may lead to the maintenance of pre-existing functions in the recipient organism that had been lost over the course of time in an evolutionary process termed as maintenance transfer (Husnik et al. 2018). The role of HGT in the acquisition of virulence and multiple drug resistance genes within prokaryotes is well established (Juhas 2015, Deng et al. 2019, Wyres et al. 2019). However, its involvement in the transfer of genetic material from prokaryotes to eukaryotes or within eukaryotes remains controversial (Keeling et al. 2008, Lacroix et al. 2016, Lacroix et al. 2018). Studies postulate that, HGT is an essential evolutionary mechanism by which unicellular eukaryotes acquire functional genetic material from bacteria that reside in close proximity such as the host gastrointestinal or genito-urinary tract (Ricard et al. 2006, Eme et al. 2017). It is also proposed that, some protozoan parasites residing in the same host environment with bacteria can receive foreign DNA fragments through phagocytic feeding on bacteria (Doolittle 1998). The acquisition of exogenous genetic material facilitates the adaptation, pathogenicity, and survival of protozoa in the face of strong selective pressures (Field et al. 2000, Hedges et al. 2001, Ricard et al. 2006, Keeling 2009, Schönknecht et al. 2013, Husnik et al. 2018, Sibbald, Eme et al. 2020, Van Etten et al. 2020). These protozoa may have acquired genes encoding bacterial redox enzymes via HGT to facilitate their survival in deteriorating environments (Nixon et al. 2002). Since protozoa have evolved to survive in oxidative and xenobiotic environments together with bacteria, these enzymes have remained central to their overall survival and were not lost after HGT (Gomaa et al. 2021). The proportion of bacteria-like genes proposed as potential candidates of HGT as apparent from whole genome sequencing of protozoa is summarized in table 2. A previous study that utilized conservative phylogenetic methods of the whole genome sequences of
E. histolytica and
T. vaginalis inferred 68 and 153 cases of HGT, respectively. Most of the transferred genes were found to encode enzymes involved in intermediary metabolism in prokaryotes, and the study concluded that prokaryotes that reside in close proximity to
E. histolytica and
T. vaginalis were potential gene donors (Alsmark, Sicheritz-Ponten et al. 2009).
During the process of transformation (i), free genetic fragments from other organisms that reside in close proximity with the recipient organisms are horizontally transferred. The foreign genetic material is incorporated into the genome of the recipient organisms. In conjugation (ii), the genetic material in the form of plasmids is transferred from a specific donor organism after close contact is established with the recipient organisms. The facilitator of a transduction process (iii) is a phage organism that directly transfers its genetic material by infecting the recipient organism. In all cases, the horizontally acquired genetic material is passed on vertically to the respective progeny. If the acquired genetic material enhances adaptation and fitness in the corresponding species, it is maintained over several generations in the lineage. In phagocytic feeding (IV), a phagotrophic organism obtains foreign genetic material by engulfing other species that reside in close proximity. The engulfed species are trapped in food vacuoles which then undergo digestion with subsequent internalization of the favorable foreign genetic material.
Prokaryotes dwelling in close proximity with some protists are potential sources of genes that encode proteins involved in important metabolic and adaptive functions required for growth and survival. In an innovation transfer, a protist that originally lacks such genes acquires them from a bacterial donor. From the example illustrated above in (i), a metabolically less endowed protist incorporates a bacterial gene responsible for expressing proteins involved in the detoxification of toxins. This serves as an adaptive advantage for the survival of the protist in an otherwise toxin-enriched environment that usually favors bacterial species. During the course of evolution, some vital genes that are lost may result in an adaptive disadvantage to the protist. In a maintenance transfer, as illustrated in (ii), a lost gene may be regained subsequently from bacterial sources to maintain the adaptation of the protist.
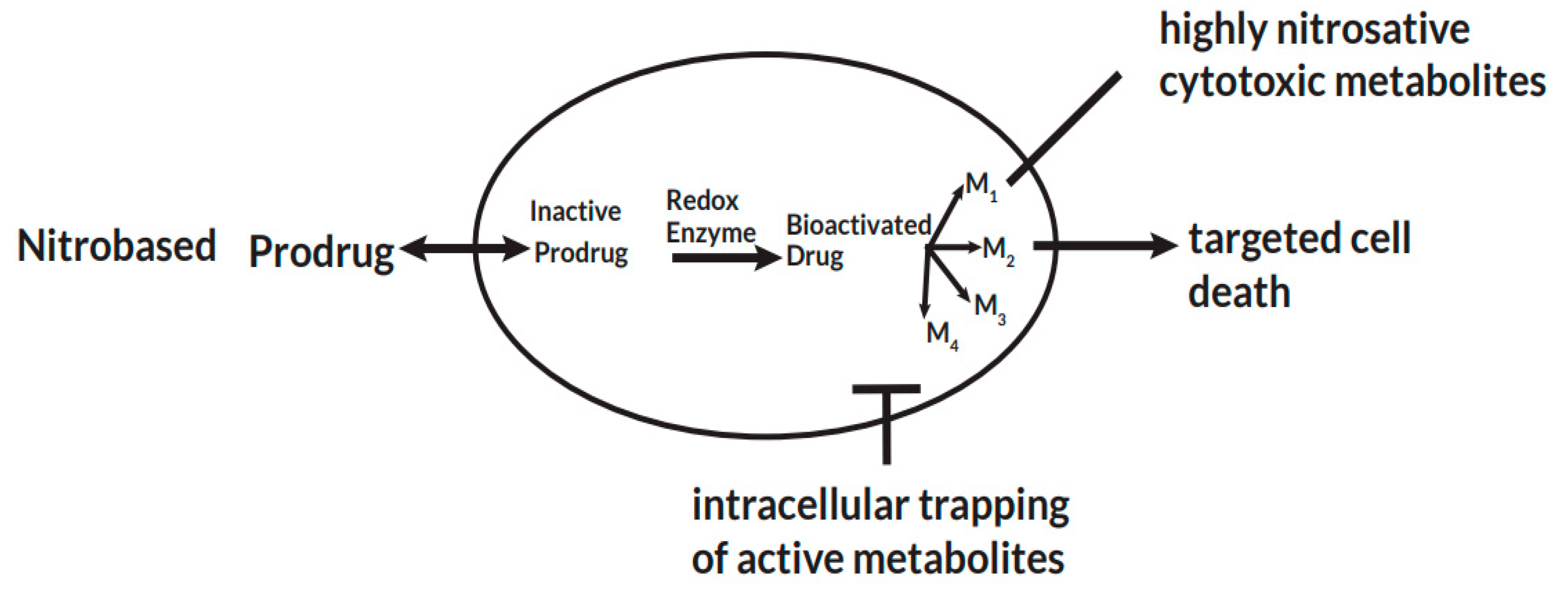
By targeting the pathogen-specific redox function of PFOR, some nitro-based antimicrobial prodrugs are bio-activated through electron transfer reactions intracellularly in invading pathogens. Substances containing one or more nitro groups, such as the nitro-heterocyclic and nitro-aromatic compounds, are used as prodrugs against these pathogens because they undergo bio-activation to form potent nitro radicals (Fleck et al. 2014). The anionic potential of the radical intermediates results in intracellular trapping and subsequent accumulation of more active metabolites. As a result, these metabolic radicals exert their cytotoxic effects selectively in invading pathogens with no significant effect on the host cells (Chung et al. 2011, Nepali et al. 2019).
Figure 3.
Pathogen specific bio-activation of nitro-based prodrugs.
Figure 3.
Pathogen specific bio-activation of nitro-based prodrugs.
After a prodrug is taken up intracellular by a pathogen, pathogen-specific redox enzymes catalyze electron transfer reactions that lead to the bio-activation of the prodrug. The activated drug undergoes a series of transformation reactions to radical cytotoxic metabolites that remain trapped intracellular and exert their cytotoxic effects. This leads to selective cell death by disrupting nucleic acid and protein synthesis of the pathogen.
Table 2.
Proportion of bacterial-like genes estimated from whole genome sequencing of some protozoan parasites.
Table 2.
Proportion of bacterial-like genes estimated from whole genome sequencing of some protozoan parasites.
| Protozoan parasite |
Total number of genes |
Percentage of bacterial-like genes |
Source |
| Plasmodium falciparum |
5,268 (212) |
4.02% |
(Gardner et al. 2002, Huang et al. 2004) |
| Blastocystis hominis |
6,020 (133) |
2.21% |
(Denoeud et al. 2011) |
| Entamoeba histolytica |
9,938 (96) |
0.97% |
(Loftus et al. 2005) |
| Toxoplasma gondii |
8,300 (72) |
0.87% |
(Huang et al. 2004, Bontell et al. 2009) |
| Trypanosoma brucei |
9,068 (47) |
0.52% |
(Berriman et al. 2005) |
| Cryptosporidium parvum |
5,519 (24) |
0.43% |
(Huang et al. 2004) |
| Dictyostelium discoideum |
13,522 (49) |
0.36% |
(Andersson 2011) |
| Naegleria gruberi |
15,727 (45) |
0.29% |
(Fritz-Laylin et al. 2010) |
| Tetrahymena thermophilia |
26,460 (74) |
0.28% |
(Xiong et al. 2015) |
| Trichomonas vaginalis |
59,681 (152) |
0.25% |
(Carlton et al. 2007) |
This table shows the varying proportion of bacterial-like genes in the whole genome sequencing of some protozoan species proposed as potential candidates for HGT from bacteria. Of all the protozoa of economic importance to humans that have been fully sequenced, P. falciparum seems to be the parasite with the highest proportion of bacterial-genes. The numbers in parenthesis are the absolute number of bacterial-like genes.
2.1. Pyruvate-ferredoxin oxidoreductase
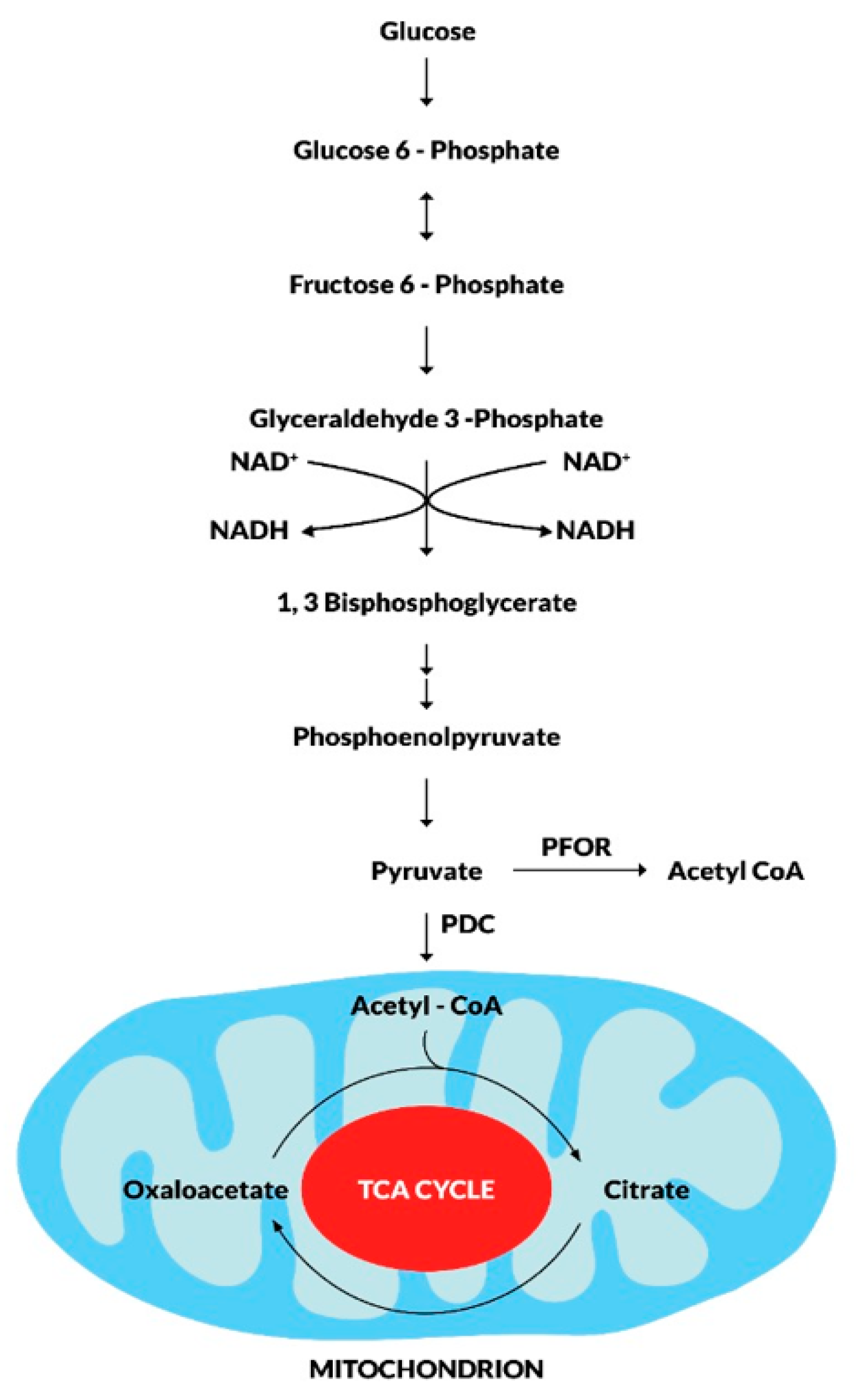
The ability to metabolize pyruvate through the process of oxidative decarboxylation remains a fundamental biochemical reaction in all living organisms (Inui et al. 1999). The pyruvate dehydrogenase multi-enzyme complex (PDC), an enzyme present in the mitochondria of almost all aerobic organisms, catalyzes this reaction (Hucho 1975). However, in most anaerobic and microaerophilic organisms, the enzyme PFOR, which is absent in higher eukaryotes, is responsible for catalyzing this reaction (Chabrière et al. 1999, Charon et al. 1999). PFOR is an ancient, oxygen sensitive, iron-sulfur protein that belongs to the 2-oxo acid ferredoxin oxidoreductase (OFOR) superfamily. It is found in diverse cellular locations in some anaerobic organisms (Emelyanov et al. 2011). Archaea, most anaerobic bacteria, and some anaerobic eukaryotes possess this enzyme (Blamey et al. 1993). The fermentative decarboxylation of pyruvate to acetyl-CoA and carbon dioxide (CO2) by PFOR generates electrons that are transferred to either ferredoxin or flavodoxin (Marczak et al. 1983, Moreno et al. 1985). Due to the reversibility of this reaction, whereby pyruvate will be regenerated, PFOR is also called pyruvate synthase.
According to previous studies, the innate function of PFOR is to protect anaerobic organisms against oxidative stress by catalyzing the transfer of electrons that eventually reduce and inactivate free oxygen radicals during unfavorable oxidative conditions (Raj, et al. 2014). In an alternative pathway, the reduced ferredoxin derived from the decarboxylation of pyruvate can be re-oxidized by ferredoxin: NAD oxidoreductase (FNOR). This involves the transfer of electrons from ferredoxin to NAD(P) (Ctrnacta et al. 2006). The resulting NAD(P)H then transfers the electrons to oxygen radicals by another reaction catalyzed by NAD(P)H oxidase to form water molecules (Mastronicola et al. 2016). In this way, PFOR contributes its redox function together with other enzymes as an antioxidant detoxification system.
Figure 4.
Metabolic fate of pyruvate in mitochondriate and some amitochondriate organisms.
Figure 4.
Metabolic fate of pyruvate in mitochondriate and some amitochondriate organisms.
During the fundamental metabolic pathway of glycolysis, glucose is converted to pyruvate along a series of phosphorylation and dehydrogenation reactions. The fate of pyruvate is mainly dependent on the presence of mitochondrion. In mitochondriate organisms, pyruvate is further dehydrogenated to acetyl CoA by the pyruvate dehydrogenase enzyme complex (PDC). Acetyl CoA enters the tricarboxylic acid cycle as the primary substrate for oxidative reactions that result in the production of CO2 and ATP molecules for intermediary metabolism. In some amitochondriate organisms, however, pyruvate undergoes fermentative decarboxylation to produce Acetyl CoA in a reversible redox reaction catalyzed by the pyruvate-ferredoxin oxidoreductase (PFOR).
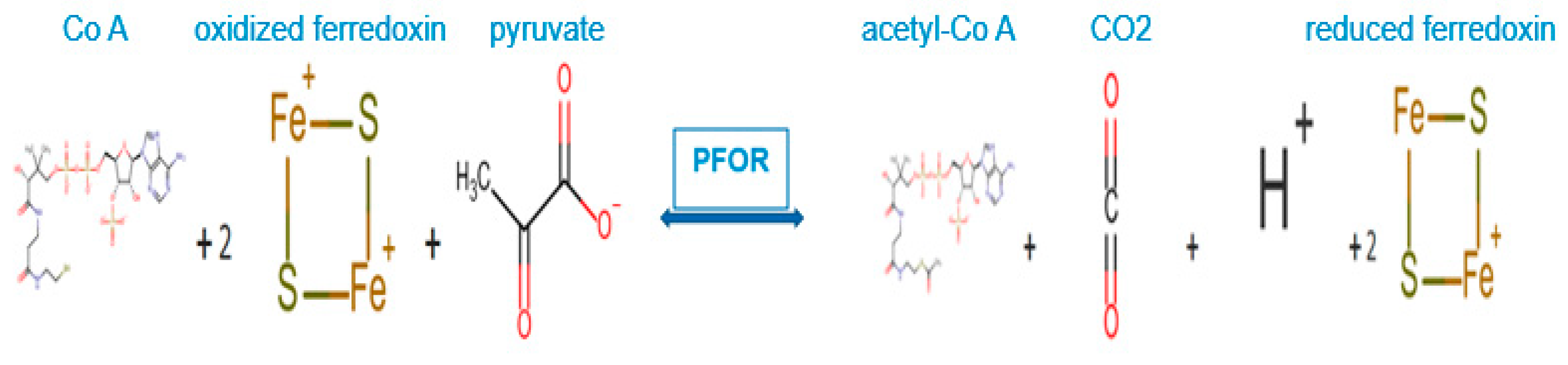
Figure 5.
PFOR-mediated anaerobic metabolism of pyruvate.
Figure 5.
PFOR-mediated anaerobic metabolism of pyruvate.
In the fermentative decarboxylation of pyruvate, PFOR catalyzes the reduction of two molecules of ferredoxin using Co-enzyme A as a co-factor to produce acetyl-CoA, carbon dioxide and hydrogen ions. The reduced ferredoxin can be utilized as an electron donor for further redox reactions.
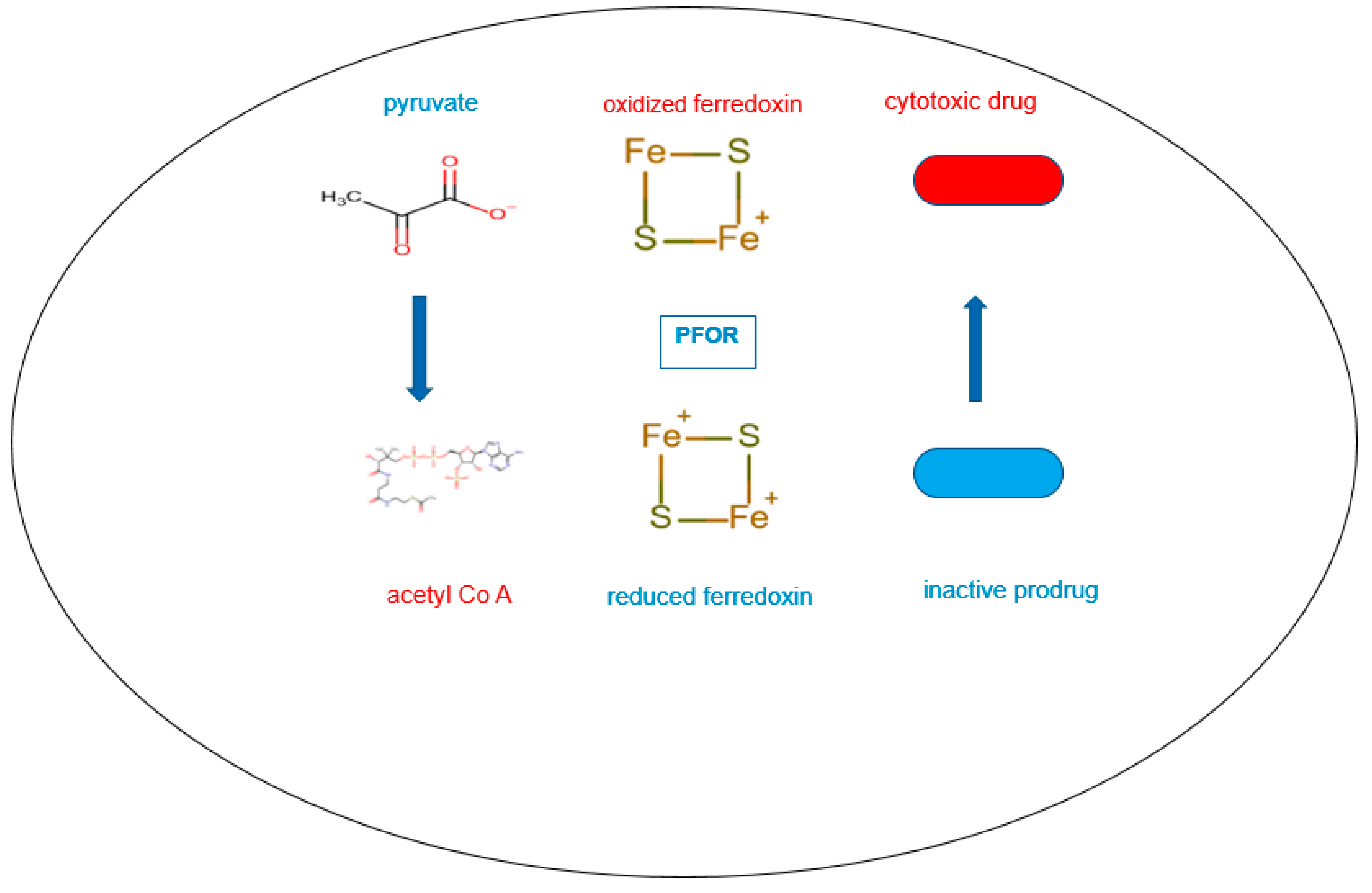
In the presence of nitro-based prodrugs, this same redox machinery is used as a drug target to make pathogens vulnerable to nitro-radical-induced cytotoxic damage. In this instance, the resulting reduced ferredoxin serves as an electron donor by transferring electrons to nitro-prodrugs. These then become pharmacologically activated and subsequently transformed into nitroso and hydroxylamine derivatives that cause cytotoxic damage (Horner et al. 1999, Ragsdale 2003).
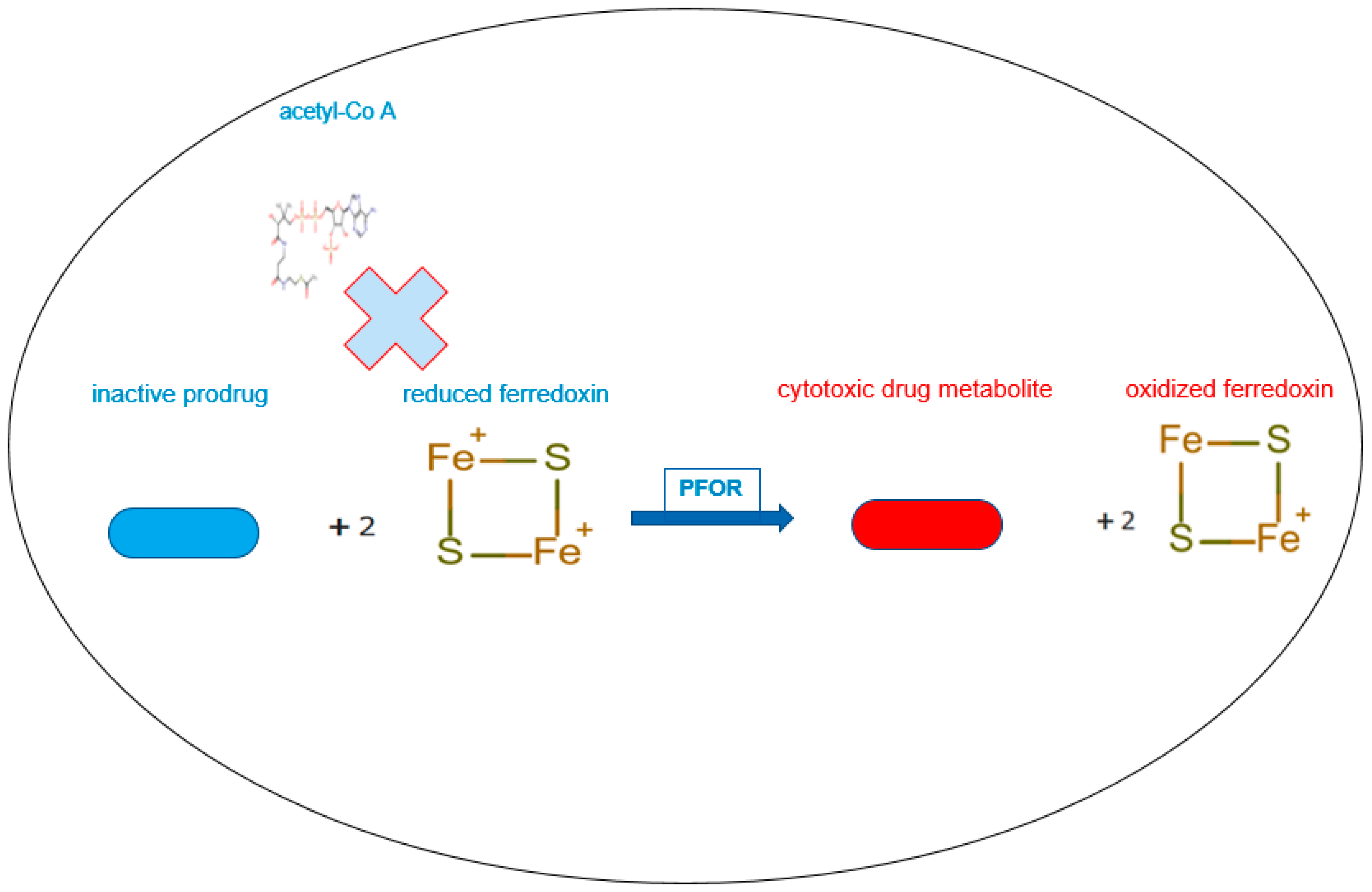
Figure 6.
PFOR-mediated re-oxidation of ferredoxin and bio-activation of nitro-based prodrugs.
Figure 6.
PFOR-mediated re-oxidation of ferredoxin and bio-activation of nitro-based prodrugs.
In the presence of sufficient concentrations of the inactive prodrug, reduced ferredoxin donates electrons to the prodrug instead of acetyl-Co A by the catalytic activity of PFOR. In this reverse redox reaction, ferredoxin is re-oxidized and the prodrug is activated to its cytotoxic radical intermediate.
Figure 7.
A summary of the PFOR-mediated fermentative decarboxylation of pyruvate and reductive activation of nitro-based prodrugs.
Figure 7.
A summary of the PFOR-mediated fermentative decarboxylation of pyruvate and reductive activation of nitro-based prodrugs.
This figure depicts the reversible redox reaction catalyzed by PFOR in the presence of very high concentrations of the inactive prodrug. In the forward reaction, PFOR catalyzes the decarboxylation of pyruvate to acetyl-Co A using oxidized ferredoxin as electron acceptor. The resulting reduced ferredoxin then donates electrons to the inactive prodrug to reductively bio-activate it. This cycle continues until the concentration of the radical intermediates of the prodrug reaches therapeutic levels within the pathogen to disrupt nucleic acid and protein synthesis.
2.1.1. Activation of metronidazole by the PFOR of G. duodenalis and E. histolytica
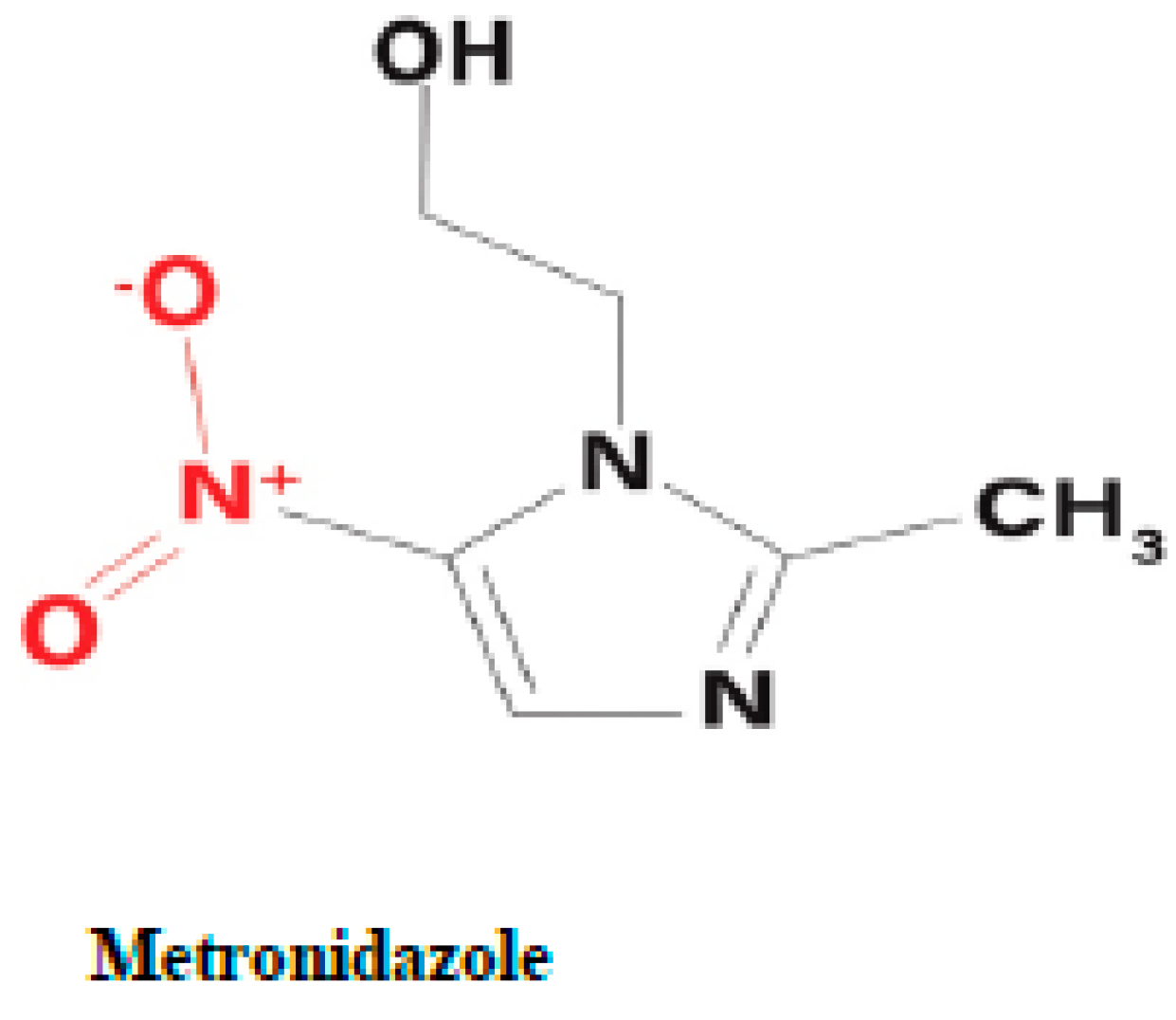
Metronidazole is a synthetic nitro-imidazole derivative effective against most obligate anaerobic bacteria and protozoan parasites such as
G. duodenalis,
E. histolytica,
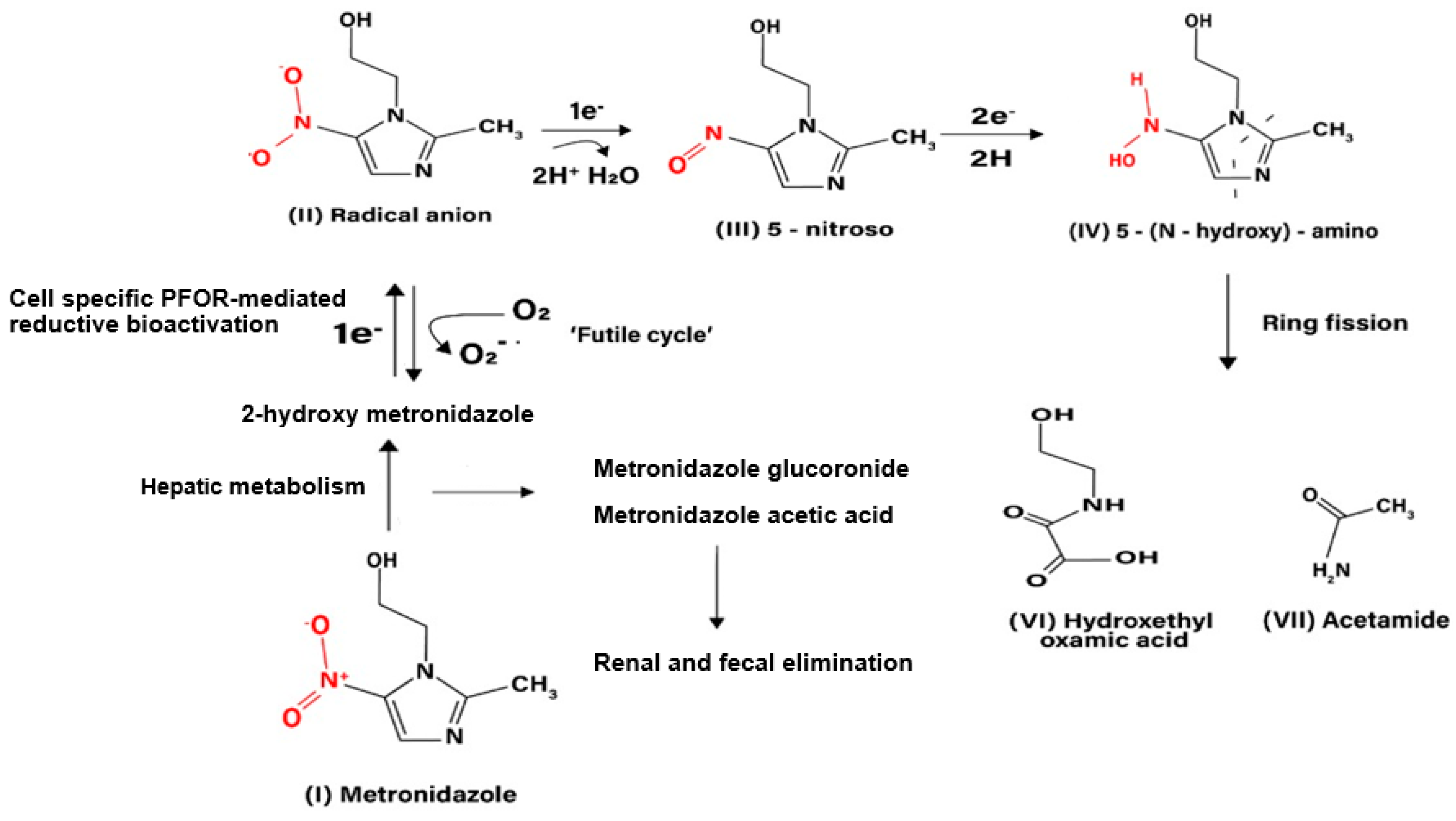
T. vaginalis (Upcroft et al. 2001, Pal et al. 2009). It exerts its antimicrobial effect by interfering with nucleic acid synthesis resulting in DNA damage in the target pathogens (Lamp et al. 1999). Electrons from the initial redox reaction catalyzed by PFOR are transferred to ferredoxin, which has a sufficiently low redox potential to reductively activate metronidazole to its toxic radical metabolites. Through a series of electron transfer reactions, different radical intermediates are formed that act primarily as alkylating agents that disrupt DNA synthesis and growth of invading pathogens (Dingsdag et al. 2018). The different radical metabolites of metronidazole have been illustrated in
Figure 8 below. Strains of
G. duodenalis and
E. histolytica with decreased or absent expression of
pfor have been shown to exhibit increased metronidazole resistance (Dan et al. 2000). However, some recent studies suggest the involvement of alternative enzymes that also play central roles in the activation of, and resistance to, metronidazole (Leitsch et al. 2011, Ansell et al. 2017, Müller et al. 2019, Lopes-Oliveira et al. 2020). For example,
T. vaginalis has been shown in some studies to display a complete resistance to metronidazole only after both PFOR and NAD-dependent malic enzyme are inactivated in the hydrogenosome (Rasoloson et al. 2002, Hrdý, Cammack et al. 2005).
The initial step in the bio-activation of metronidozole involves the transfer of one electron from ferredoxin to metronidazole to form its radical anion. In the presence of molecular oxygen the radical anion is re-oxidized to the parent compound in a futile cycle, hence, the overall bio-activation reaction is stalled at this stage. In anaerobic conditions, the radical anion recieves a further electron to form the 5-nitroso intermediate. In a subsequent reduction reaction involving the transfer of two electrons, the 5-nitroso intermediate is converted to a 5-(N–hydroxy)-amino intermediate, which then undergoes a spotaneous ring fision to produce hydroxethyl oxamic acid and acetamide. These fission products are the proposed end products of the bioactivation process that contribute immensely to the toxicity of metronidazole in invading pathogens.
2.1.2. Interaction between nitazoxanide and the PFOR of C. parvum
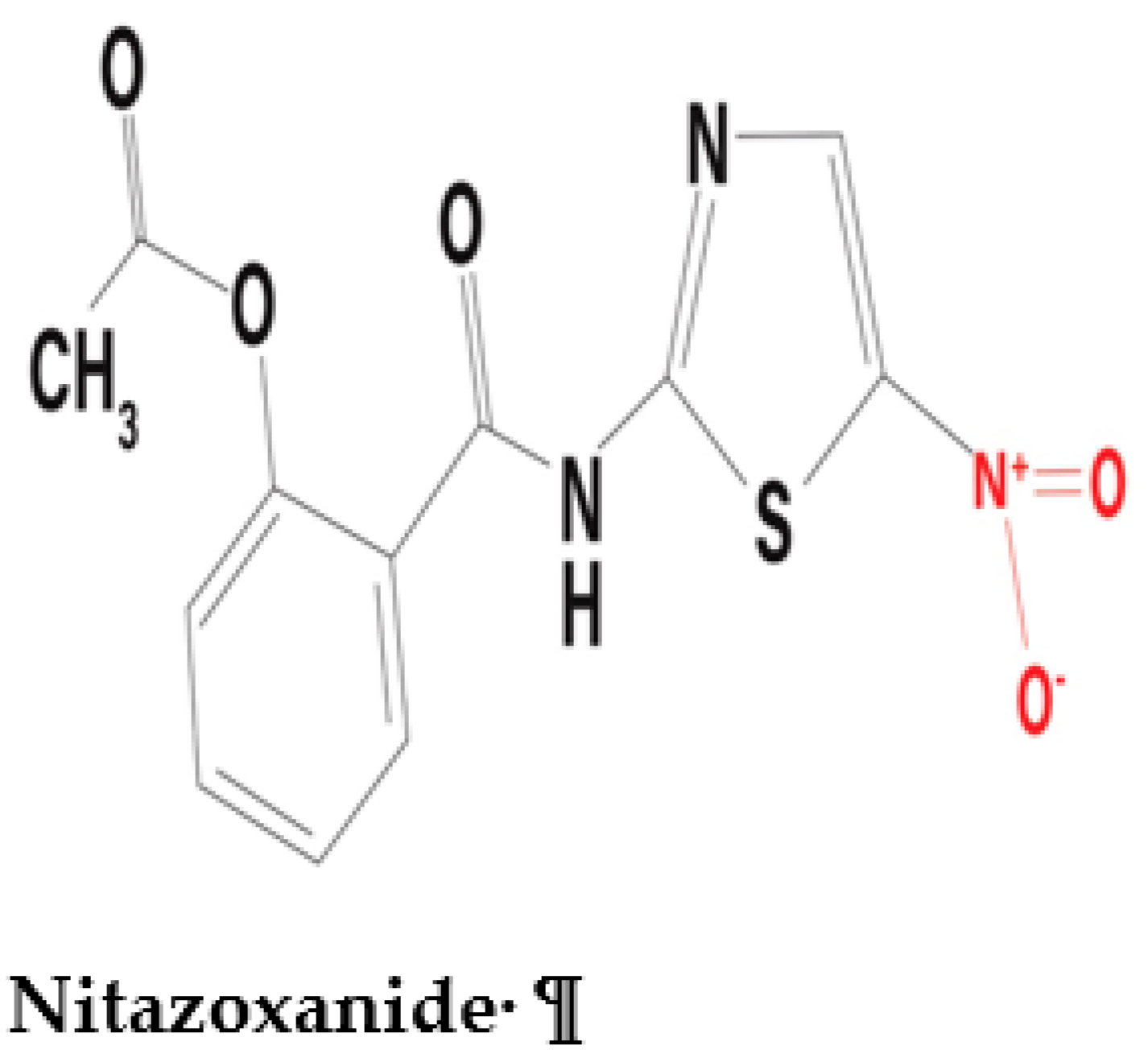
Nitazoxanide is a nitrothiazolyl-salicylamide derivative which exhibits broad-spectrum antimicrobial activity against a variety of pathogens including anaerobic and microaerophilic bacteria, protozoa, helminths, and viruses (Shakya et al. 2018, Bharti et al. 2021). Recent preclinical studies have demonstrated potential antineoplastic activity of nitazoxanide that can be further explored in the field of oncology (Pal et al. 2020, Abd El-Fadeal et al. 2021, Kiehl et al. 2021). The antiviral activity of nitazoxanide against the SARS-CoV-2 infection is currently being explored (Meneses et al. 2020, Lokhande et al. 2021, Rocco et al. 2021). Interestingly, nitazoxanide interacts with PFOR in a completely different manner than metronidazole. Some in-vitro studies postulate an interference of the redox function of PFOR by nitazoxanide (Hoffman et al. 2007, Scior et al. 2015). It deactivates the oxidative decarboxylation of pyruvate along with the electron transfer machinery that produces reduced ferredoxin in the presence of the coenzyme thiamine pyrophosphate (TPP) (Hoffman et al. 2007). This is the generally accepted mechanism by which it inactivates the metabolism, growth and survival of C. parvum (Gargala 2008). A similar mechanism of action is postulated for the interaction between the PFOR of some anaerobic bacteria and nitazoxanide (Warren et al. 2012, Kennedy et al. 2016).
4. Aim
To investigate the distribution and evolutionary relationships of genes encoding redox enzymes involved in the activation of nitro-based prodrugs in protozoan and bacterial species.
5. Objectives
To analyze the phylogenetic distribution of redox enzymes of selected protozoan parasites using proteins encoded by housekeeping genes as controls.
To use comparative bioinformatics approaches to test the hypothesis that these redox enzymes were most likely acquired through horizontal gene transfer from bacterial origin.
To use proteome-wide analysis and gene expression analysis to identify other genes that were putatively acquired through horizontal gene transfer.
6. Expected Outcomes
Since the control proteins are encoded by housekeeping genes, their distribution along the taxonomic tree is expected to be in consonance with the phylogeny of eukaryotes depicted by Adl SM (Adl et al. 2012) and the phylogenomics-based reconstruction of the protozoan species tree by Ocaña KA (Ocaña et al. 2011). A significant phylogenetic incongruence observed in the protein sequences of the redox enzymes could suggest that pfor was acquired through HGT. Distinct monophyletic relationships inferred from redox proteins between bacteria and protozoa would be highly suggestive of HGT events from bacteria. Through comparative analysis of the proteomes of closely related protozoa that do not possess the redox enzymes and putative bacterial gene donors, further information on other genes that putatively share similar pattern of horizontal acquisition with pfor will be obtained. By applying gene enrichment analysis of proteins exclusively present in protozoa and bacteria, the functional relationship of these genes to pfor will be ascertained.
7. Methodology
7.1. Source of protein sequences
Protein sequences for the PFOR of the selected protozoan parasites with their respective gene and accession numbers were retrieved from the UniProt™ protein database. (
https://www.uniprot.org/uniprot/). Three control proteins of each parasite: glyceraldehayde-3-phosphate dehydrogenase (GAPDH) (EC 1.2.1.12), tubulin alpha chain (TUBa), and DNA-directed RNA polymerase II subunit beta I (RPB1) (EC 2.7.7.6) were retrieved using the same procedure. These proteins were selected because they are encoded by housekeeping genes and are well conserved in protozoa.
Table 3.
Gene and accession numbers of the redox enzymes.
Table 3.
Gene and accession numbers of the redox enzymes.
| Parasite |
Enzyme |
Gene number |
Accession number |
Redox drug |
| G. duodenalis |
PFOR |
Q24982 |
AAA74894.1 |
Metronidazole |
| E. histolytica |
PFOR |
N9TJX2 |
ENY63830.1 |
Metronidazole |
| C. parvum |
PFOR |
A1DRF1 |
ABK91849.1 |
Nitazoxanide |
Table showing the gene and accession numbers of the redox enzymes with the corresponding pro-drug substrates of selected protozoan parasites that were obtained from the UniProt™ Protein Database
Table 4.
Gene and accession numbers of the control proteins.
Table 4.
Gene and accession numbers of the control proteins.
| Parasite |
Control protein1 |
Gene/Accession number |
Control protein 2 |
Gene/Accession number |
Control protein 3 |
Gene/Accession number |
| G. duodenalis |
GAPDH |
P53429 AAB18421.1 |
TUB |
A8BPC0EDO78046.1 |
RPB1 |
Q8MUU2 AAM77743.1 |
| E. histolytica |
GAPDH |
C4LVR9BAN37492.1 |
TUB |
P31017AAA57315.1 |
RPB1 |
Q6IUR3 AAT40981.1 |
| C. parvum |
GAPDH |
Q7YYQ9BAJ77164.1 |
TUB |
Q9UAC3AAD20239.1 |
RPB1 |
A0A7G2HJ78 CAD98371.1 |
Table showing the gene and accession numbers of the control proteins of selected protozoan parasites that were obtained from the UniProt™ protein database.
7.2. Sequence alignment
Sequence similarity queries were performed using the proteins of interest against the NCBI non-redundant protein sequence database with the basic local alignment search tool program (BLASTP version 2.12.0+) from the National Center for Biotechnology Information (NCBI) website (
https://blast.ncbi.nlm.nih.gov/Blast) (Altschul et al. 1997). Initially, the queries were performed using the control proteins of each parasite. The same procedure was performed for the corresponding redox enzymes using a “BLAST pyramid” approach with the following algorithm parameters. Expected threshold: 1e
-20 , Word size: 6, Matrix: BLOSUM62, Gap Costs: Existence 11, Extension 1 and Compositional adjustments: Conditional compositional score matrix adjustments (Altschul et al. 2005).
7.3. Inclusion-Exclusion approach of BLAST search
The total number of distinct species that possess the control proteins and the corresponding redox enzymes were derived from the taxonomy reports of the BLAST search results. In the BLAST pyramid approach, the first BLAST search was performed by excluding the protein sequence of the protozoan species of interest whilst including the sequences of all other species belonging to its genus. In the next query, the genus was excluded whilst including the next available taxonomic level. This was followed by subsequent inclusion-exclusion queries in increasing order along the taxonomic hierarchy until the eukaryotic domain. In a final step, all eukaryotes were excluded whilst including all prokaryotes. The proportion of organisms in each taxon that had hits for the redox enzymes was represented as percentage deviation. Since the total number of distinct species in each taxon was not available in the NCBI database, the percentage deviation was determined by comparing the total number of species that had hits for the redox enzymes to the total number of organisms that had hits for GAPDH.
Table 5.
Illustration of BLAST search pyramid approach.
Table 5.
Illustration of BLAST search pyramid approach.
 |
Eukaryota |
Procaryotae |
| Alveolata |
Eukaryota |
| Apicomplexa |
Alveolata |
| Conoidasida |
Apicomplexa |
| Eucoccidiorida |
Conoidasida |
| Cryptosporidiidae |
Eucoccidiorida |
| Cryptosporidium |
Cryptosporidiidae |
| Cryptosporidium parvum |
Cryptosporidium |
| EXCLUDE |
INCLUDE |
This table illustrates how the BLAST searches are run using C. parvum protein sequences as query. In the first step, a BLAST search is done with hits excluding the species C. parvum while including all other species of the genus Cryptosporidium. In the next step, all species of the genus Cryptosporidium are excluded whilst including organisms of the next taxonomical hierarchy. This is done sequentially until all eukaryotic organisms are excluded to filter hits for prokaryotes only.
7.4. Phylogenetic analyses
Protein sequences of the PFOR and control proteins of representative species from corresponding eukaryotic taxa and bacterial species were manually selected after sequence similarity searches using the same parameters as for BLASTP. The sequences were downloaded from the NCBI website in fasta format. The fasta files were transferred to Notepad++ to simplify the names of the protein sequences using regular expressions. Multiple sequence alignments were performed with the Molecular Evolutionary Genetics Analysis (MEGA) version X software using the Muscle algorithm with default parameters (Kumar et al. 2018). The evolutionary distances were computed using the Poisson correction method. This was followed by trimming the ends of the alignments. The protein sequence alignments were then exported in Mega format to construct the phylogenetic trees using the neighbor-joining algorithm (Saitou et al. 1987) and the Poisson substitution model. The bootstrap approach of confidence limits on phylogenies was implemented with 1,000 replications for both algorithms (Felsenstein 1985).
7.5. Screening selected proteomes against HMM profile libraries
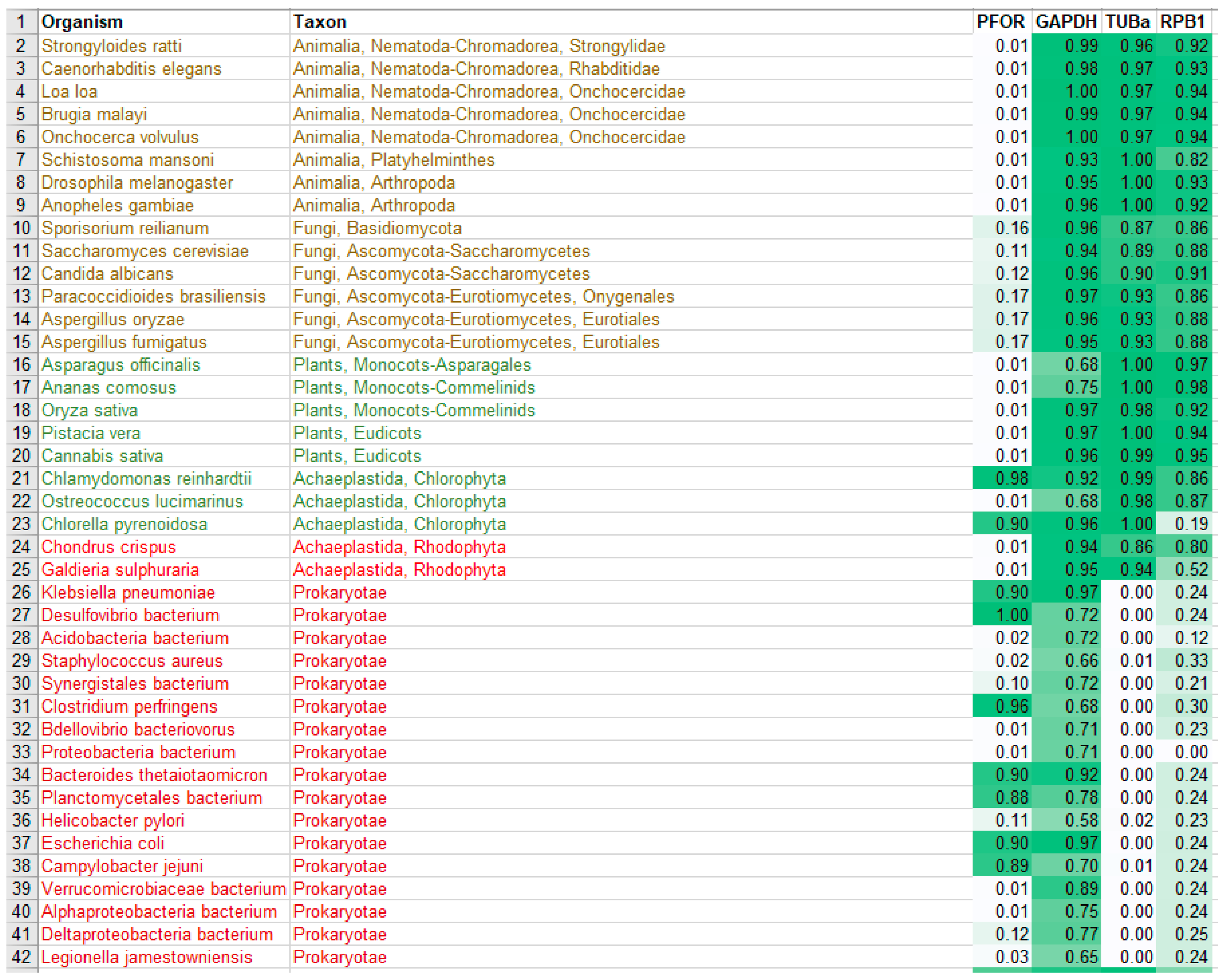
All the steps were executed with self-made Perl scripts on a linux computer. The multiple sequence alignment of each of the five proteins in fasta format from MEGA were converted into a position-dependent scoring matrix using the command hmmbuild of the HMMer package (Eddy 2009, Eddy 2011). The resulting profiles were concatenated and converted to a HMM profile libraries using the command hmmpress. Complete proteomes of representative organisms from each eukaryotic taxon and from selected bacterial organisms were downloaded in fasta format from the ensemblgenomes database (ftp://ftp.ensemblgenomes.org/pub/). The HMM profile libraries were used to screen the downloaded proteomes with hmmscan of the HMMer package. The number of hits per profile against a given proteome was derived using a cutoff E-value of < 1e-20. The highest scores obtained were used to construct a heat map on an Excel sheet to illustrate the distribution of the redox enzymes amongst the protozoan and bacterial species.
7.6. Distribution of redox enzymes and control proteins in cladograms and density maps
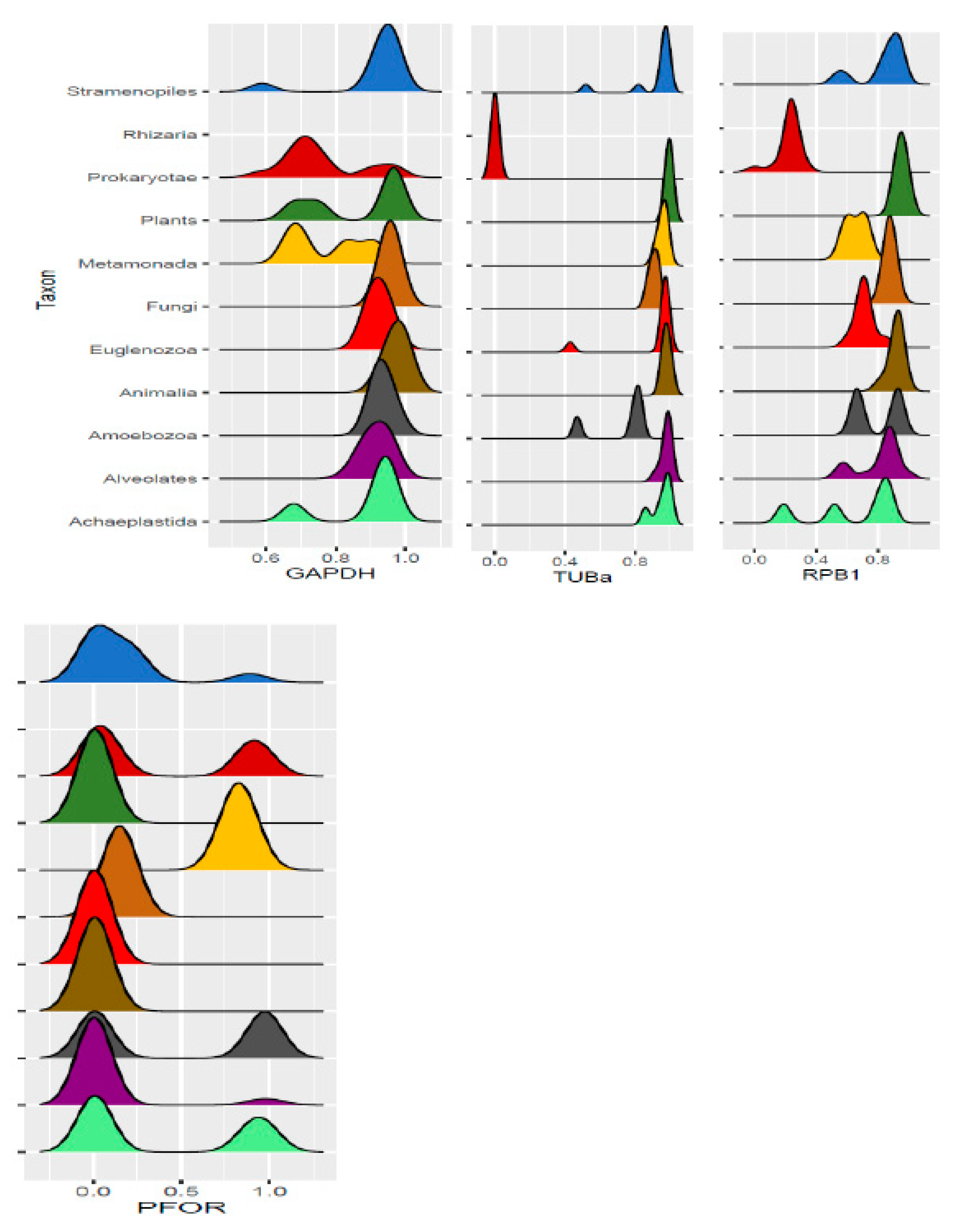
Eukaryotic species with whole proteome sequences were grouped by their respective phylogenetic clades and the presence or absence of PFOR and the control proteins was analyzed by normalizing the scores obtained from the screening of the HMM profile libraries. The clades of the protozoa of interest were further expanded by including all available species in their respective genus and other closely related species with whole proteomes in the ensemblgenomes database. In an alternative analysis, all scores from the HMM screening were transferred to R studio to generate density plots. In the density plots, the relative distribution of the redox enzymes and control proteins in each eukaryotic taxon was represented.
7.7. Identification of other genes that were putatively acquired through HGT
Whole proteome BLAST searches using the proteomes of the protozoan species of interest as the query were performed against the proteomes of closely related protozoan species as identified based on the cladograms. The results were compared to the proteome BLAST against Desulfovibrio vulgaris. This bacterium was selected as a putative gene donor because (i) it had the highest similarity score for PFOR from the whole proteome screening and (ii) it resides in the gastrointestinal tract of mammals as well as insects (Hong et al. 2021, Peydaei et al. 2021). All proteins that had high similarity scores in D. vulgaris but not to the closely related protozoan control species were selected for gene enrichment analysis.
7.8. Gene enrichment analysis
The gene IDs of the proteins that were exclusively present in the selected protozoan species and
D. vulgaris were transferred to the Gene Ontology (GO) website
(http://geneontology.org/) for gene enrichment analysis. The analyses were performed using the PANTHER Overrepresentation Test (released on 13-10-2022) and the GO database DOI: 10.5281/zenodo.6799722 (PANTHER version 17.0, released on 01-07-2022). The PANTHER Pathway for biological process was used for the annotation data set and Fisher's exact test with false discovery rate (FDR) correction was selected for the statistical analysis. Only results showing statistically significant FDR values and p-values < 0.05 with corresponding fold enrichment were evaluated.
8. Results
8.1. BLAST Pyramid
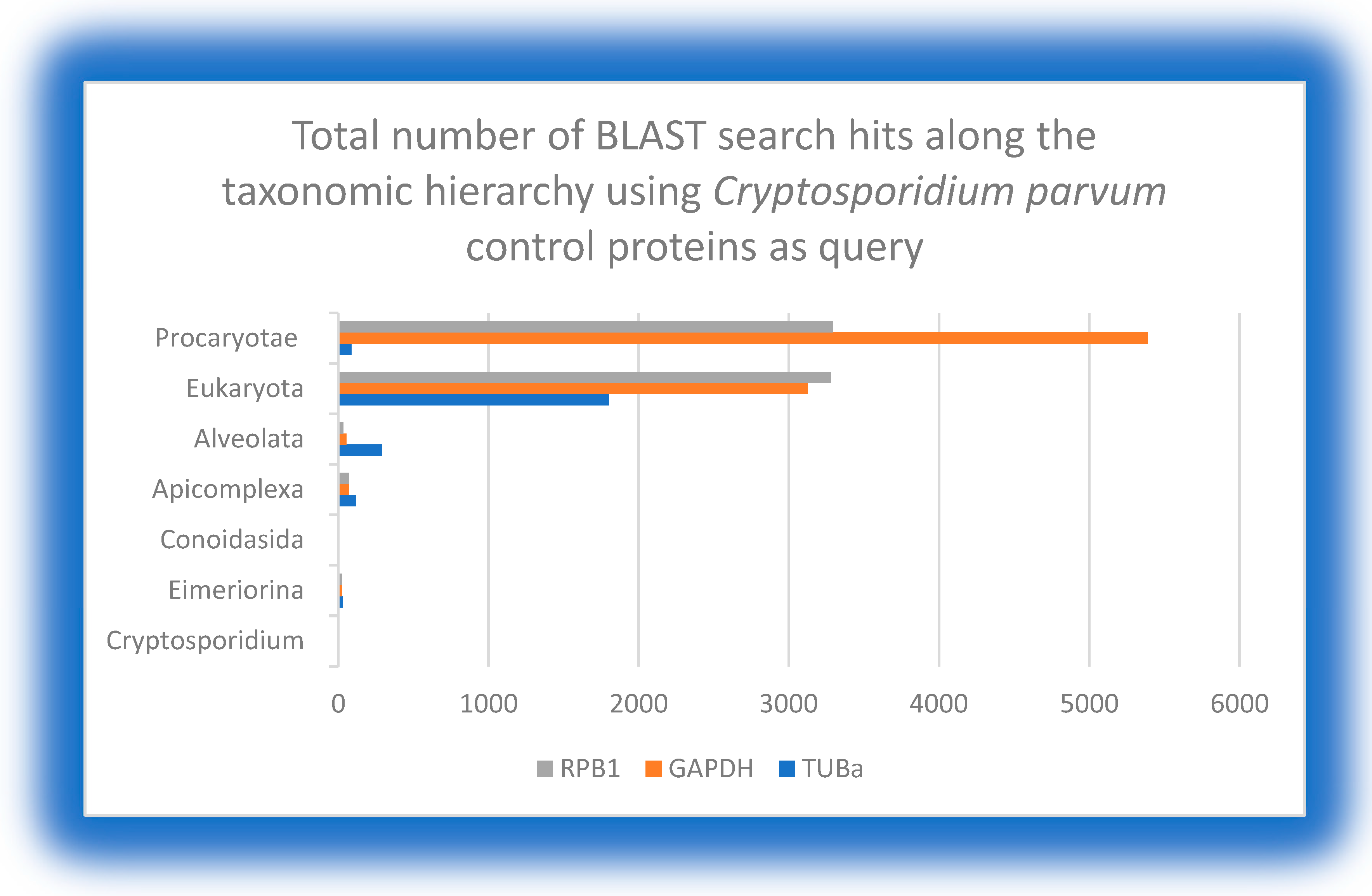
The three selected control proteins are encoded by housekeeping genes in eukaryotic organisms. As expected, the BLAST search using the corresponding proteins from each of the five protozoan species as the query showed positive hits with a steady increase in the total number of distinct organisms in each level along the taxonomic hierarchy from species to domain. The total number of hits in the eukaryotic domain compared to the prokaryotic domain showed a further increase for GAPDH but not for TUBa and RPB1. Since the total number of prokaryotic organisms is larger than that of eukaryotic organisms, it can be concluded that GAPDH is more conserved in both domains and represents a better control protein compared to TUBa and RPB1. This was to be expected, as bacteria do not possess microtubules and have different types of nucleic acid polymerases. Therefore, GAPDH was used for further comparative analysis against PFOR.
Figure 9.
The distribution of control proteins along the taxonomic hierarchy using query proteins of C. parvum shows that GAPDH is relatively more conserved in both eukaryotes and prokaryotes. .
Figure 9.
The distribution of control proteins along the taxonomic hierarchy using query proteins of C. parvum shows that GAPDH is relatively more conserved in both eukaryotes and prokaryotes. .
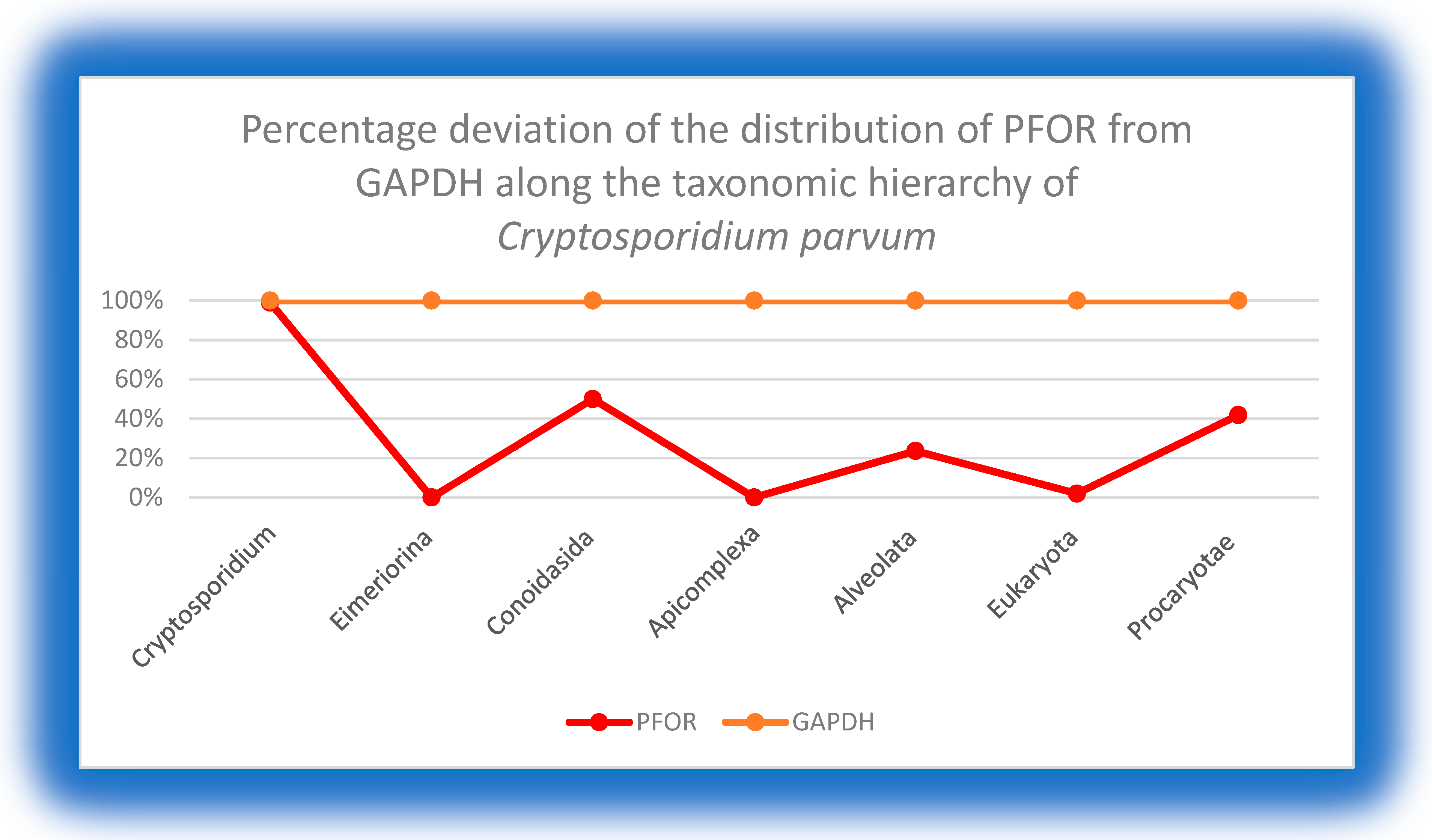
8.2. Percentage deviation of PFOR from GAPDH
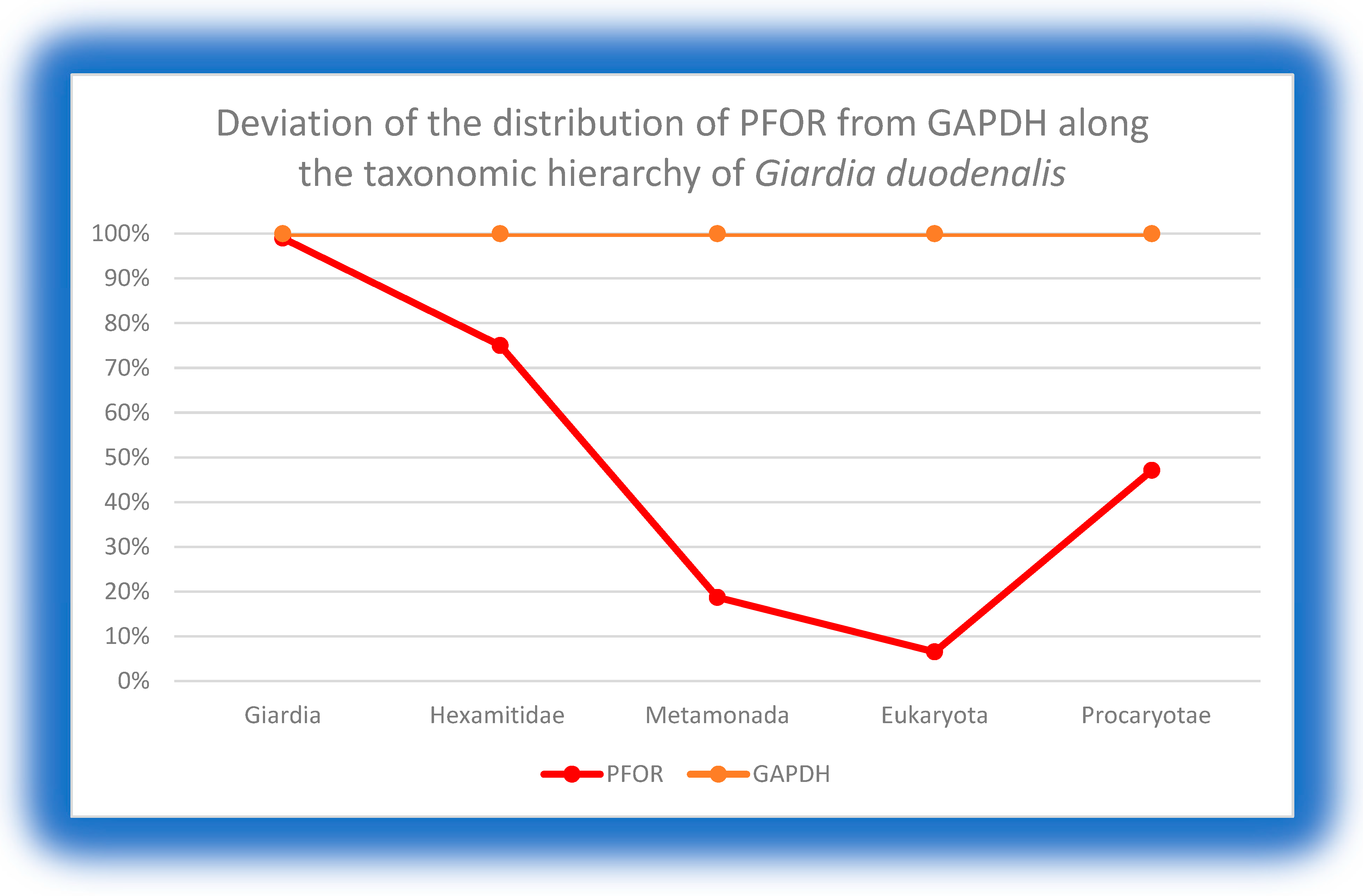
At the genus level of Giardia, Entamoeba and Cryptosporidium, there was no deviation observed in the total number of hits for PFOR compared to GAPDH in the NBCI database. From these findings, it can be extrapolated that all species in the genus Giardia, Entamoeba and Cryptosporidium possess PFOR. Above the genus level, the total number of organisms that had hits for PFOR compared to GAPDH was reduced.
Figure 10.
Graph showing no significant deviation in the total number of hits for PFOR compared to GAPDH at the genus level of Giardia. Above the genus level to the eukaryotic domain, there was a gradual increase in the percentage deviation. Approximately 50% of all prokaryotes that express GAPDH showed no hits for PFOR.
Figure 10.
Graph showing no significant deviation in the total number of hits for PFOR compared to GAPDH at the genus level of Giardia. Above the genus level to the eukaryotic domain, there was a gradual increase in the percentage deviation. Approximately 50% of all prokaryotes that express GAPDH showed no hits for PFOR.
Figure 11.
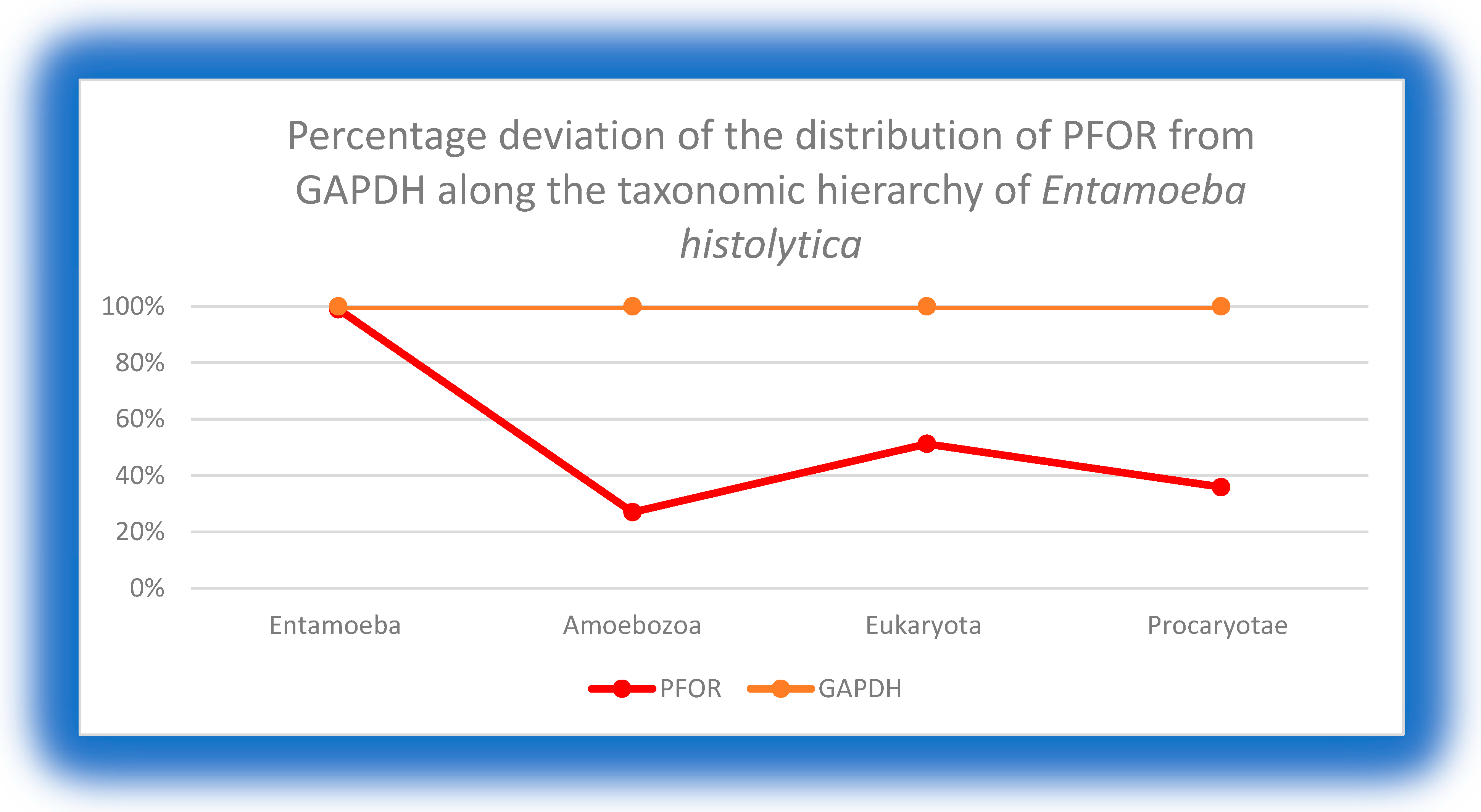
This graph shows an equal expression of GAPDH and PFOR in all the species of the genus Entamoeba. There is an approximately 72% deviation at the level of Amoebozoa, 50% in the eukaryotic domain and 64% in the prokaryotic domain. .
Figure 11.
This graph shows an equal expression of GAPDH and PFOR in all the species of the genus Entamoeba. There is an approximately 72% deviation at the level of Amoebozoa, 50% in the eukaryotic domain and 64% in the prokaryotic domain. .
Figure 12.
This graph also shows equal expression of GAPDH and PFOR in all the species of the genus Cryptosporidium. In the taxa above the genus level, relatively fewer organisms had positive hits for PFOR compared to GAPDH.
Figure 12.
This graph also shows equal expression of GAPDH and PFOR in all the species of the genus Cryptosporidium. In the taxa above the genus level, relatively fewer organisms had positive hits for PFOR compared to GAPDH.
8.3. Phylogenetic trees
The multiple alignments of 43 GAPDH and 33 PFOR protein sequences of representative eukaryotic and prokaryotic species were used to generate the corresponding phylogenies as shown in fig. 16-18. The percentages of replicate phylogenies in which the species were clustered in the bootstrap test of 1000 replicates are shown next to the branches. The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used for the inference. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site.
Figure 13.
The phylogeny of the selected eukaryotic and prokaryotic species based on the protein sequences of GAPDH shows a pattern of relatedness that is in consonance with the expected phylogeny of eukaryotes. The topology was inferred using the Neighbor-Joining method. The optimal unrooted tree with the sum of branch length = 4.58236819 is shown from a total of 659 positions in the final dataset.
Figure 13.
The phylogeny of the selected eukaryotic and prokaryotic species based on the protein sequences of GAPDH shows a pattern of relatedness that is in consonance with the expected phylogeny of eukaryotes. The topology was inferred using the Neighbor-Joining method. The optimal unrooted tree with the sum of branch length = 4.58236819 is shown from a total of 659 positions in the final dataset.
Figure 14.
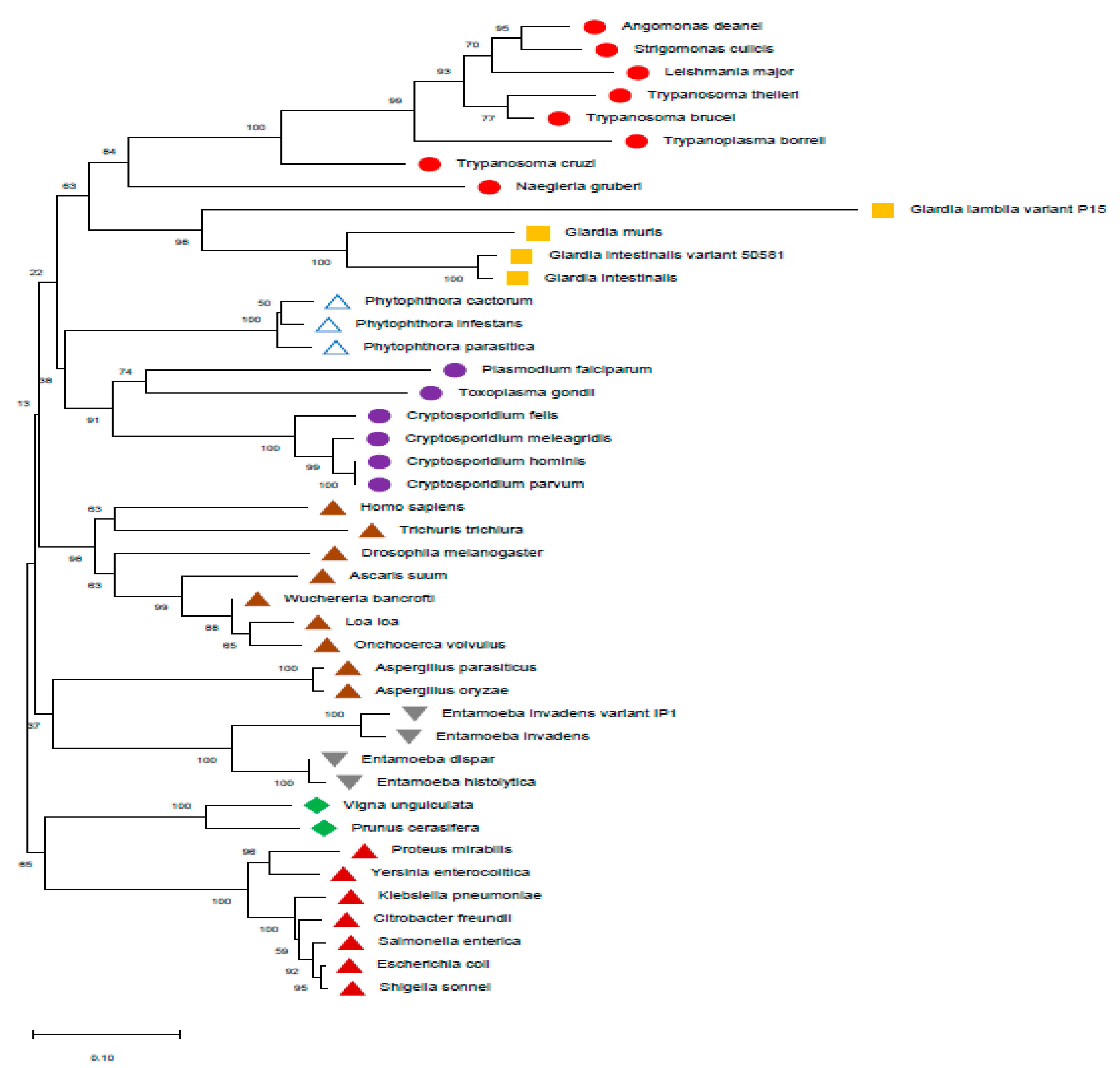
The phylogeny based on the protein sequences of PFOR was inferred using the maximum likelihood method. The unrooted tree with the highest log likelihood (-48017.90) is shown from a total of 1331 positions in the final dataset. The PFOR of Entamoeba invadens shows a monophyletic relationship with Giardia and that of Euglena gracilis is monophyletic with Cryptosporidium. This phylogenetic incongruence is however not entirely conclusive of a bacterial origin of pfor, since no protozoa is monophyletic with the selected bacterial species.
Figure 14.
The phylogeny based on the protein sequences of PFOR was inferred using the maximum likelihood method. The unrooted tree with the highest log likelihood (-48017.90) is shown from a total of 1331 positions in the final dataset. The PFOR of Entamoeba invadens shows a monophyletic relationship with Giardia and that of Euglena gracilis is monophyletic with Cryptosporidium. This phylogenetic incongruence is however not entirely conclusive of a bacterial origin of pfor, since no protozoa is monophyletic with the selected bacterial species.
8.4. Heat maps, density plots and cladograms
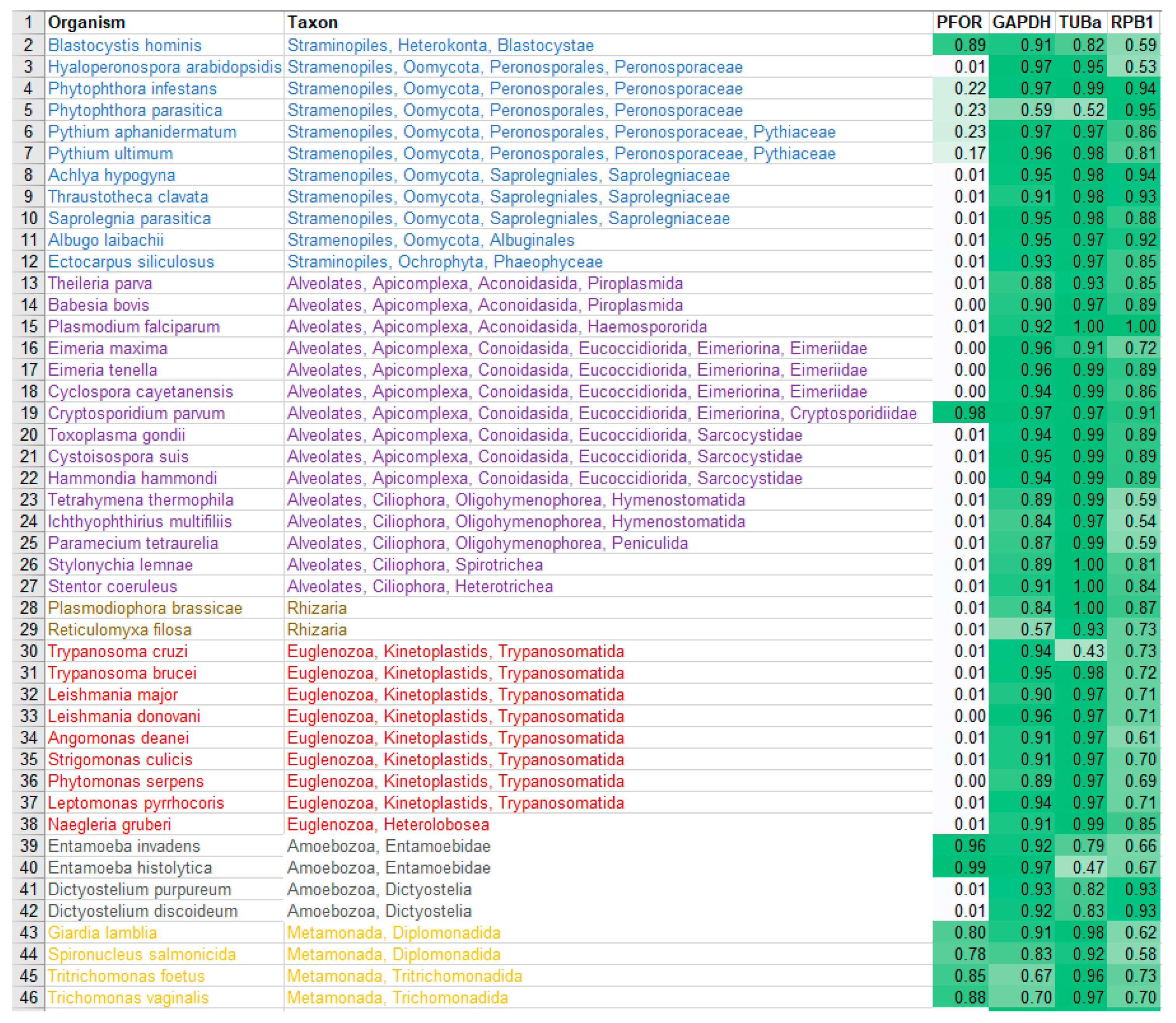
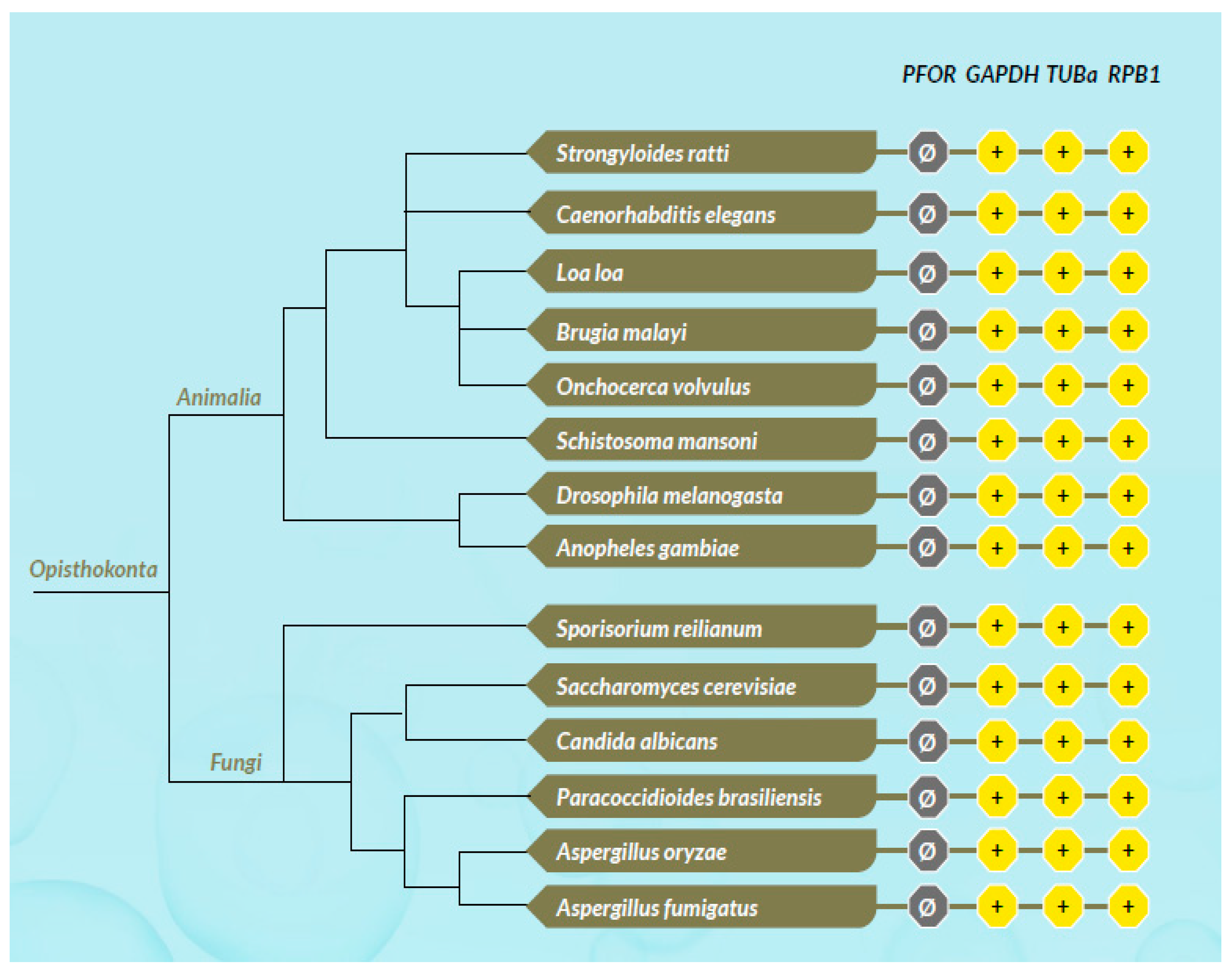
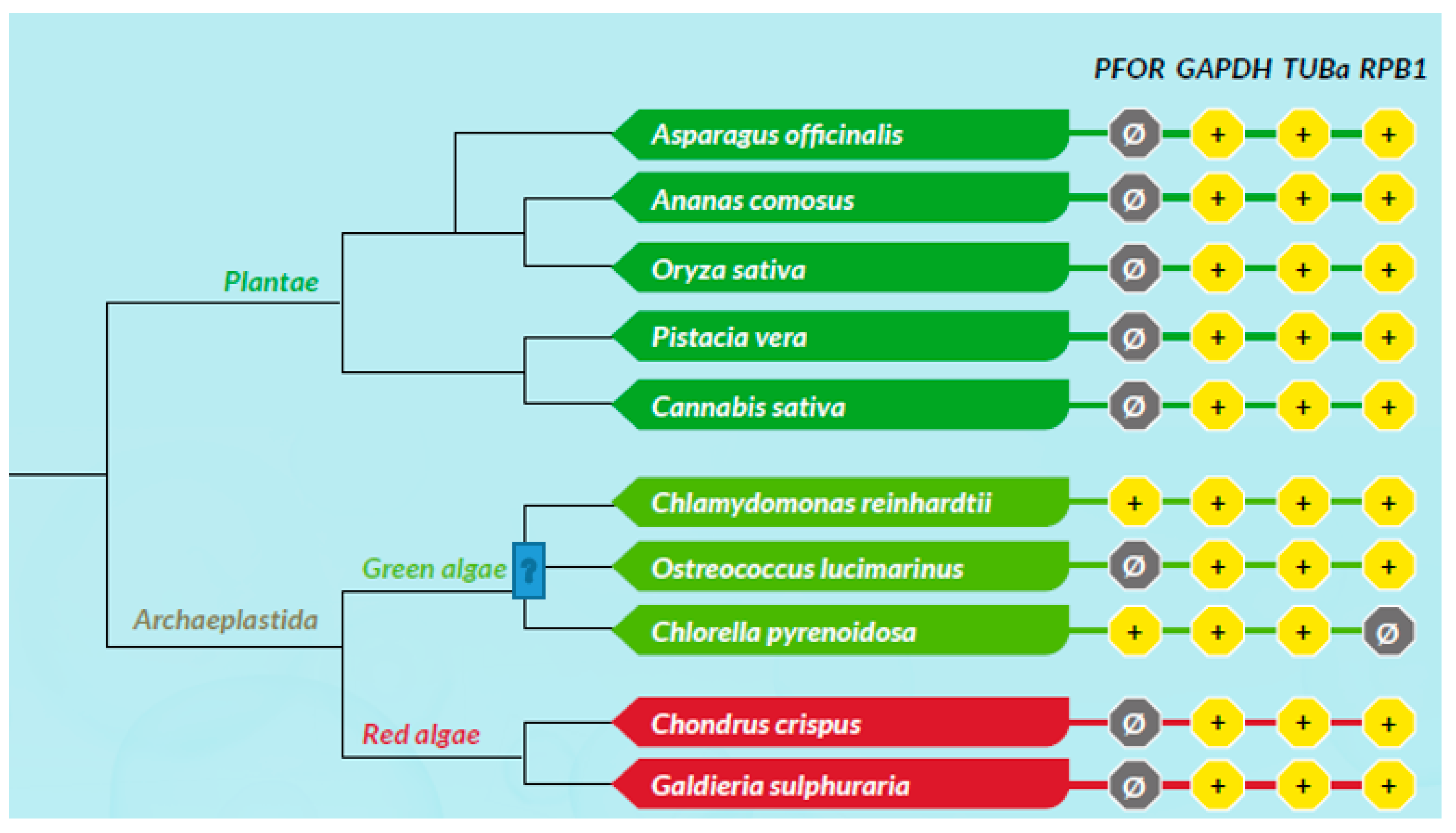
All scores from the proteome screening against the HMM profile libraries were normalized to 1 as the highest score obtained for each HMM, and a minimum cut-off significance value of 0.25 was used to represent a positive hit. As expected, there was a positive hit for the control proteins in almost all the selected eukaryotic species, which indicates a high degree of sensitivity of the screening program. All selected prokaryotic species recorded positive hits for GAPDH but, as expected, had relatively low hits for RPB1 and virtually no hit for TUBa. In the density plots, it was observed that GAPDH was relatively very well conserved in all eukaryotic taxa as well as the prokaryotic domain. Even though TUBa and RPB1 were well distributed in the eukaryotic taxa, there was a poor representation in the prokaryotic domain. The results from the proteome analysis approach correlated considerably well with the results from the BLAST pyramid approach. As expected, all animal, plant and fungal species showed no positive hits for PFOR. These results represent very good negative controls and hence, a high degree of specificity of the screening program.
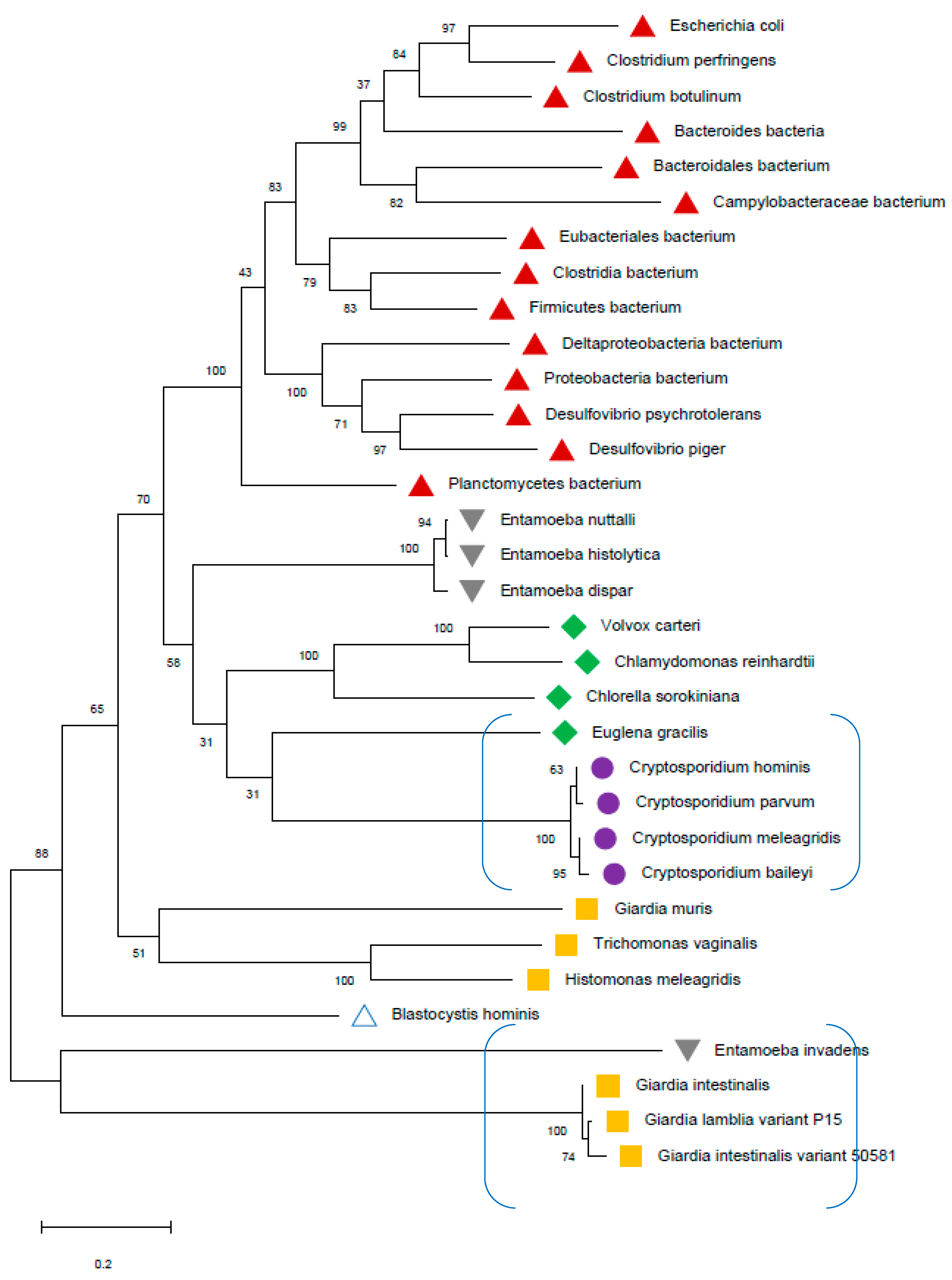
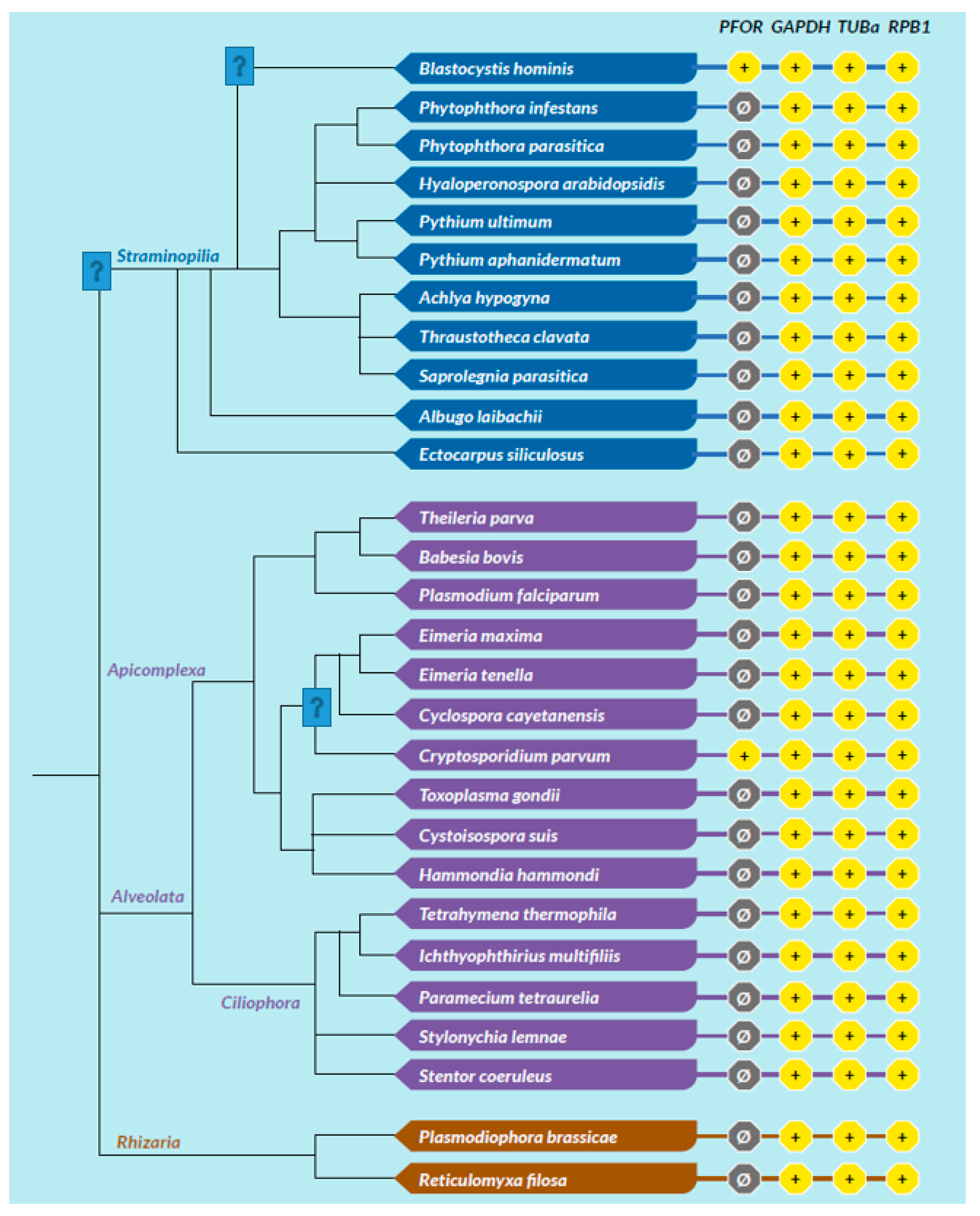
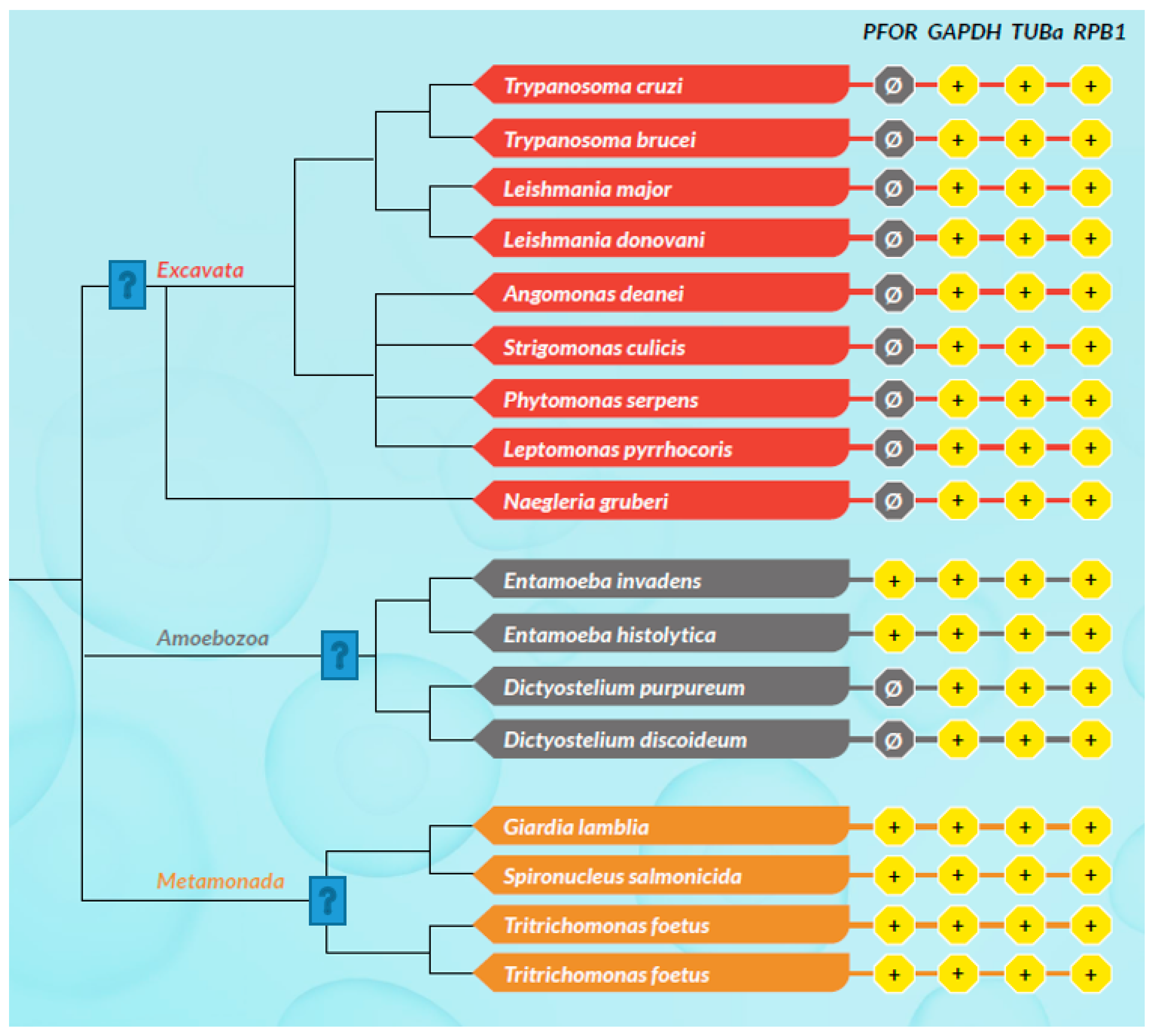
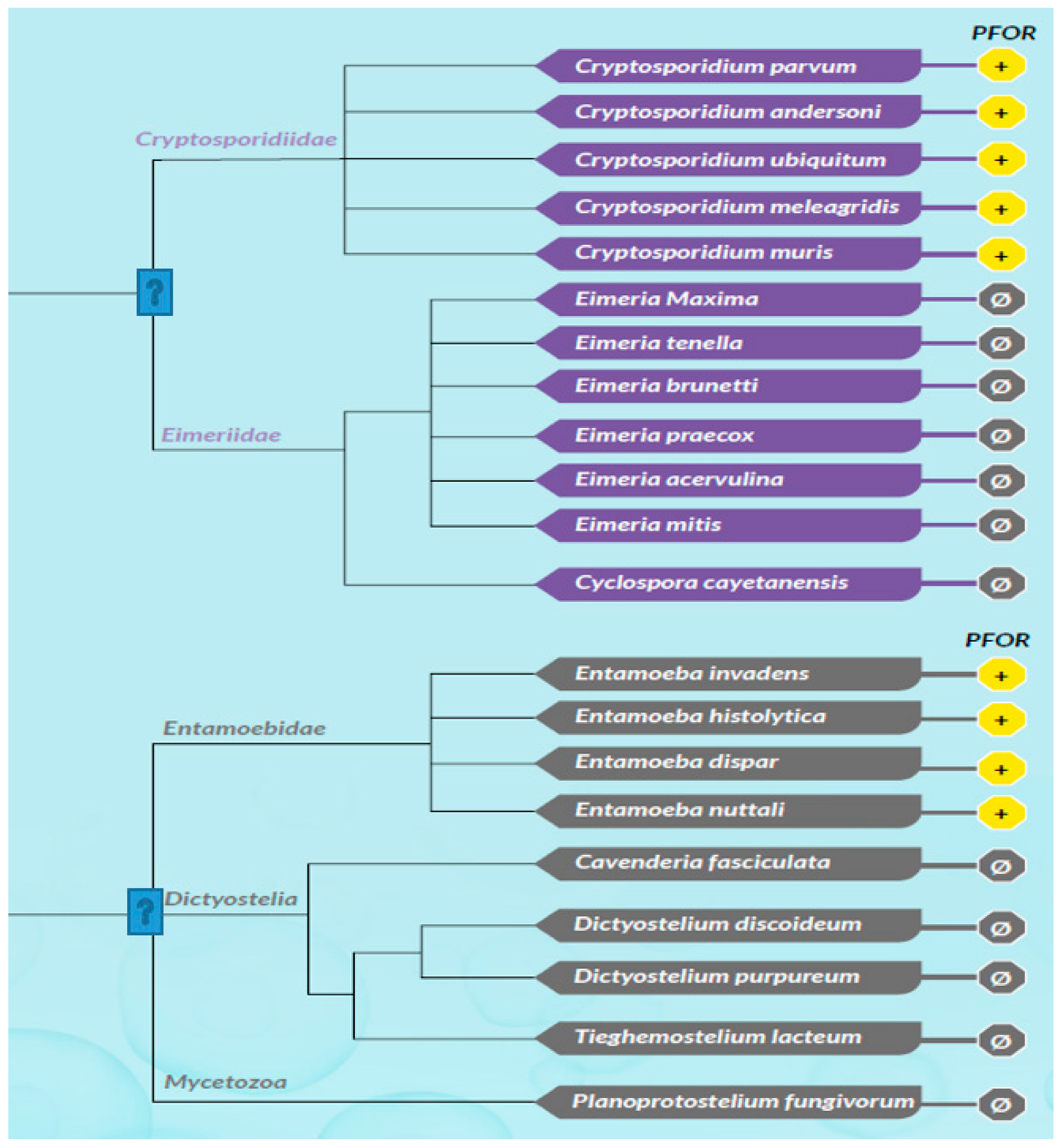
All selected species in the stramenopile group had no hits for PFOR with the exception of B. hominis. All species in the alveolate group had no hits for PFOR except C. parvum that showed a significant hit. The only two rhizarian species with available proteome sequences in the ensembl genomes database had no hit for PFOR. All the selected protozoa in the phylum Metamonada recorded hits for PFOR. All species in the euglenozoa group had no hit for PFOR. Although Entamoeba and Dictyostelium are closely related amoebozoans, the two species in the genus Entamoeba showed positive hits for PFOR whilst the species in the genus Dictyostelium had no significant hit. All the species in the genus Cryptosporidium recorded significant hits for PFOR, but the closely related species in the genus Eimeria and Cyclospora cayetanensis had no significant hits. A similar pattern was depicted in all the species of the genus Entamoeba versus those of Dictyostelium, Cavenderia, Tiegemostilium, and Planoprotostelium. For the metamonada group, there was unfortunately no further species for the expanded cladogram. About 41% of the selected bacterial species showed significant hits for PFOR.
Table 6.
Heat map of normalized scores from the whole proteome analysis of the selected protozoan species.
Table 6.
Heat map of normalized scores from the whole proteome analysis of the selected protozoan species.
Table 7.
Heat map of normalized scores from the whole proteome analysis the selected animal, plant
and bacterial species.
Table 7.
Heat map of normalized scores from the whole proteome analysis the selected animal, plant
and bacterial species.
Figure 15.
Density plots showing the distribution of control proteins compared to that of NTR and PFOR in the major eukaryotic taxa and the prokaryotic domain. .
Figure 15.
Density plots showing the distribution of control proteins compared to that of NTR and PFOR in the major eukaryotic taxa and the prokaryotic domain. .
Figure 16.
In the SAR clade, it can be observed that, B. hominis is the only straminopile that has positive hit for PFOR. Furthermore, only Cryptosporidium shows a positive hit for PFOR in the alveolate clade. It is important to note that even the closely related Eimeria and Cyclospora have no hits for PFOR.
Figure 16.
In the SAR clade, it can be observed that, B. hominis is the only straminopile that has positive hit for PFOR. Furthermore, only Cryptosporidium shows a positive hit for PFOR in the alveolate clade. It is important to note that even the closely related Eimeria and Cyclospora have no hits for PFOR.
Figure 17.
This cladogram shows exclusively no expression of PFOR in all the selected protozoa in the discoba clade. For the phylum Amoebozoa, PFOR is positive in the genus Entamoeba but not Dictyostelium, despite their close relatedness. All available organisms with proteomes in the metamonad clade show positive hits for PFOR. .
Figure 17.
This cladogram shows exclusively no expression of PFOR in all the selected protozoa in the discoba clade. For the phylum Amoebozoa, PFOR is positive in the genus Entamoeba but not Dictyostelium, despite their close relatedness. All available organisms with proteomes in the metamonad clade show positive hits for PFOR. .
Figure 18.
As expected, animals and fungi do not express PFOR, hence there was no significant hit recorded in all species. The controls proteins, however, are positive for all selected species.
Figure 18.
As expected, animals and fungi do not express PFOR, hence there was no significant hit recorded in all species. The controls proteins, however, are positive for all selected species.
Figure 19.
All plant species had no hits for PFOR. For the selected archaeplastida species, Chlamydomonas and Chlorella show hits for PFOR.
Figure 19.
All plant species had no hits for PFOR. For the selected archaeplastida species, Chlamydomonas and Chlorella show hits for PFOR.
Figure 20.
An expanded coverage of all organisms with available whole proteome sequences from the ensemblgenomes database shows a differential expression of PFOR in the genus Cryptosporidium against the closely related genera Eimeria and Cyclospora, and the genus Entamoeba against the closely related genera Dictyostelium, Cavenderia, Tiegemostilium and Planoprotostelium. .
Figure 20.
An expanded coverage of all organisms with available whole proteome sequences from the ensemblgenomes database shows a differential expression of PFOR in the genus Cryptosporidium against the closely related genera Eimeria and Cyclospora, and the genus Entamoeba against the closely related genera Dictyostelium, Cavenderia, Tiegemostilium and Planoprotostelium. .
8.5. Proteome BLAST and gene enrichment analysis
8.5.1. Results for E. histolytica
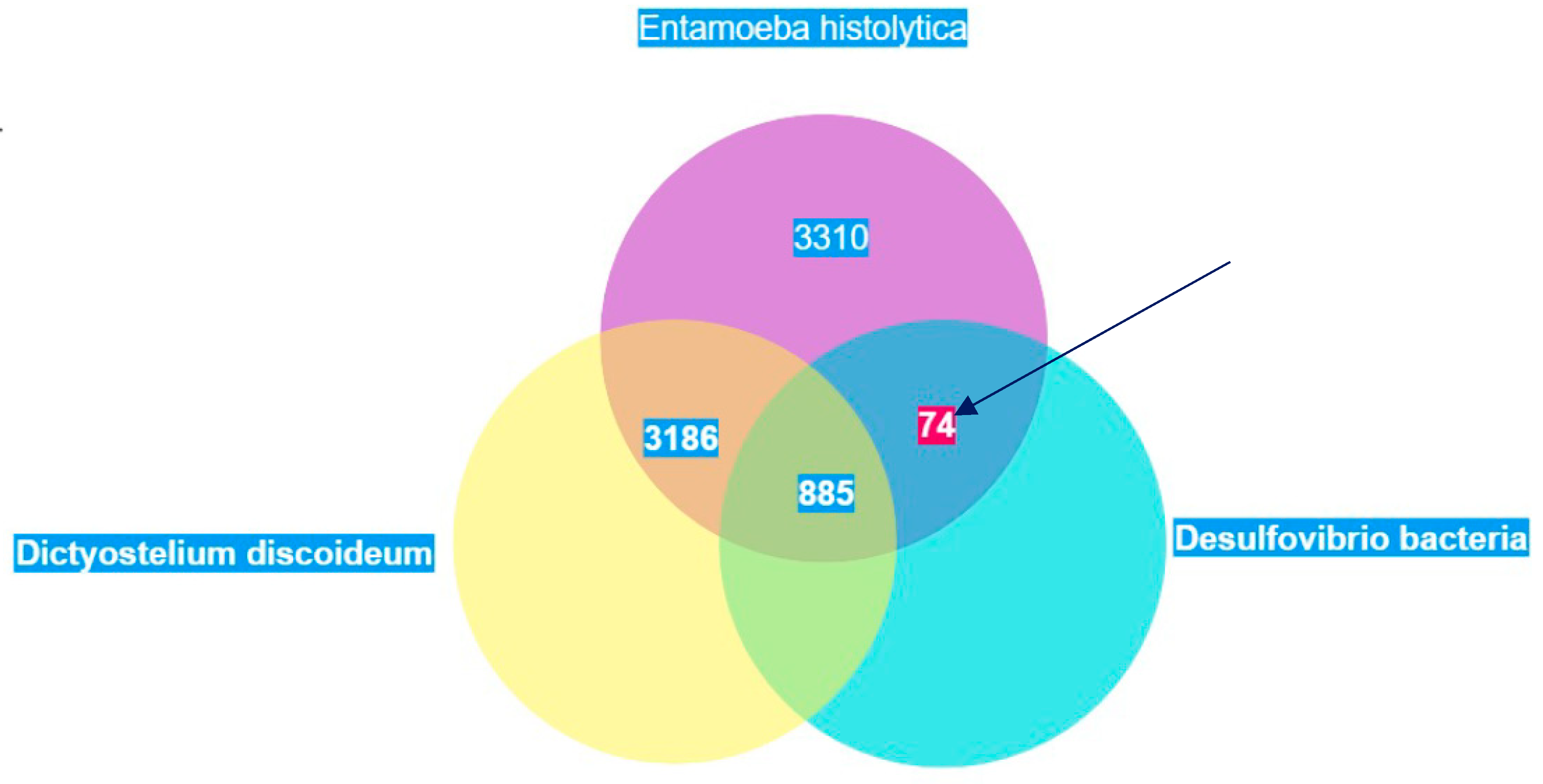
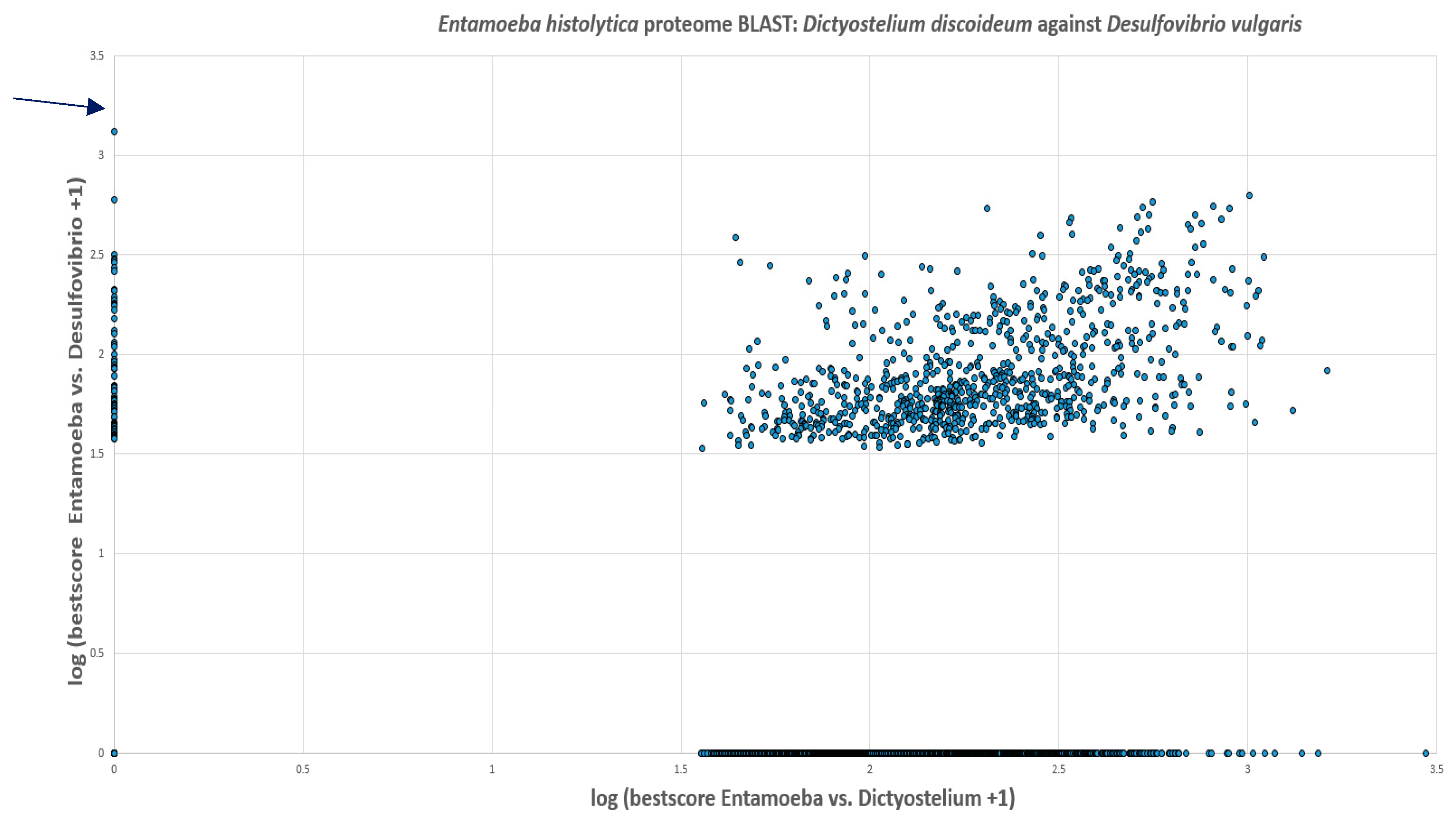
The predicted proteome of E. histolytica from the ensembl genomes database had 7,455 proteins. The BLAST search showed that 44.4% of these proteins had no hits in the proteomes of D. discoideum and D. vulgaris. 11.9% had hits in both D. discoideum and D. vulgaris. 42.7% of the proteins in E. histolytica showed hits in the proteome of D. discoideum whilst only 1% had hits in D. vulgaris. Out of this 1%, PFOR had the highest score of 1,329. The scores of all proteins were increased by one and the logarithmic values were plotted with Entamoeba versus Desulfovibrio on the vertical axis and Entamoeba versus Dictyostelium on the horizontal axis as shown in Figure 27.
Figure 21.
Illustration of the total number of proteins in Entamoeba that have significant hits in Dictyostelium and Desulfovibrio. Seventy-four proteins occur exclusively in Entamoeba and Desulfovibrio. These proteins are potential candidates of horizontal gene transfer.
Figure 21.
Illustration of the total number of proteins in Entamoeba that have significant hits in Dictyostelium and Desulfovibrio. Seventy-four proteins occur exclusively in Entamoeba and Desulfovibrio. These proteins are potential candidates of horizontal gene transfer.
Figure 22.
Every spot represents one protein of E. histolytica. Proteins with hits in only D. vulgaris are located on the vertical axis and those with hits in only D. discoideum are on the horizontal axis. E. histolytica proteins with no hits in either organism are located at the origin whilst those with hits in both D. discoideum and D. vulgaris are located in the middle of the graph. PFOR is marked with an arrow. Log is to base 10.
Figure 22.
Every spot represents one protein of E. histolytica. Proteins with hits in only D. vulgaris are located on the vertical axis and those with hits in only D. discoideum are on the horizontal axis. E. histolytica proteins with no hits in either organism are located at the origin whilst those with hits in both D. discoideum and D. vulgaris are located in the middle of the graph. PFOR is marked with an arrow. Log is to base 10.
8.5.2. Results for C. parvum
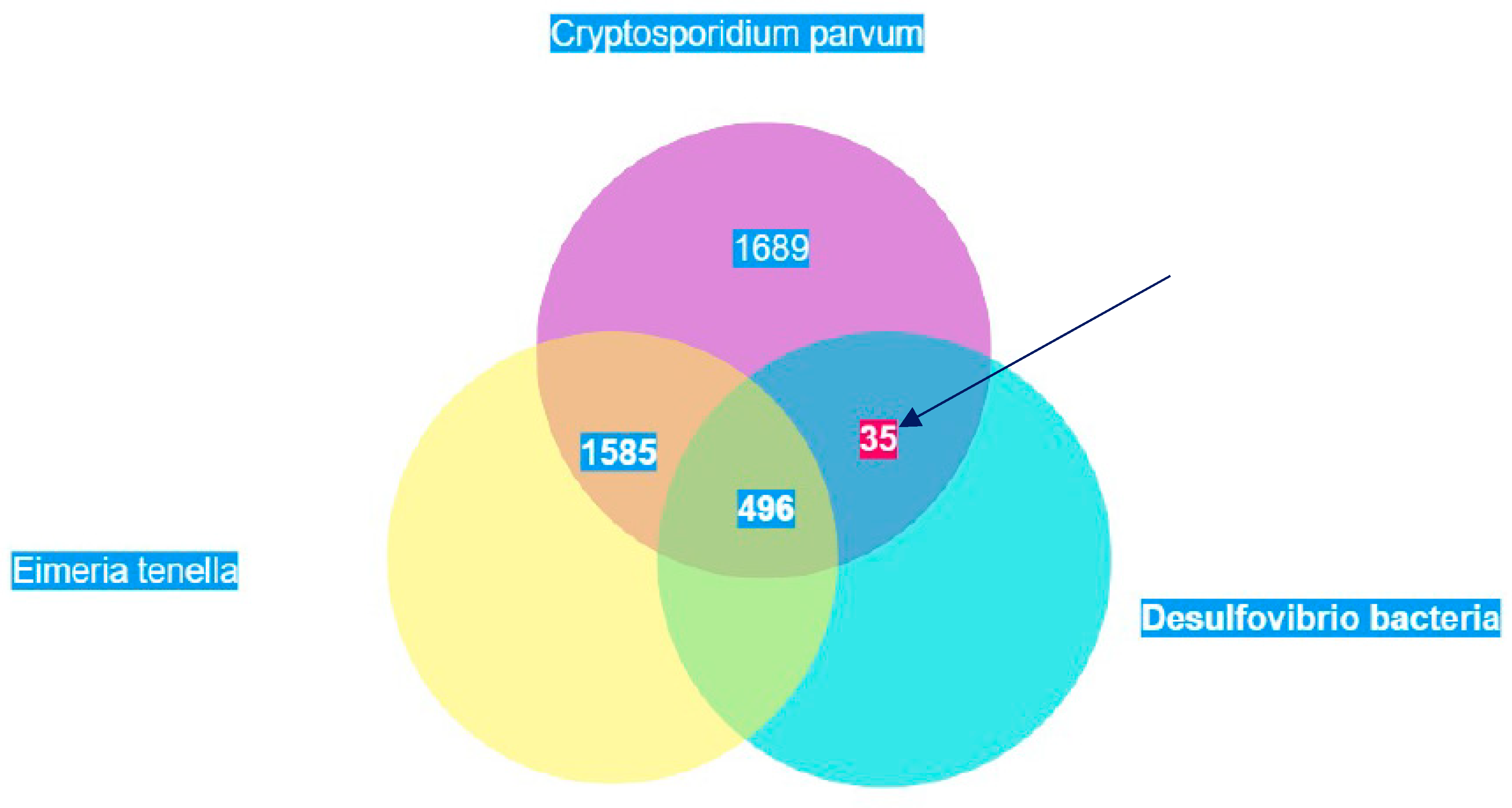
Of the 3,805 predicted proteins in the proteome of C. parvum, the BLAST search showed that 44.4% had no hits in the proteomes of E. tenella and D. vulgaris. 13% of the proteins had hits in E. tenella and D. vulgaris. 41.7% of the proteins in C. parvum showed hits in the proteome of E. tenella whilst 0.9% had hits in D. vulgaris. Out of this 0.9%, PFOR had the highest score of 1,202. The scores of all proteins were increased by one and the logarithmic values were plotted with Entamoeba versus Desulfovibrio on the vertical axis and Entamoeba versus Dictyostelium on the horizontal axis as shown in Figure 29.
Figure 23.
Illustration of the total number of proteins in Cryptosporidium that have significant hits in Eimeria and Desulfovibrio. Thirty-five proteins occur exclusively in Cryptosporidium and Desulfovibrio. These proteins are potential candidates of horizontal gene transfer.
Figure 23.
Illustration of the total number of proteins in Cryptosporidium that have significant hits in Eimeria and Desulfovibrio. Thirty-five proteins occur exclusively in Cryptosporidium and Desulfovibrio. These proteins are potential candidates of horizontal gene transfer.
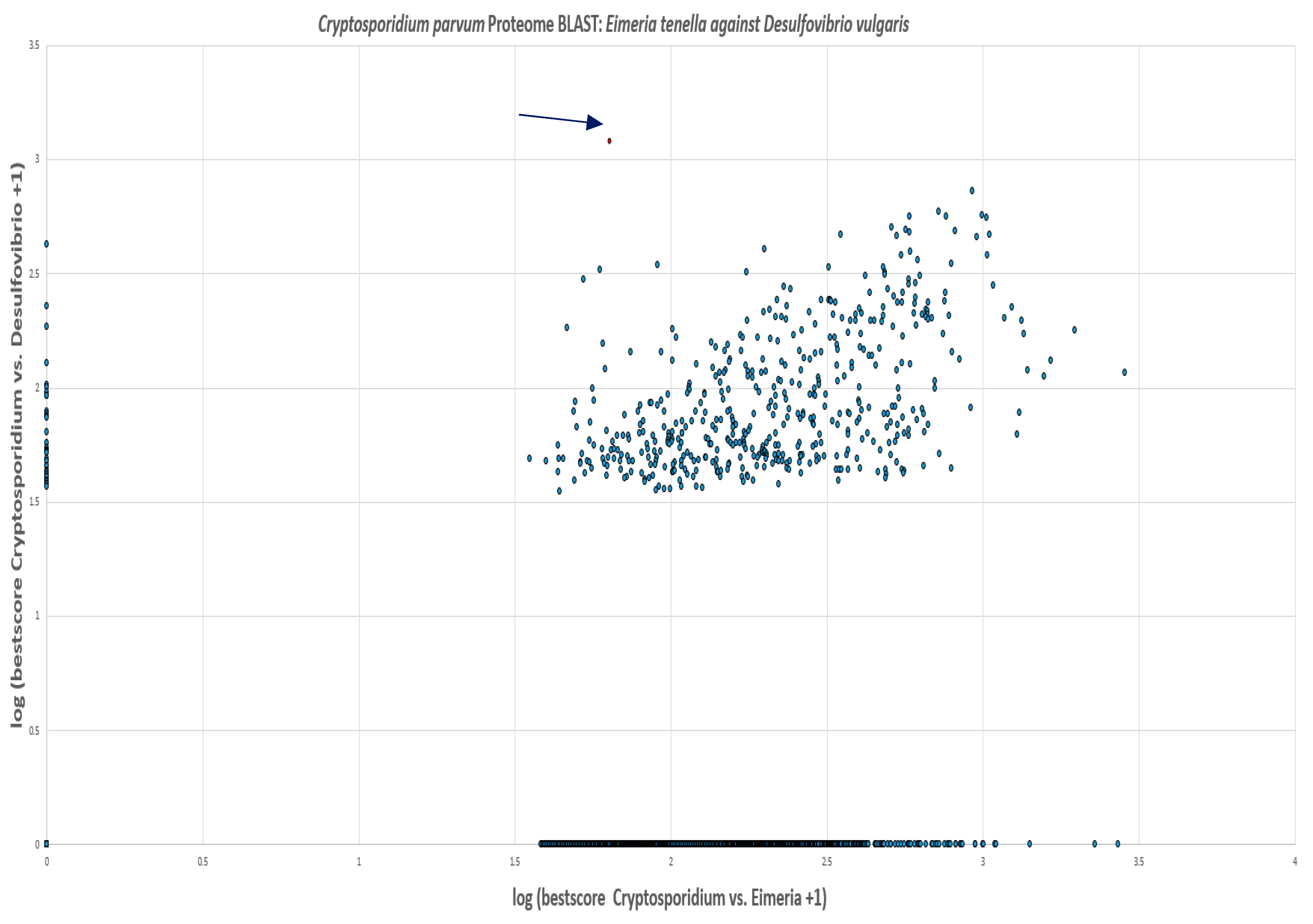
Figure 24.
Every spot represents one protein of C. parvum. Proteins with hits in only D. vulgaris are located on the vertical axis and those with hits in only E. tenella are on the horizontal axis. C. parvum proteins with no hits in either organism are located at the origin whilst those with hits in both E. tenella and D. vulgaris are located in the middle of the graph. PFOR is marked with an arrow. Log is to base 10.
Figure 24.
Every spot represents one protein of C. parvum. Proteins with hits in only D. vulgaris are located on the vertical axis and those with hits in only E. tenella are on the horizontal axis. C. parvum proteins with no hits in either organism are located at the origin whilst those with hits in both E. tenella and D. vulgaris are located in the middle of the graph. PFOR is marked with an arrow. Log is to base 10.
8.5.3. Gene enrichment analysis
Due to the unavailability of a reference genome for
C. parvum at the GO website
(http://geneontology.org/), the gene enrichment analysis was limited to
E. histolytica. The gene IDs of proteins that had significantly high similarity based on the proteome BLAST scores for
E. histolytica against
D. vulgaris were analyzed.
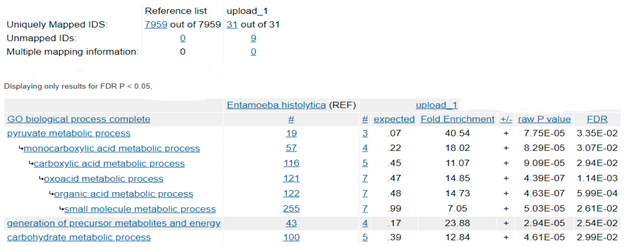
Biological process was used as annotation data category to analyze the gene list of 40 query proteins against the reference genome of E. histolytica with 7,959 genes. Nine of the 40 query genes were unmapped, hence the remaining 31 genes were used for the gene enrichment analysis. 19 genes from the reference list and 3 genes from the query list mapped to the pyruvate metabolic pathway. Based on the reference list, 0.239% of genes were expected to map to the pyruvate metabolic pathway. Since the query list contained 31 genes, the number of genes expected to map to this pathway was 0.074.
The Fold-Enrichment of the genes observed in the query list to the expected outcome was 40.54, which indicates that the pyruvate metabolic pathway is overrepresented in the query list. The raw p-value for the statistical analysis as determined by Fisher’s exact test and the False Discovery Rate as calculated by the Benjamini-Hochberg procedure were 7.75-0.5 and 3.35-0.2 respectively. These findings suggest that the number of genes from the query list that were mapped to the pyruvate metabolic pathway did not occur by chance, as determined by comparison to the reference genome of E. histolytica. This small p-value also indicates that the result is non-random and potentially interesting, and worth looking at in closer detail.
The other genes that were also significantly enriched were mapped to the carbohydrate metabolic process and pathways involved in the generation of precursor metabolites and energy. The list of all the genes that were significantly enriched is shown in Table 8.
Table 8.
Results of the gene enrichment analysis from the gene ontology website.
Table 8.
Results of the gene enrichment analysis from the gene ontology website.
 |
| Mapped IDs of enriched genes |
Expressed proteins |
| XP_657019 |
Pyruvate: ferredoxin oxidoreductase |
| XP_651627 |
Type A flavoprotein |
| XP_657332 |
Pyruvate phosphate dikinase |
| XP_651155 |
Alkyl sulfatase |
| XP_653620 |
Phosphotransferase |
| XP_654182 |
Phosphoglycerate mutase |
| XP_656980 |
Branched-chain amino acid aminotransferase |
| XP_655990 |
acetate kinase |
| XP_656909 |
Polysaccharide deacetylase |
| XP_651609 |
Ribose 5-phosphate isomerase |
| XP_656356 |
Carbohydrate degrading enzyme |
The table above shows only results for the statistically significant gene enrichment data with p and FDR values < 0.05. The second and third columns contain the number of genes in the reference list and the query list that map to the corresponding metabolic processes. The fourth column contains the expected number of genes that map to the corresponding metabolic processes based on the reference list. The GO biological processes with the highest enrichment of 40.54 was obtained from genes that mapped to the pyruvate metabolic pathway. A plus sign indicates over-representation and a negative sign indicates under-representation. The table below shows the list of all genes that were significantly enriched.
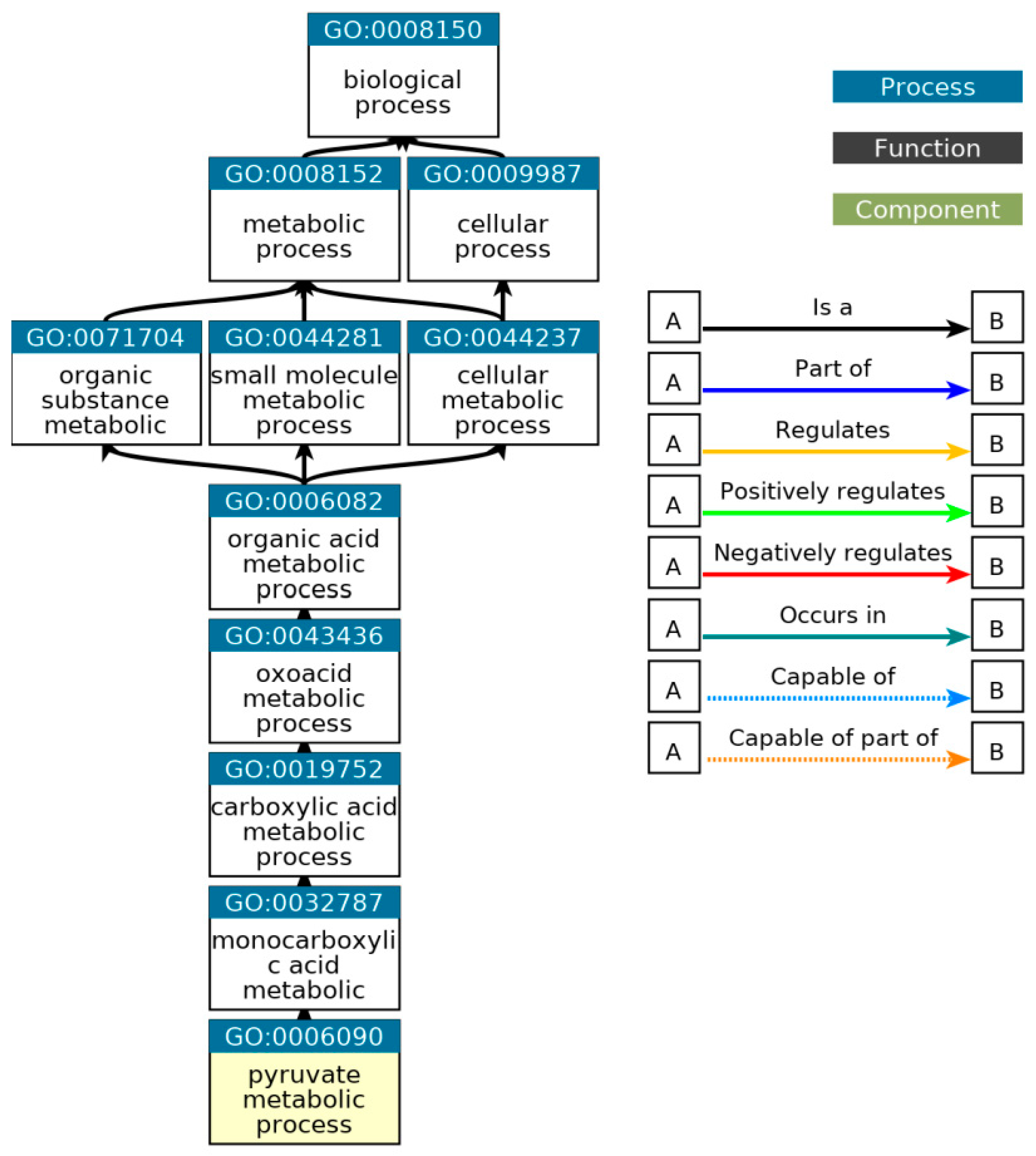
Figure 25.
Ancestor chart for the pathways related to the pyruvate metabolic process, GO: 0006090. Source:
(http://geneontology.org/).
Figure 25.
Ancestor chart for the pathways related to the pyruvate metabolic process, GO: 0006090. Source:
(http://geneontology.org/).
Table 9.
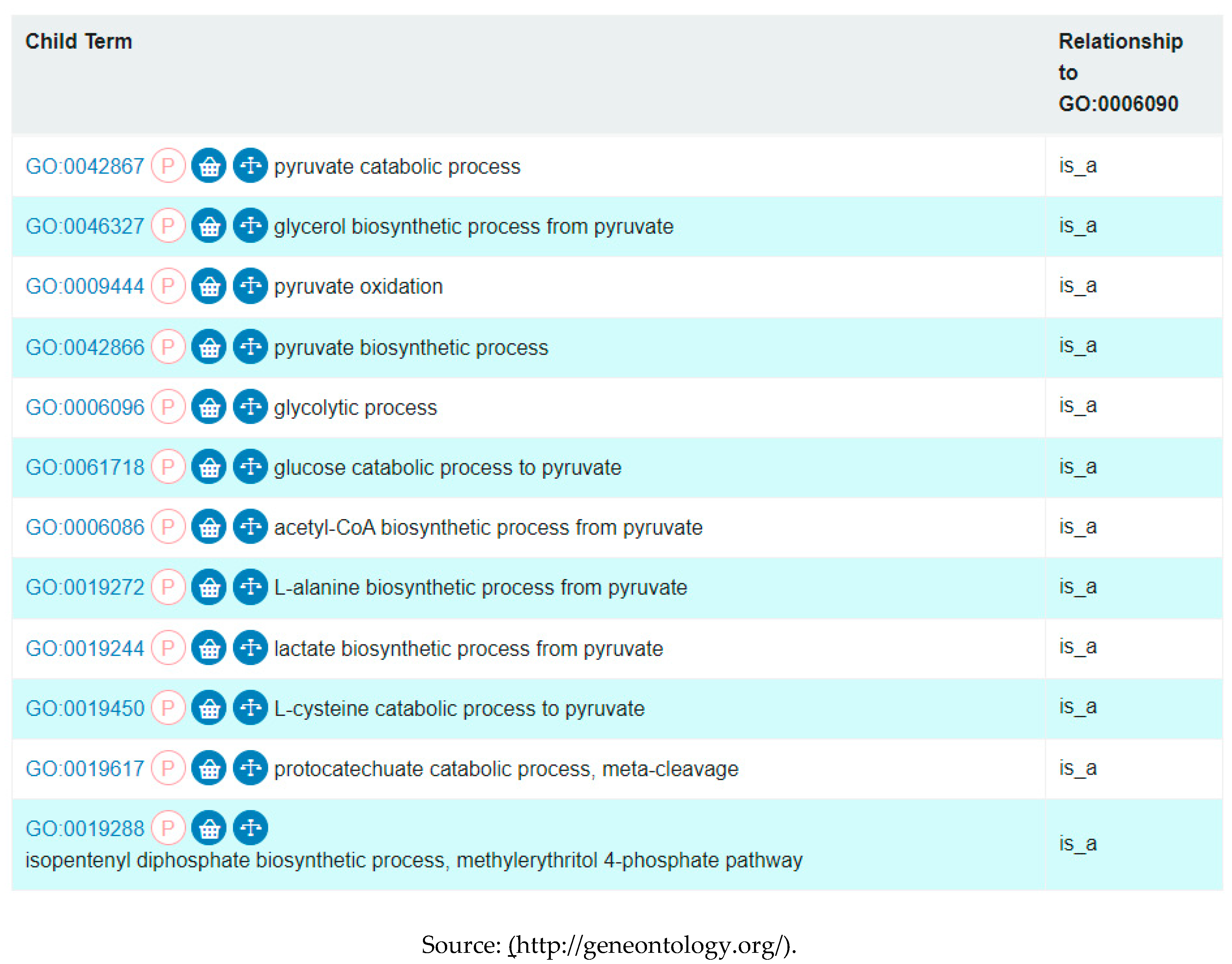
List of all GO terms that are direct descendants of the pyruvate metabolic process.
Table 9.
List of all GO terms that are direct descendants of the pyruvate metabolic process.
9. Discussion
Although all the three selected control proteins are encoded by housekeeping genes in eukaryotic organisms, the outcome of the protein BLAST searches demonstrated that GAPDH is clearly more conserved in both eukaryotes and prokaryotes. Since the total number of distinct organisms in each taxon was unavailable in the NCBI database, it was justifiable to use the total number of distinct organisms that possessed GAPDH as the reference for the comparative analysis in the BLAST pyramid approach. The outcome of the whole proteome analysis showed a significant correlation with that of the BLAST pyramid analysis, since – as expected – all selected prokaryotic species showed positive hits for GAPDH but insignificant hits for TUBa and RPB1. The positive hits for the control proteins in all the selected eukaryotic species obtained from the proteome screening against the HMM profile libraries indicated a high degree of sensitivity of the proteome screening approach. On the other hand, the insignificant hits for PFOR observed in all animal, plant and fungal species indicated a high degree of specificity. The whole proteome analysis served as an independent and a more robust approach that validated the results obtained from the BLAST pyramid analysis. The results from the whole proteome screening of the selected species against the HMM profile libraries that were translated onto the density plots illustrated further the relative distribution of the control proteins and redox enzymes in the various eukaryotic taxa and the prokaryotic domain.
The significant percentage deviation of the distribution of PFOR from that of GAPDH along the taxonomic tree of the selected protozoan species confirmed the relatively limited occurrence of the redox enzymes in eukaryotes. The observed restricted pattern of the BLAST pyramid results further confirmed that the distribution of these enzymes is highly restricted to few organisms in the eukaryotic domain, which is potentially interesting and worth looking at in closer detail. Due to the absence of deviation observed in the total number of hits for PFOR compared to GAPDH in the genera Giardia, Entamoeba and Cryptosporidium, it can be inferred that all species in these genera possess PFOR. The expanded cladograms of Entamoeba and Cryptosporidium that were derived from the whole proteome analysis confirmed the findings from the BLAST pyramid analysis, since all the selected species in these genera with available whole proteome sequences from the ensemblgenomes database had significant hits for PFOR. Taking a broader view of all the selected protozoan species in the alveolate clade, only C. parvum showed a significant hit for PFOR. This is an interesting observation in support of a possible horizontal acquisition of pfor because the expanded cladogram of the genus Cryptosporidium showed that the closely related species in the genera Eimeria and Cyclospora had no significant hits for PFOR. A similar pattern observed in the expanded cladogram of all the available species of the genus Entamoeba and those of the closely related genera Dictyostelium, Cavenderia, Tiegemostilium and Planoprotostelium further substantiated the hypothesis that pfor had most likely been horizontally acquired. However, due to the limited availability of additional species with whole proteome sequences in the metamonad clade, it was not possible to generate an expanded cladogram for the genus Giardia and its closely related genera to support the horizontal transfer hypothesis of pfor in G. duodenalis. Consequently, all further analyses to support the hypothesis were limited to C. parvum and E. histolytica.
The phylogenic tree generated from the protein sequences of PFOR using the maximum likelihood method showed some degree of incongruence with the expected phylogeny of eukaryotes since the PFOR of Entamoeba invadens was monophyletic with Giardia species, and that of Euglena gracilis was monophyletic with Cryptosporidium species. This phylogenetic disturbance indicates multiple sources of the acquisition of pfor in protozoa; however, the various bacterial and protozoan species were clustered separately in distinct clades. This pattern is not convincing as strong evidence suggestive of multiple HGT events from bacteria, since none of the protozoan species was monophyletic with the bacterial species.
The source of horizontally transferred genes in protozoa is based on the proximity hypothesis, which explains that bacterial species dwelling in the same host environment as protozoa serve as potential gene donors.
Table 10 summarizes how the relationship between the lifecycle of the selected protozoa and the expression of PFOR supports the proximity hypothesis. Even though
B. hominis belongs to the stramenopile taxon,
B. hominis exclusively expresses PFOR. The most plausible explanation for this observation is that,
B. hominis is a strict anaerobe that requires PFOR for optimal adaptation to the host gastrointestinal tract, whereas the other species in the stramenopile taxon are free-living protists. None of the selected protozoa in the alveolate taxon, apart from species belonging to the genera
Cryptosporidium and
Eimeria, dwells predominantly in their host's gastrointestinal tract. Based on their lifecycles, species in the genera
Cryptosporidium and
Eimeria are expected to possess PFOR since they are closely related apicomplexan parasites that inhabit a similar host environment. However, the presence of PFOR was restricted to
Cryptosporidium. A plausible hypothesis to explain the absence of PFOR in
Eimeria could be a gene loss event that occurred after the extant protozoa from the common ancestral lineage to
Eimeria and
Cryptosporidium had acquired
pfor. Since
Eimeria species strive in the host gastrointestinal tract without the possession of PFOR, they evolved to express alternative enzymes for intermediary metabolism, growth and survival. Taking a detailed look at the taxon amoebozoa,
Entamoeba species are closely related to
Dictyostelium. However,
Entamoeba species are anaerobic parasites that dwell predominantly in the host gastrointestinal tract, whereas
Dictyostelium species are free-living organisms. Based on the proximity hypothesis,
E. histolytica is expected to possess PFOR but not
D. discoideum. Entamoeba species probably diverged from
Dictyostelium species before horizontally acquiring
pfor. These findings are in consonance with multiple independent horizontal transfer events of
pfor in
Cryptosporidium and
Entamoeba.
Species in the genera Giardia, Trichomonas and Tritrichomonas are flagellated anaerobic protozoa of the taxon metamonada. They inhabit and invade the gastrointestinal or genitourinary system of their hosts; hence, they are expected to possess PFOR, as was seen in the results. Unfortunately, due to the unavailability of whole proteome sequences of other closely related protozoa in this taxon, it was not possible to perform further analyses to explore the hypothesis of horizontal acquisition of pfor in these genera. The two species with available whole proteome sequences in the taxon rhizaria are free-living protists; therefore, they are not expected to have PFOR, as was observed in the results.
From this table it can be postulated that except Eimeria, all other organisms that reside in close proximity with bacteria in the gastro-intestinal or genitourinary tract express PFOR. A gene loss event may explain why Eimeria does not express PFOR. Even though Blastocystis is grouped with the other straminopiles that do not express PFOR, it possesses PFOR. This is probably because it dwells in the gastrointestinal tract whiles the others are free living.
In view of the proximity hypothesis that supports the horizontal transfer of genes from bacteria, the bacterium with the highest normalized similarity score of 1.0 for PFOR,
D. vulgaris, qualifies as the most suitable putative gene donor or representative of an extant bacterial lineage of gene donors.
D. vulgaris is a gram-negative, anaerobic, non-spore forming, curved rod-shaped, sulfate-reducing bacterium capable of producing hydrogen sulfide in the host gastrointestinal tract and as such dwells in close proximity with some invading protozoan parasites. The comparative whole proteome BLAST analysis of
E. histolytica and
C. parvum against their corresponding closely related control species,
D. discoideum and
E. tenella, and the bacterium
D. vulgaris demonstrated significantly high hits for some proteins that are exclusively present in
D. vulgaris but absent in the closely related protozoan species. Owing to unavailability of a reference genome for
C. parvum in the database of the GO website
(http://geneontology.org/) for gene enrichment analysis, only the exclusively occurring genes in
E. histolytica and
D. vulgaris were analyzed further.
Out of the forty selected genes in E. histolytica and D. vulgaris, eleven genes demonstrated relatively high enrichment with significant p-values as determined by Fisher’s exact test and False Discovery Rates as calculated by the Benjamini-Hochberg procedure. The outcome of the gene enrichment analysis shows that these genes encode proteins that function in biological processes related to small molecule metabolism, generation of precursor metabolites, and carbohydrate metabolism. The pyruvate metabolic process is a biological process related to the small molecule metabolic pathway, under which three out of the forty exclusively expressed proteins are functionally categorized. The findings suggest that the eleven genes that were significantly enriched did not occur by chance as compared to the reference genome of E. histolytica. Furthermore, the small p-values indicate that the outcome of the gene enrichment analysis was non-random and worth looking at in further detail.
The significantly enriched genes can be analyzed further using syntenic approaches to give substantial information on how they are structurally related according to their relative position at the chromosomal level. If it can be established that these genes are located close to each other at the chromosomal level, this would provide a relatively stronger evidence in support of the hypothesis that they were most likely horizontally transferred together with pfor as a genetic fragment from bacteria. Moreover, if these findings can be reproduced in the other PFOR-containing protozoan species, this will provide more evidence in support of multiple independent horizontal gene transfer events.
10. Conclusion
The pathogen specific redox enzyme, PFOR, plays significant roles in the energy metabolism and growth of pathogens that dwell predominantly in oxygen-deprived niches. From the BLAST pyramid analysis as well as the whole proteome analysis, it can be inferred that the expression of PFOR is limited to few protozoan species in the eukaryotic domain whereas a relatively large proportion of prokaryotes possess PFOR. Protozoa belonging to the genera Cryptosporidium, Giardia, Entamoeba and Blastocystis are anaerobic parasites that reside predominantly in close proximity with bacterial species in the host gastrointestinal tract (Higuera, Villamizar et al. 2020). Whereas, those belonging to the genera Trichomonas and Tritrichomonas are sexually transmitted extracellular anaerobic parasites that also dwell in close proximity with bacteria in the host genito-urinary tract (Benchimol, de Andrade Rosa et al. 2008, Chapwanya, Usman et al. 2016).
A plausible explanation for the restricted occurrence of PFOR in the above-mentioned protozoa is based on the hypothesis that bacterial species serve as potential sources for the acquisition of genes that enhance the optimal adaptation of these protozoa in hostile host environments. The expanded cladograms of Entamoeba and Cryptosporidium with their closely related genera substantiated this hypothesis. Although the phylogeny based on the protein sequences of PFOR demonstrated some degree of phylogenetic incongruence, the PFOR of none of the protozoa was monophyletic with that of the bacteria. The observed monophyletic relationship of the PFOR of E. invadens with Giardia species, and that of E. gracilis with Cryptosporidium species, is suggestive that these two distinct groups of protozoa may have acquired pfor independently from common ancestral lineages. Regardless of the low bootstrap values, these findings support multiple independent horizontal transfer events. The exclusively expressed proteins obtained from E. histolytica and the putative bacterial gene donor, D. vulgaris, showed an over-representation of eleven genes involved in small molecule metabolism, generation of precursor metabolites, and carbohydrate metabolism. If these results obtained from E. histolytica and D. vulgaris can be reproduced in other PFOR-possessing protozoan species, it would provide a more validated evidence to support the horizontal transfer of pfor from bacteria. Subsequent syntenic analyses of the significantly enriched genes would be required to provide further information regarding the positional relatedness of these genes at the chromosomal level.
11. Final remarks
Since metronidazole is an established, well tolerated drug for treating infections caused by PFOR-possessing pathogens, it can be considered as a potential drug candidate for the treatment of infections caused by Cryptosporidium, Spironucleus and Blastocystis.
In this project, there was a considerable selection bias from using non-redundant protein sequences as the sole database for the BLAST searches and a relatively skewed coverage of pathogenic protozoa against free-living protozoa. Moreover, unavailability of the total number of distinct organisms in each taxon and possible bacterial contamination with PFOR protein sequences from the gastrointestinal tract were challenging factors.
The unavailability of whole proteome sequences of other closely related species in the phylum Metamonada in the ensemblgenomes database posed a significant limitation in performing further analyses for the distribution of PFOR.
The unavailability of reference genome for C. parvum in the Gene Ontology database was a major limiting factor in performing comparative studies with the results of the gene enrichment analysis from the exclusively present proteins in E. histolytica and D. vulgaris.
Acknowledgement
I would like to thank my supervisors, Pascal Mäser, the team leader of the parasite chemotherapy unit and Daniela Brites of the tuberculosis research unit of the Swiss Tropical and Public Health Institute for the opportunity to undertake this research project. I extremely appreciate not only their academic supervision but also their invaluable friendly support.
List of abbreviations
| BLASTP |
Basic local alignment search tool program |
| CHMP |
Committee for Medicinal Products for Human Use |
| FNOR |
Ferredoxin: NAD oxidoreductase |
| GAPDH |
Glyceraldehayde-3-phosphate dehydrogenase |
| HGT |
Horizontal gene transfer |
| LGT |
Lateral gene transfer |
| MEGA |
Molecular Evolutionary Genetics Analysis |
| NCBI |
National Center for Biotechnology Information |
| PDC |
Pyruvate dehydrogenase multi-enzyme complex |
| PFOR |
Pyruvate-ferredoxin oxidoreductase |
| RPB1 |
DNA-directed RNA polymerase II subunit beta I |
| TPP |
Thiamine pyrophosphate |
| TUBa |
Tubulin alpha chain |
References
- Abd El-Fadeal, N. M., M. S. Nafie, K. E.-K. M, A. El-Mistekawy, H. M. F. Mohammad, A. M. Elbahaie, A. A. Hashish, S. Y. Alomar, S. Y. Aloyouni, M. El-Dosoky, K. M. Morsy and S. A. Zaitone (2021). "Antitumor Activity of Nitazoxanide against Colon Cancers: Molecular Docking and Experimental Studies Based on Wnt/β-Catenin Signaling Inhibition." Int J Mol Sci 22(10). [CrossRef]
- Ackerley, D. F., C. F. Gonzalez, M. Keyhan, R. Blake, 2nd and A. Matin (2004). "Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction." Environ Microbiol 6(8): 851-860. [CrossRef]
- Adl, S. M., A. G. Simpson, C. E. Lane, J. Lukeš, D. Bass, S. S. Bowser, M. W. Brown, F. Burki, M. Dunthorn, V. Hampl, A. Heiss, M. Hoppenrath, E. Lara, L. Le Gall, D. H. Lynn, H. McManus, E. A. Mitchell, S. E. Mozley-Stanridge, L. W. Parfrey, J. Pawlowski, S. Rueckert, L. Shadwick, C. L. Schoch, A. Smirnov and F. W. Spiegel (2012). "The revised classification of eukaryotes." J Eukaryot Microbiol 59(5): 429-493. [CrossRef]
- Alsmark, U. C., T. Sicheritz-Ponten, P. G. Foster, R. P. Hirt and T. M. Embley (2009). "Horizontal gene transfer in eukaryotic parasites: a case study of Entamoeba histolytica and Trichomonas vaginalis." Methods Mol Biol 532: 489-500. [CrossRef]
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman (1997). "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs." Nucleic Acids Res 25(17): 3389-3402. [CrossRef]
- Altschul, S. F., J. C. Wootton, E. M. Gertz, R. Agarwala, A. Morgulis, A. A. Schäffer and Y. K. Yu (2005). "Protein database searches using compositionally adjusted substitution matrices." Febs j 272(20): 5101-5109. [CrossRef]
- Andersson, J. O. (2011). "Evolution of patchily distributed proteins shared between eukaryotes and prokaryotes: Dictyostelium as a case study." J Mol Microbiol Biotechnol 20(2): 83-95. [CrossRef]
- Ansell, B. R., L. Baker, S. J. Emery, M. J. McConville, S. G. Svärd, R. B. Gasser and A. R. Jex (2017). "Transcriptomics Indicates Active and Passive Metronidazole Resistance Mechanisms in Three Seminal Giardia Lines." Front Microbiol 8: 398. [CrossRef]
- Benchimol, M. (2009). "Hydrogenosomes under microscopy." Tissue Cell 41(3): 151-168. [CrossRef]
- Benchimol, M., J. C. Almeida and W. de Souza (1996). "Further studies on the organization of the hydrogenosome in Tritrichomonas foetus." Tissue Cell 28(3): 287-299. [CrossRef]
- Benchimol, M., I. de Andrade Rosa, R. da Silva Fontes and A. J. Burla Dias (2008). "Trichomonas adhere and phagocytose sperm cells: adhesion seems to be a prominent stage during interaction." Parasitol Res 102(4): 597-604. [CrossRef]
- Bendesky, A., D. Menéndez and P. Ostrosky-Wegman (2002). "Is metronidazole carcinogenic?" Mutat Res 511(2): 133-144. [CrossRef]
- Bharti, C., S. Sharma, N. Goswami, H. Sharma, S. A. Rabbani and S. Kumar (2021). "Role of nitazoxanide as a repurposed drug in the treatment and management of various diseases." Drugs Today (Barc) 57(7): 455-473. [CrossRef]
- Blamey, J. M. and M. W. Adams (1993). "Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus." Biochim Biophys Acta 1161(1): 19-27. [CrossRef]
- Bontell, I. L., N. Hall, K. E. Ashelford, J. P. Dubey, J. P. Boyle, J. Lindh and J. E. Smith (2009). "Whole genome sequencing of a natural recombinant Toxoplasma gondii strain reveals chromosome sorting and local allelic variants." Genome Biol 10(5): R53. [CrossRef]
- Boxma, B., R. M. de Graaf, G. W. van der Staay, T. A. van Alen, G. Ricard, T. Gabaldón, A. H. van Hoek, S. Y. Moon-van der Staay, W. J. Koopman, J. J. van Hellemond, A. G. Tielens, T. Friedrich, M. Veenhuis, M. A. Huynen and J. H. Hackstein (2005). "An anaerobic mitochondrion that produces hydrogen." Nature 434(7029): 74-79. [CrossRef]
- Bradic, M., S. D. Warring, G. E. Tooley, P. Scheid, W. E. Secor, K. M. Land, P. J. Huang, T. W. Chen, C. C. Lee, P. Tang, S. A. Sullivan and J. M. Carlton (2017). "Genetic Indicators of Drug Resistance in the Highly Repetitive Genome of Trichomonas vaginalis." Genome Biol Evol 9(6): 1658-1672. [CrossRef]
- Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, J. R. Wortman, S. L. Bidwell, U. C. Alsmark, S. Besteiro, T. Sicheritz-Ponten, C. J. Noel, J. B. Dacks, P. G. Foster, C. Simillion, Y. Van de Peer, D. Miranda-Saavedra, G. J. Barton, G. D. Westrop, S. Müller, D. Dessi, P. L. Fiori, Q. Ren, I. Paulsen, H. Zhang, F. D. Bastida-Corcuera, A. Simoes-Barbosa, M. T. Brown, R. D. Hayes, M. Mukherjee, C. Y. Okumura, R. Schneider, A. J. Smith, S. Vanacova, M. Villalvazo, B. J. Haas, M. Pertea, T. V. Feldblyum, T. R. Utterback, C. L. Shu, K. Osoegawa, P. J. de Jong, I. Hrdy, L. Horvathova, Z. Zubacova, P. Dolezal, S. B. Malik, J. M. Logsdon, Jr., K. Henze, A. Gupta, C. C. Wang, R. L. Dunne, J. A. Upcroft, P. Upcroft, O. White, S. L. Salzberg, P. Tang, C. H. Chiu, Y. S. Lee, T. M. Embley, G. H. Coombs, J. C. Mottram, J. Tachezy, C. M. Fraser-Liggett and P. J. Johnson (2007). "Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis." Science 315(5809): 207-212. [CrossRef]
- Chabrière, E., M. H. Charon, A. Volbeda, L. Pieulle, E. C. Hatchikian and J. C. Fontecilla-Camps (1999). "Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate." Nat Struct Biol 6(2): 182-190. [CrossRef]
- Chapwanya, A., A. Y. Usman and P. C. Irons (2016). "Comparative aspects of immunity and vaccination in human and bovine trichomoniasis: a review." Trop Anim Health Prod 48(1): 1-7. [CrossRef]
- Charon, M. H., A. Volbeda, E. Chabriere, L. Pieulle and J. C. Fontecilla-Camps (1999). "Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase." Curr Opin Struct Biol 9(6): 663-669. [CrossRef]
- Chung, M. C., P. L. Bosquesi and J. L. dos Santos (2011). "A prodrug approach to improve the physico-chemical properties and decrease the genotoxicity of nitro compounds." Curr Pharm Des 17(32): 3515-3526. [CrossRef]
- Ctrnacta, V., J. G. Ault, F. Stejskal and J. S. Keithly (2006). "Localization of pyruvate:NADP+ oxidoreductase in sporozoites of Cryptosporidium parvum." J Eukaryot Microbiol 53(4): 225-231. [CrossRef]
- Dan, M., A. L. Wang and C. C. Wang (2000). "Inhibition of pyruvate-ferredoxin oxidoreductase gene expression in Giardia lamblia by a virus-mediated hammerhead ribozyme." Mol Microbiol 36(2): 447-456. [CrossRef]
- Daubin, V. and G. J. Szöllősi (2016). "Horizontal Gene Transfer and the History of Life." Cold Spring Harb Perspect Biol 8(4): a018036. [CrossRef]
- Deng, Y., H. Xu, Y. Su, S. Liu, L. Xu, Z. Guo, J. Wu, C. Cheng and J. Feng (2019). "Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis." BMC Genomics 20(1): 761. [CrossRef]
- Denoeud, F., M. Roussel, B. Noel, I. Wawrzyniak, C. Da Silva, M. Diogon, E. Viscogliosi, C. Brochier-Armanet, A. Couloux, J. Poulain, B. Segurens, V. Anthouard, C. Texier, N. Blot, P. Poirier, G. C. Ng, K. S. Tan, F. Artiguenave, O. Jaillon, J. M. Aury, F. Delbac, P. Wincker, C. P. Vivarès and H. El Alaoui (2011). "Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite." Genome Biol 12(3): R29. [CrossRef]
- Dingsdag, S. A. and N. Hunter (2018). "Metronidazole: an update on metabolism, structure-cytotoxicity and resistance mechanisms." J Antimicrob Chemother 73(2): 265-279. [CrossRef]
- Docampo, R., S. N. Moreno and R. P. Mason (1987). "Free radical intermediates in the reaction of pyruvate:ferredoxin oxidoreductase in Tritrichomonas foetus hydrogenosomes." J Biol Chem 262(26): 12417-12420. [CrossRef]
- Doolittle, W. F. (1998). "You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes." Trends Genet 14(8): 307-311. [CrossRef]
- Dubini, A., F. Mus, M. Seibert, A. R. Grossman and M. C. Posewitz (2009). "Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity." J Biol Chem 284(11): 7201-7213. [CrossRef]
- Dyall, S. D., W. Yan, M. G. Delgadillo-Correa, A. Lunceford, J. A. Loo, C. F. Clarke and P. J. Johnson (2004). "Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex." Nature 431(7012): 1103-1107. [CrossRef]
- Embley, T. M., M. van der Giezen, D. S. Horner, P. L. Dyal and P. Foster (2003). "Mitochondria and hydrogenosomes are two forms of the same fundamental organelle." Philos Trans R Soc Lond B Biol Sci 358(1429): 191-201; discussion 201-192. [CrossRef]
- Eme, L., E. Gentekaki, B. Curtis, J. M. Archibald and A. J. Roger (2017). "Lateral Gene Transfer in the Adaptation of the Anaerobic Parasite Blastocystis to the Gut." Curr Biol 27(6): 807-820. [CrossRef]
- Emelyanov, V. V. and A. V. Goldberg (2011). "Fermentation enzymes of Giardia intestinalis, pyruvate:ferredoxin oxidoreductase and hydrogenase, do not localize to its mitosomes." Microbiology (Reading) 157(Pt 6): 1602-1611. [CrossRef]
- Felsenstein, J. (1985). "Confidence limits on phylogenies: An approach using the bootstrap." Evolution 39(4): 783-791. [CrossRef]
- Field, J., B. Rosenthal and J. Samuelson (2000). "Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica." Mol Microbiol 38(3): 446-455. [CrossRef]
- Fleck, L. E., E. J. North, R. E. Lee, L. R. Mulcahy, G. Casadei and K. Lewis (2014). "A screen for and validation of prodrug antimicrobials." Antimicrob Agents Chemother 58(3): 1410-1419. [CrossRef]
- Freeman, C. D., N. E. Klutman and K. C. Lamp (1997). "Metronidazole. A therapeutic review and update." Drugs 54(5): 679-708. [CrossRef]
- Fritz-Laylin, L. K., S. E. Prochnik, M. L. Ginger, J. B. Dacks, M. L. Carpenter, M. C. Field, A. Kuo, A. Paredez, J. Chapman, J. Pham, S. Shu, R. Neupane, M. Cipriano, J. Mancuso, H. Tu, A. Salamov, E. Lindquist, H. Shapiro, S. Lucas, I. V. Grigoriev, W. Z. Cande, C. Fulton, D. S. Rokhsar and S. C. Dawson (2010). "The genome of Naegleria gruberi illuminates early eukaryotic versatility." Cell 140(5): 631-642. [CrossRef]
- Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser and B. Barrell (2002). "Genome sequence of the human malaria parasite Plasmodium falciparum." Nature 419(6906): 498-511. [CrossRef]
- Gargala, G. (2008). "Drug treatment and novel drug target against Cryptosporidium." Parasite 15(3): 275-281. [CrossRef]
- Gomaa, F., D. R. Utter, C. Powers, D. J. Beaudoin, V. P. Edgcomb, H. L. Filipsson, C. M. Hansel, S. D. Wankel, Y. Zhang and J. M. Bernhard (2021). "Multiple integrated metabolic strategies allow foraminiferan protists to thrive in anoxic marine sediments." Sci Adv 7(22). [CrossRef]
- Hackstein, J. H., A. Akhmanova, B. Boxma, H. R. Harhangi and F. G. Voncken (1999). "Hydrogenosomes: eukaryotic adaptations to anaerobic environments." Trends Microbiol 7(11): 441-447. [CrossRef]
- Hedges, S. B., H. Chen, S. Kumar, D. Y. Wang, A. S. Thompson and H. Watanabe (2001). "A genomic timescale for the origin of eukaryotes." BMC Evol Biol 1: 4. [CrossRef]
- Higuera, A., X. Villamizar, G. Herrera, J. C. Giraldo, A. L. Vasquez, P. Urbano, O. Villalobos, C. Tovar and J. D. Ramírez (2020). "Molecular detection and genotyping of intestinal protozoa from different biogeographical regions of Colombia." PeerJ 8: e8554. [CrossRef]
- Hoffman, P. S., G. Sisson, M. A. Croxen, K. Welch, W. D. Harman, N. Cremades and M. G. Morash (2007). "Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni." Antimicrob Agents Chemother 51(3): 868-876. [CrossRef]
- Horner, D. S., R. P. Hirt and T. M. Embley (1999). "A single eubacterial origin of eukaryotic pyruvate: ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes." Mol Biol Evol 16(9): 1280-1291. [CrossRef]
- Hrdý, I., R. Cammack, P. Stopka, J. Kulda and J. Tachezy (2005). "Alternative pathway of metronidazole activation in Trichomonas vaginalis hydrogenosomes." Antimicrob Agents Chemother 49(12): 5033-5036. [CrossRef]
- Hrdy, I., R. P. Hirt, P. Dolezal, L. Bardonová, P. G. Foster, J. Tachezy and T. M. Embley (2004). "Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I." Nature 432(7017): 618-622. [CrossRef]
- Hrdý, I. and M. Müller (1995). "Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis." J Mol Evol 41(3): 388-396. [CrossRef]
- Huang, J., N. Mullapudi, T. Sicheritz-Ponten and J. C. Kissinger (2004). "A first glimpse into the pattern and scale of gene transfer in Apicomplexa." Int J Parasitol 34(3): 265-274. [CrossRef]
- Hucho, F. (1975). "The pyruvate dehydrogenase multienzyme complex." Angew Chem Int Ed Engl 14(9): 591-601. [CrossRef]
- Hug, L. A., A. Stechmann and A. J. Roger (2010). "Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes." Mol Biol Evol 27(2): 311-324. [CrossRef]
- Husnik, F. and J. P. McCutcheon (2018). "Functional horizontal gene transfer from bacteria to eukaryotes." Nat Rev Microbiol 16(2): 67-79. [CrossRef]
- Inui, H. and Y. Nakano (1999). "[Vitamin B1]." Nihon Rinsho 57(10): 2187-2192.
- Johnson, P. J., C. J. Lahti and P. J. Bradley (1993). "Biogenesis of the hydrogenosome in the anaerobic protist Trichomonas vaginalis." J Parasitol 79(5): 664-670. [CrossRef]
- Juhas, M. (2015). "Horizontal gene transfer in human pathogens." Crit Rev Microbiol 41(1): 101-108. [CrossRef]
- Keeling, P. J. (2009). "Functional and ecological impacts of horizontal gene transfer in eukaryotes." Curr Opin Genet Dev 19(6): 613-619. [CrossRef]
- Keeling, P. J. and J. D. Palmer (2008). "Horizontal gene transfer in eukaryotic evolution." Nat Rev Genet 9(8): 605-618. [CrossRef]
- Kennedy, A. J., A. M. Bruce, C. Gineste, T. E. Ballard, I. N. Olekhnovich, T. L. Macdonald and P. S. Hoffman (2016). "Synthesis and Antimicrobial Evaluation of Amixicile-Based Inhibitors of the Pyruvate-Ferredoxin Oxidoreductases of Anaerobic Bacteria and Epsilonproteobacteria." Antimicrob Agents Chemother 60(7): 3980-3987. [CrossRef]
- Kiehl, I. G. A., E. Riccetto, A. C. C. Salustiano, M. V. Ossick, K. L. Ferrari, H. B. Assalin, O. Ikari and L. O. Reis (2021). "Boosting bladder cancer treatment by intravesical nitazoxanide and bacillus calmette-guérin association." World J Urol 39(4): 1187-1194. [CrossRef]
- Kulda, J. (1999). "Trichomonads, hydrogenosomes and drug resistance." Int J Parasitol 29(2): 199-212. [CrossRef]
- Kumar, S., G. Stecher, M. Li, C. Knyaz and K. Tamura (2018). "MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms." Mol Biol Evol 35(6): 1547-1549. [CrossRef]
- Kusdian, G. and S. B. Gould (2014). "The biology of Trichomonas vaginalis in the light of urogenital tract infection." Mol Biochem Parasitol 198(2): 92-99. [CrossRef]
- Lacroix, B. and V. Citovsky (2016). "Transfer of DNA from Bacteria to Eukaryotes." mBio 7(4). [CrossRef]
- Lacroix, B. and V. Citovsky (2018). "Beyond Agrobacterium-Mediated Transformation: Horizontal Gene Transfer from Bacteria to Eukaryotes." Curr Top Microbiol Immunol 418: 443-462. [CrossRef]
- Lamp, K. C., C. D. Freeman, N. E. Klutman and M. K. Lacy (1999). "Pharmacokinetics and pharmacodynamics of the nitroimidazole antimicrobials." Clin Pharmacokinet 36(5): 353-373. [CrossRef]
- Leitsch, D., A. G. Burgess, L. A. Dunn, K. G. Krauer, K. Tan, M. Duchêne, P. Upcroft, L. Eckmann and J. A. Upcroft (2011). "Pyruvate:ferredoxin oxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia." J Antimicrob Chemother 66(8): 1756-1765. [CrossRef]
- Lewis, W. H., A. E. Lind, K. M. Sendra, H. Onsbring, T. A. Williams, G. F. Esteban, R. P. Hirt, T. J. G. Ettema and T. M. Embley (2020). "Convergent Evolution of Hydrogenosomes from Mitochondria by Gene Transfer and Loss." Mol Biol Evol 37(2): 524-539. [CrossRef]
- Lindmark, D. G. and M. Müller (1973). "Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism." J Biol Chem 248(22): 7724-7728. [CrossRef]
- Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillén, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser and N. Hall (2005). "The genome of the protist parasite Entamoeba histolytica." Nature 433(7028): 865-868. [CrossRef]
- Lokhande, A. S. and P. V. Devarajan (2021). "A review on possible mechanistic insights of Nitazoxanide for repurposing in COVID-19." Eur J Pharmacol 891: 173748. [CrossRef]
- Lopes-Oliveira, L. A. P., M. Fantinatti and A. M. Da-Cruz (2020). "In vitro-induction of metronidazole-resistant Giardia duodenalis is not associated with nucleotide alterations in the genes involved in pro-drug activation." Mem Inst Oswaldo Cruz 115: e200303. [CrossRef]
- Makiuchi, T. and T. Nozaki (2014). "Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa." Biochimie 100: 3-17. [CrossRef]
- Marczak, R., T. E. Gorrell and M. Müller (1983). "Hydrogenosomal ferredoxin of the anaerobic protozoon, Tritrichomonas foetus." J Biol Chem 258(20): 12427-12433. [CrossRef]
- Martinez, V. and E. Caumes (2001). "[Metronidazole]." Ann Dermatol Venereol 128(8-9): 903-909.
- Mastronicola, D., M. Falabella, E. Forte, F. Testa, P. Sarti and A. Giuffrè (2016). "Antioxidant defence systems in the protozoan pathogen Giardia intestinalis." Mol Biochem Parasitol 206(1-2): 56-66. [CrossRef]
- Meneses Calderón, J., M. D. R. Figueroa Flores, L. Paniagua Coria, J. C. Briones Garduño, J. Meneses Figueroa, M. J. Vargas Contretas, L. De la Cruz Ávila, S. Díaz Meza, R. Ramírez Chacón, S. Padmanabhan and H. Mendieta Zerón (2020). "Nitazoxanide against COVID-19 in three explorative scenarios." J Infect Dev Ctries 14(9): 982-986. [CrossRef]
- Moreno, S. N. and R. Docampo (1985). "Mechanism of toxicity of nitro compounds used in the chemotherapy of trichomoniasis." Environ Health Perspect 64: 199-208. [CrossRef]
- Müller, J., S. Braga, M. Heller and N. Müller (2019). "Resistance formation to nitro drugs in Giardia lamblia: No common markers identified by comparative proteomics." Int J Parasitol Drugs Drug Resist 9: 112-119. [CrossRef]
- nergy metabolism of anaerobic protists-a special case in eukaryotic evolution." Evolutionary relationships among protozoa.
- Müller, M. (1993). "The hydrogenosome." J Gen Microbiol 139(12): 2879-2889. [CrossRef]
- Narikawa, S. (1986). "Distribution of metronidazole susceptibility factors in obligate anaerobes." J Antimicrob Chemother 18(5): 565-574. [CrossRef]
- Nepali, K., H. Y. Lee and J. P. Liou (2019). "Nitro-Group-Containing Drugs." J Med Chem 62(6): 2851-2893. [CrossRef]
- Nixon, J. E., A. Wang, J. Field, H. G. Morrison, A. G. McArthur, M. L. Sogin, B. J. Loftus and J. Samuelson (2002). "Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica." Eukaryot Cell 1(2): 181-190. [CrossRef]
- Noth, J., D. Krawietz, A. Hemschemeier and T. Happe (2013). "Pyruvate:ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii." J Biol Chem 288(6): 4368-4377. [CrossRef]
- Ocaña, K. A. and A. M. Dávila (2011). "Phylogenomics-based reconstruction of protozoan species tree." Evol Bioinform Online 7: 107-121. [CrossRef]
- Pal, A. K., M. Nandave and G. Kaithwas (2020). "Chemoprophylactic activity of nitazoxanide in experimental model of mammary gland carcinoma in rats." 3 Biotech 10(8): 338. [CrossRef]
- Pal, D., S. Banerjee, J. Cui, A. Schwartz, S. K. Ghosh and J. Samuelson (2009). "Giardia, Entamoeba, and Trichomonas enzymes activate metronidazole (nitroreductases) and inactivate metronidazole (nitroimidazole reductases)." Antimicrob Agents Chemother 53(2): 458-464. [CrossRef]
- E. Boucher and I. B. Lambert (2002). "Regulation of the nfsA Gene in Escherichia coli by SoxS." J Bacteriol 184(1): 51-58. [CrossRef]
- Peterson, F. J., R. P. Mason, J. Hovsepian and J. L. Holtzman (1979). "Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes." J Biol Chem 254(10): 4009-4014. [CrossRef]
- Rada, P., P. Doležal, P. L. Jedelský, D. Bursac, A. J. Perry, M. Šedinová, K. Smíšková, M. Novotný, N. C. Beltrán, I. Hrdý, T. Lithgow and J. Tachezy (2011). "The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis." PLoS One 6(9): e24428. [CrossRef]
- Ragsdale, S. W. (2003). "Pyruvate ferredoxin oxidoreductase and its radical intermediate." Chem Rev 103(6): 2333-2346. [CrossRef]
- Raj, D., E. Ghosh, A. K. Mukherjee, T. Nozaki and S. Ganguly (2014). "Differential gene expression in Giardia lamblia under oxidative stress: significance in eukaryotic evolution." Gene 535(2): 131-139. [CrossRef]
- Rasoloson, D., S. Vanacova, E. Tomkova, J. Razga, I. Hrdy, J. Tachezy and J. Kulda (2002). "Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis." Microbiology (Reading) 148(Pt 8): 2467-2477. [CrossRef]
- Rau, J. and A. Stolz (2003). "Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent Azo reductases." Appl Environ Microbiol 69(6): 3448-3455. [CrossRef]
- Ricard, G., N. R. McEwan, B. E. Dutilh, J. P. Jouany, D. Macheboeuf, M. Mitsumori, F. M. McIntosh, T. Michalowski, T. Nagamine, N. Nelson, C. J. Newbold, E. Nsabimana, A. Takenaka, N. A. Thomas, K. Ushida, J. H. Hackstein and M. A. Huynen (2006). "Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment." BMC Genomics 7: 22. [CrossRef]
- Rocco, P. R. M., P. L. Silva, F. F. Cruz, M. A. C. Melo-Junior, P. Tierno, M. A. Moura, L. F. G. De Oliveira, C. C. Lima, E. A. Dos Santos, W. F. Junior, A. Fernandes, K. G. Franchini, E. Magri, N. F. de Moraes, J. M. J. Gonçalves, M. N. Carbonieri, I. S. Dos Santos, N. F. Paes, P. V. M. Maciel, R. P. Rocha, A. F. de Carvalho, P. A. Alves, J. L. Proença-Módena, A. T. Cordeiro, D. B. B. Trivella, R. E. Marques, R. R. Luiz, P. Pelosi and E. S. J. R. Lapa (2021). "Early use of nitazoxanide in mild COVID-19 disease: randomised, placebo-controlled trial." Eur Respir J 58(1). [CrossRef]
- Rodríguez, M. A., R. M. García-Pérez, L. Mendoza, T. Sánchez, N. Guillen and E. Orozco (1998). "The pyruvate:ferredoxin oxidoreductase enzyme is located in the plasma membrane and in a cytoplasmic structure in Entamoeba." Microb Pathog 25(1): 1-10. [CrossRef]
- Roger, A. J., C. G. Clark and W. F. Doolittle (1996). "A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis." Proc Natl Acad Sci U S A 93(25): 14618-14622. [CrossRef]
- Roldán, M. D., E. Pérez-Reinado, F. Castillo and C. Moreno-Vivián (2008). "Reduction of polynitroaromatic compounds: the bacterial nitroreductases." FEMS Microbiol Rev 32(3): 474-500. [CrossRef]
- Rosenthal, B., Z. Mai, D. Caplivski, S. Ghosh, H. de la Vega, T. Graf and J. Samuelson (1997). "Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoan parasite Entamoeba histolytica." J Bacteriol 179(11): 3736-3745. [CrossRef]
- Rotte, C., F. Stejskal, G. Zhu, J. S. Keithly and W. Martin (2001). "Pyruvate : NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists." Mol Biol Evol 18(5): 710-720. [CrossRef]
- Saitou, N. and M. Nei (1987). "The neighbor-joining method: a new method for reconstructing phylogenetic trees." Mol Biol Evol 4(4): 406-425.
- Schönknecht, G., W. H. Chen, C. M. Ternes, G. G. Barbier, R. P. Shrestha, M. Stanke, A. Bräutigam, B. J. Baker, J. F. Banfield, R. M. Garavito, K. Carr, C. Wilkerson, S. A. Rensing, D. Gagneul, N. E. Dickenson, C. Oesterhelt, M. J. Lercher and A. P. Weber (2013). "Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote." Science 339(6124): 1207-1210. [CrossRef]
- Scior, T., J. Lozano-Aponte, S. Ajmani, E. Hernández-Montero, F. Chávez-Silva, E. Hernández-Núñez, R. Moo-Puc, A. Fraguela-Collar and G. Navarrete-Vázquez (2015). "Antiprotozoal Nitazoxanide Derivatives: Synthesis, Bioassays and QSAR Study Combined with Docking for Mechanistic Insight." Curr Comput Aided Drug Des 11(1): 21-31. [CrossRef]
- Shakya, A., H. R. Bhat and S. K. Ghosh (2018). "Update on Nitazoxanide: A Multifunctional Chemotherapeutic Agent." Curr Drug Discov Technol 15(3): 201-213. [CrossRef]
- Sibbald, S. J., L. Eme, J. M. Archibald and A. J. Roger (2020). "Lateral Gene Transfer Mechanisms and Pan-genomes in Eukaryotes." Trends Parasitol 36(11): 927-941. [CrossRef]
- Sisson, G., A. Goodwin, A. Raudonikiene, N. J. Hughes, A. K. Mukhopadhyay, D. E. Berg and P. S. Hoffman (2002). "Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori." Antimicrob Agents Chemother 46(7): 2116-2123. [CrossRef]
- Soucy, S. M., J. Huang and J. P. Gogarten (2015). "Horizontal gene transfer: building the web of life." Nat Rev Genet 16(8): 472-482. [CrossRef]
- Upcroft, P. and J. A. Upcroft (2001). "Drug targets and mechanisms of resistance in the anaerobic protozoa." Clin Microbiol Rev 14(1): 150-164. [CrossRef]
- Van Etten, J. and D. Bhattacharya (2020). "Horizontal Gene Transfer in Eukaryotes: Not if, but How Much?" Trends Genet 36(12): 915-925. [CrossRef]
- van Lis, R., C. Baffert, Y. Couté, W. Nitschke and A. Atteia (2013). "Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate:ferredoxin oxidoreductase that functions with FDX1." Plant Physiol 161(1): 57-71. [CrossRef]
- Warren, C. A., E. van Opstal, T. E. Ballard, A. Kennedy, X. Wang, M. Riggins, I. Olekhnovich, M. Warthan, G. L. Kolling, R. L. Guerrant, T. L. Macdonald and P. S. Hoffman (2012). "Amixicile, a novel inhibitor of pyruvate: ferredoxin oxidoreductase, shows efficacy against Clostridium difficile in a mouse infection model." Antimicrob Agents Chemother 56(8): 4103-4111. [CrossRef]
- Wawrzyniak, I., M. Roussel, M. Diogon, A. Couloux, C. Texier, K. S. Tan, C. P. Vivarès, F. Delbac, P. Wincker and H. El Alaoui (2008). "Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis." Int J Parasitol 38(12): 1377-1382. [CrossRef]
- Wyres, K. L., R. R. Wick, L. M. Judd, R. Froumine, A. Tokolyi, C. L. Gorrie, M. M. C. Lam, S. Duchêne, A. Jenney and K. E. Holt (2019). "Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae." PLoS Genet 15(4): e1008114. [CrossRef]
- Xiong, J., G. Wang, J. Cheng, M. Tian, X. Pan, A. Warren, C. Jiang, D. Yuan and W. Miao (2015). "Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer." Sci Rep 5: 15470. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).