1. Introduction
Gut-colonization by multi-drug resistant organisms (MDROs) has been reported as an independent risk factor for bloodstream infection (BSI) with high mortality among allogeneic hematopoietic stem cell transplant (HSCT) patients[
1,
2]. Strategies for reducing MDROs colonization and infection are challenging, and most studies have focused on the use of antimicrobial agents[
3]. Few studies have attempted to modulate the presence of MDROs in the intestinal microbiome[
4] by focusing on the individual’s diet[
5] or the use of prebiotics, probiotics and symbiotics[
6,
7]. However, this has rarely been reported in patients undergoing HSCT. Probiotics are live microorganisms that can provide benefit to the host if present in adequate doses[
8].
Usually, probiotics are not offered to immunocompromised patients, like those undergoing HSCT, due to a risk of translocation and infection. To date, only one study demonstrated the safety and feasibility of using a specific probiotic,
Lactobacillus plantarum, in pediatric neutropenic patients undergoing HSCT[
9]
. The presence of
Lactobacillus spp. in the fecal microbiota of hospitalized subjects has been associated with a lack of MDROs acquisition, indicating a potential protective role for these bacteria in HSCT[
10].
At our center, Ferreira et al.[
1] evaluated 232 HSCT patients between 2014 and 2015 and demonstrated, MDRO colonization rate of 30.5%. Bloodstream infection(BSI) occurred in 20% of the MDRO positive group, the authors observed by multivariate analysis that prior gut-colonization by gram negative MDROs was a risk factor for this outcome.
In the present study, we evaluated the use of L. plantarum (intervention) as a probiotic in patients undergoing HSCT who were colonized by MDROs. We hypothesized that this intervention would reduce MDRO colonization compared to an untreated control group and we evaluated as well the impact of the intervention on the gut microbiome.
2. Results
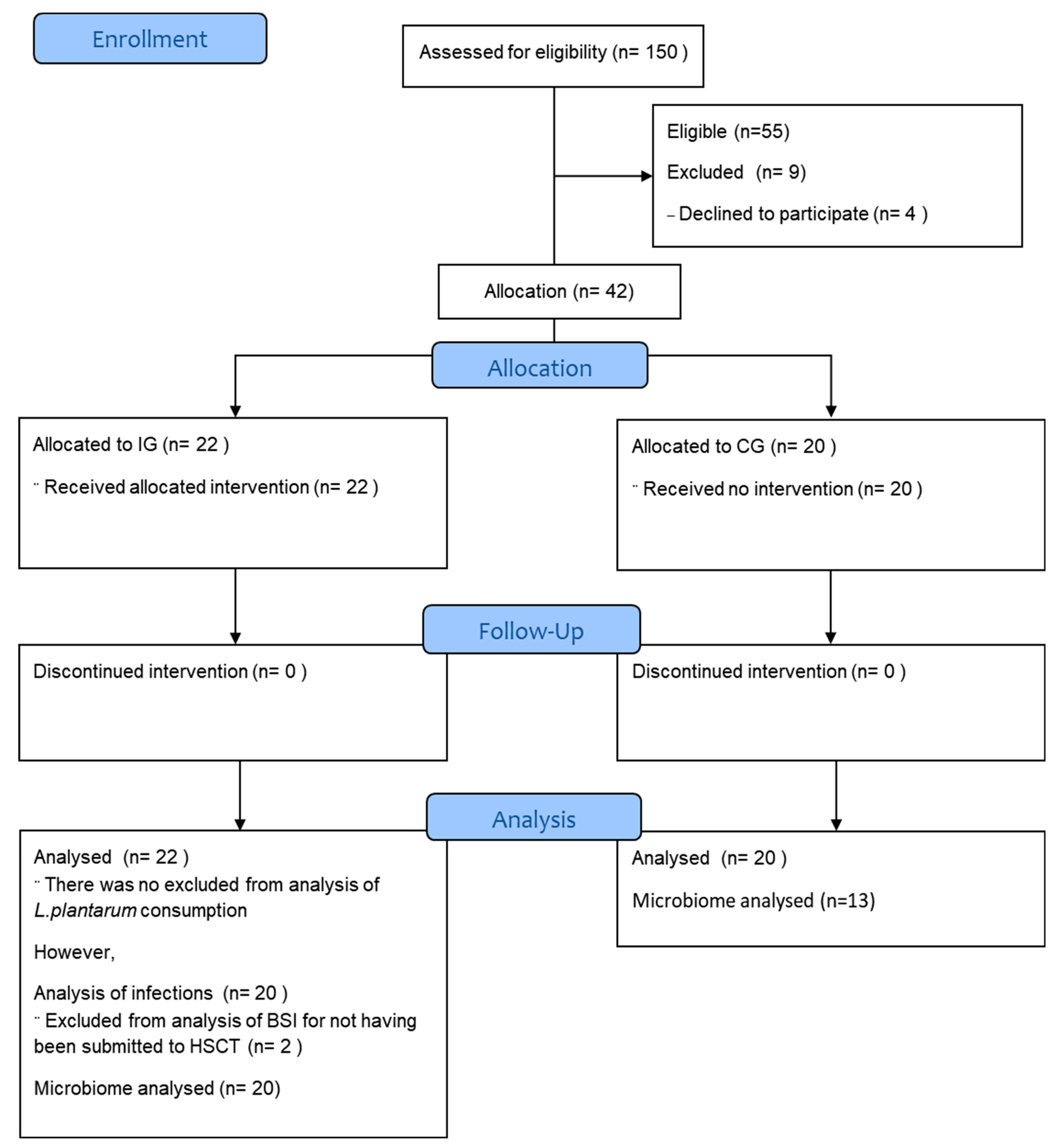
A total of 55 candidates for HSCT were selected for the study, nine were excluded for not having collected the baseline stool sample (
Figure 1). A final cohort of 42 participants was included in the study: 20 CG and 22 IG. However, we did not analyze the stool microbiome for all of them. Two participants in the IG did not undergo HSCT due to disease refractoriness and one participant was yet to undergo HSCT to the end of this study. Therefore, for the analysis of MDROs and modulation of the fecal microbiome following
L. plantarum ingestion there were 22 participants in the IG and 13 in the CG.
Table 2 shows patient and HSCT characteristics, including gastrointestinal toxicities observed during HSCT.
The analysis of L. plantarum capsules through NGS showed an abundance of Lactobacillaceae. When we compared the most abundant ASV in the samples with GenBank(NCBI), there was 99% identity with L. plantarum. Additionally, the Maldi-TOF analysis of the L. plantarum capsules showed high correspondence with L. plantarum. However, bacterial counts in culture medium demonstrated a deficit in the expected quantity of L. plantarum Colony Forming Units in the capsules, 6x108 ±3.45 x108 UFC instead of the requested 5x109 UFC. Therefore, we adjusted the number of probiotic capsules prescribed to the IG to one capsule to two twice a day.
The L. plantarum capsules were consumed an average of 86%(±11%) at 43(±29) days. Light flatulence (grade 1 in CTCAE) or moderate flatulence (grade 2) were each reported in three subjects. Of the 16 patients who reported Bristol Scale Stool type 3 before L. plantarum ingestion, only one remained at type 3 after its use, while the other 15 subsequently reported type 4 stools.
All patients included in the study were colonized by MDROs. In the evaluation before the onset of HSCT (median 6 days, IQR 7), 3/20(15.0%) individuals in the CG continued to be colonized by VRE- colonized. In the IG 6/22 patients(27.0%) continued to be colonized, three by VRE-colonized and the other three patients by Carbapenem-resistant Enterobacteriaceae, with a median consumption of L.plantarum was 17 days(IQR 7) and 47 days(IQR 36), respectively. The median number of days between the beginning of Lactobacillus and the decolonization evaluation was 19 days(IQR 47). There was not new colonization by other MDROs during the follow-up period for both groups.
Febrile neutropenia occurred in 16/20(80.0%) of the CG and 10/20(52.6%) in the IG(p=0.14). In the IG, seven patients developed MBI-LCBI and five patients had
Klebsiella pneumonia infection. Three of five-patient IG who developed MBI-LCBI by
Klebsiella pneumonia had been colonized by Carbapenem-resistant
K. pneumonia (
Table 3).
Stool samples were collected periodically from the CG (baseline n=13 and on week grafting n=10) and IG (baseline n=23, on the second week of
L. plantarum use n=22, at the time of neutropenia n=11, and one week before infection - n=4 and grafting n=10). The presence of
Lactobacillus was accessed on the different periods for the IG(
Supplementary Figure S1).
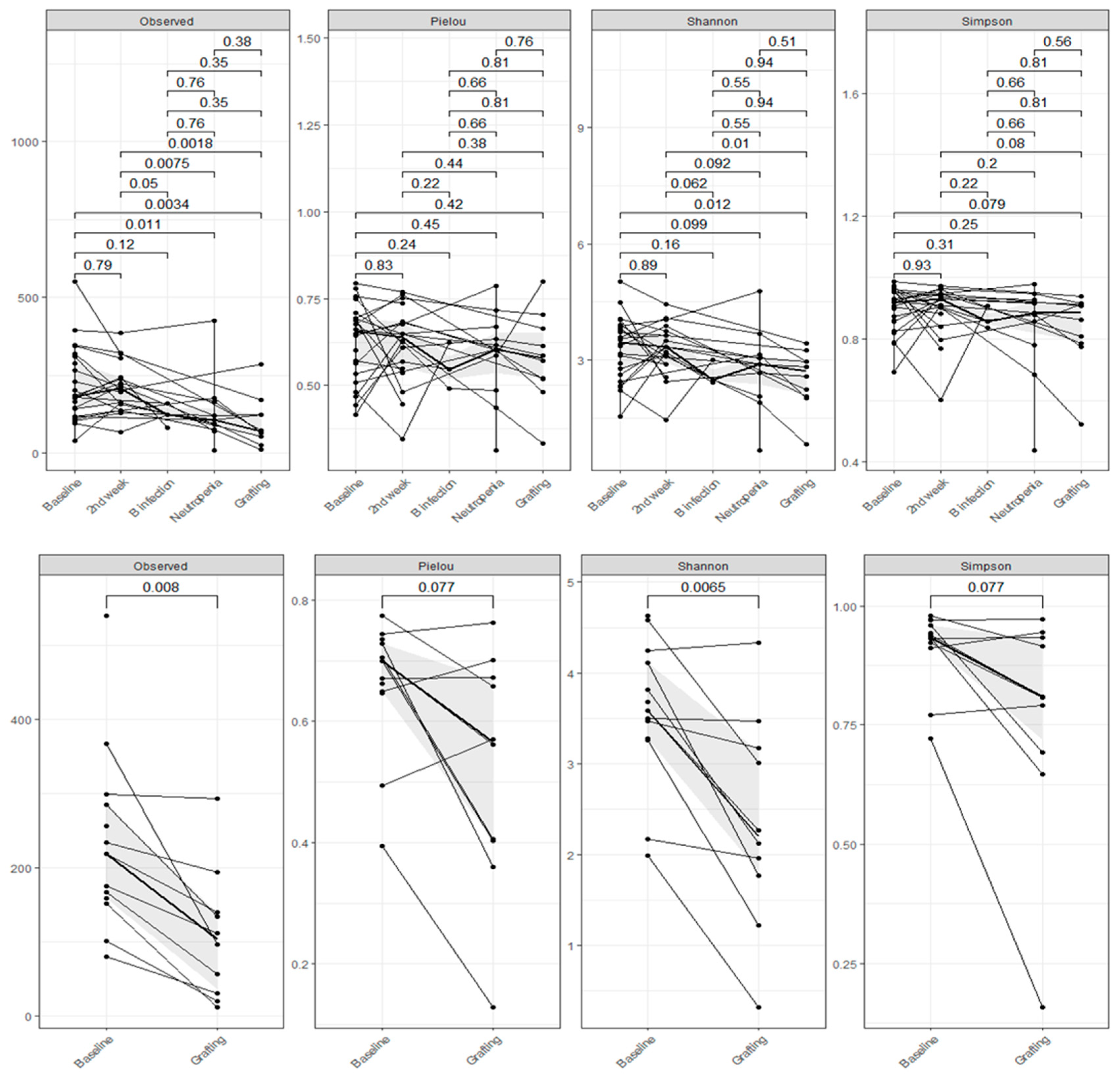
Overall, there was a reduction in alpha diversity (Observed ASVs, Shannon and Simpson) for the patients between the periods, regardless of the use of
L. plantarum, with significant values especially between baseline and second-week versus neutropenia and grafting (Wilcoxon p<0.05 and p<0.01). Additionally, we can see intense intraindividual variability in the alpha diversity between the time points. Alpha diversity by Faith was decrease between baseline and grafting in both groups (IG p=0.0026; CG p=0.018). (
Figure 2).
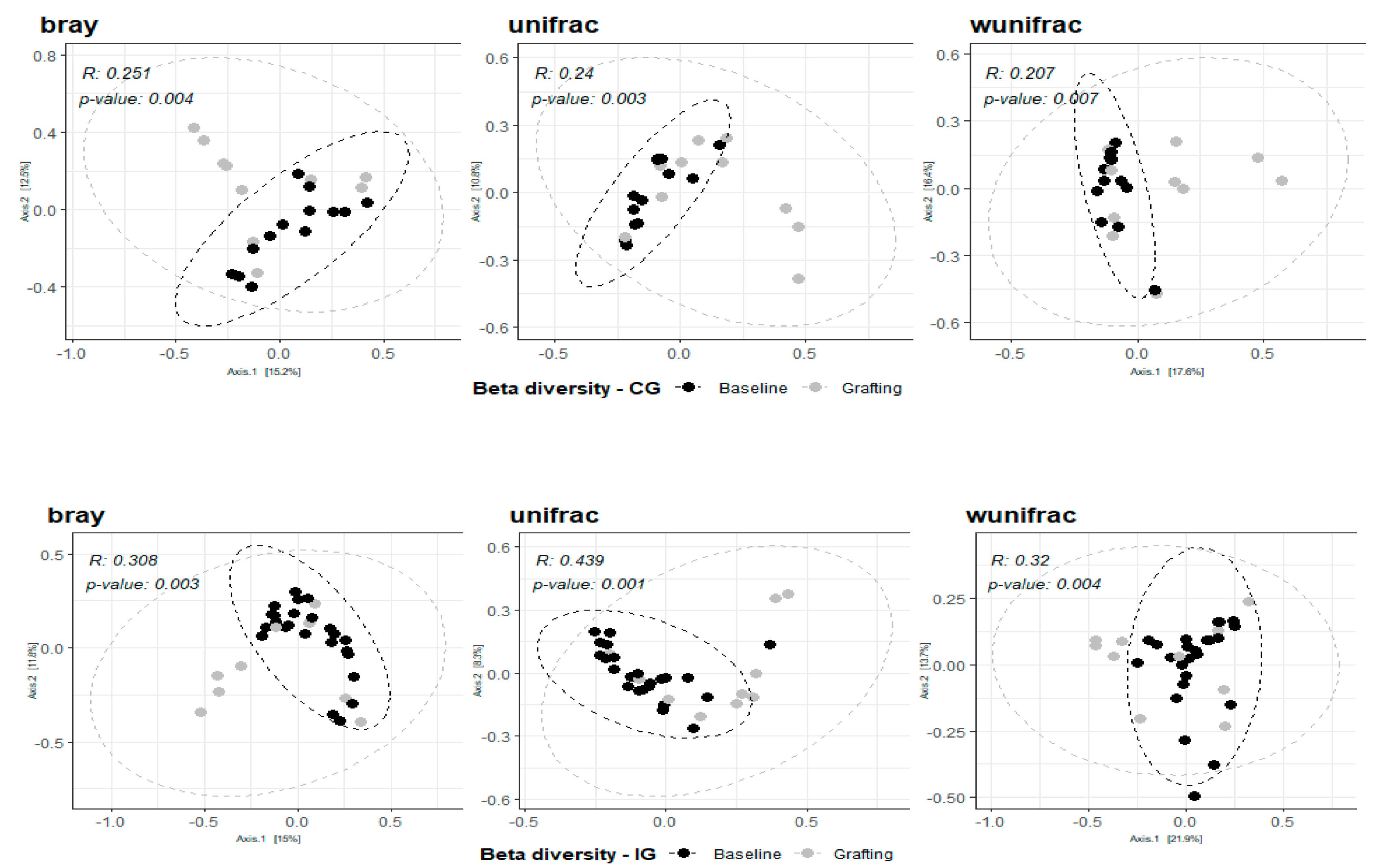
When analyzing the composition of both IG and CG, we observed a significant change in their bacteriome, qualitatively and quantitatively, between baseline and grafting (ANOSIM - bray, weighted and unweighted unifrac p< 0.05)(
Figure 3). On the other hand, when the bacterial composition of both groups was compared on these two points, no significant differences were found (IG x CG at baseline – ANOSIM unifrac
P=0.409, R=0.007; bray
P= 0.181, R=0.057; wunifrac
P= 0.37, R=0.01); (IG x CG at grafting – ANOSIM unifrac
P=0.868, R=-0.056; bray
P= 0.569, R=-0.022; wunifrac
P= 0.837, R=-0.057).
At baseline, at least 50.9% of bacteria were predominantly from the phyla Bacteroidetes, followed by Firmicutes(38.3%) and Proteobacteria(4.5%). These phyla were still predominant at grafting for both IG and CG. Proteobacteria increased in both groups between baseline and grafting (p=0.009), without significant differences between groups (p=0.41). The detection of Firmicutes and Bacteroidetes phyla was comparable in the IG and CG at baseline (p=0.18 and p=0.72, respectively) and following grafting (p=0.50 and p=0.14, respectively).
Stool samples from four IG’s patients who developed BSI were sequenced a week before the infection event. One patient with K. pneumoniae had an intestinal infection dominated by Enterobacteriaceae. Two other patients had an increase of the Bacteroides genus, one of them had BSI by E. coli ESBL-positive and the other one by carbapenem-resistant K. pneumonia.
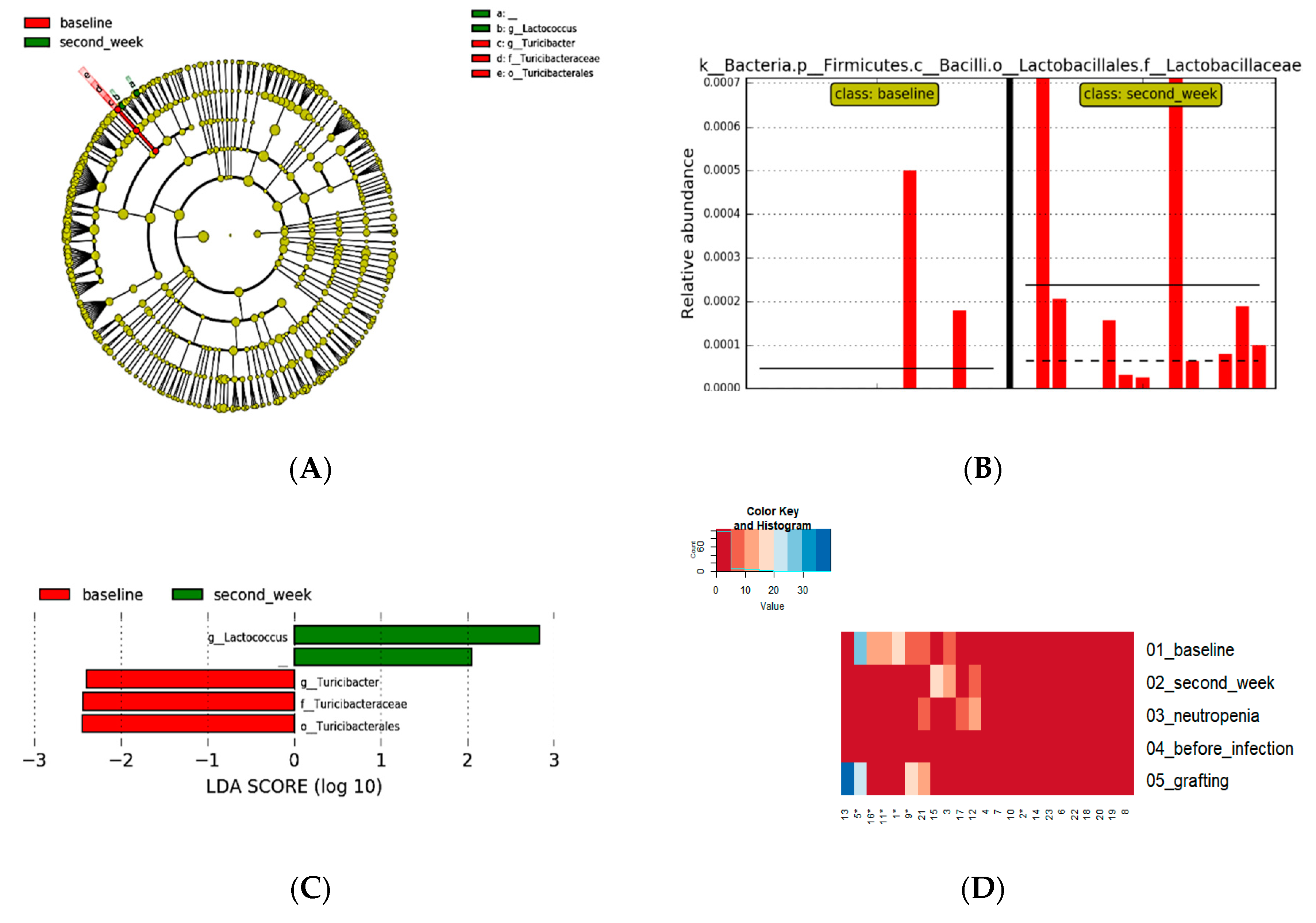
The consumption of
L. plantarum was associated with an increased presence of the
Lactobacillales(p=0.004) in second-week period. Differences between baseline and second week measured by LefSe with alpha value for the factorial Kruskal-Wallis test p<0.05 and LDA 2.0, showed an increase of
Lactococcus, with a decrease of
Turicibacter (
Figure 4).
The
Lactobacillaceae family was also identified as differently abundant when the alpha value for the factorial Kruskal-Wallis test was p<0.01 and the LDA was 2.0. Whereas the LDA score >3.0 did not present differential features with statistical significance (see Suplementary
Figure 2). Although no differently abundant bacteria were identified between IG and CG, both groups presented a decrease in the presence of
Roseburia and
Coprococcus, when baseline and grafting were compared
Figure 5).
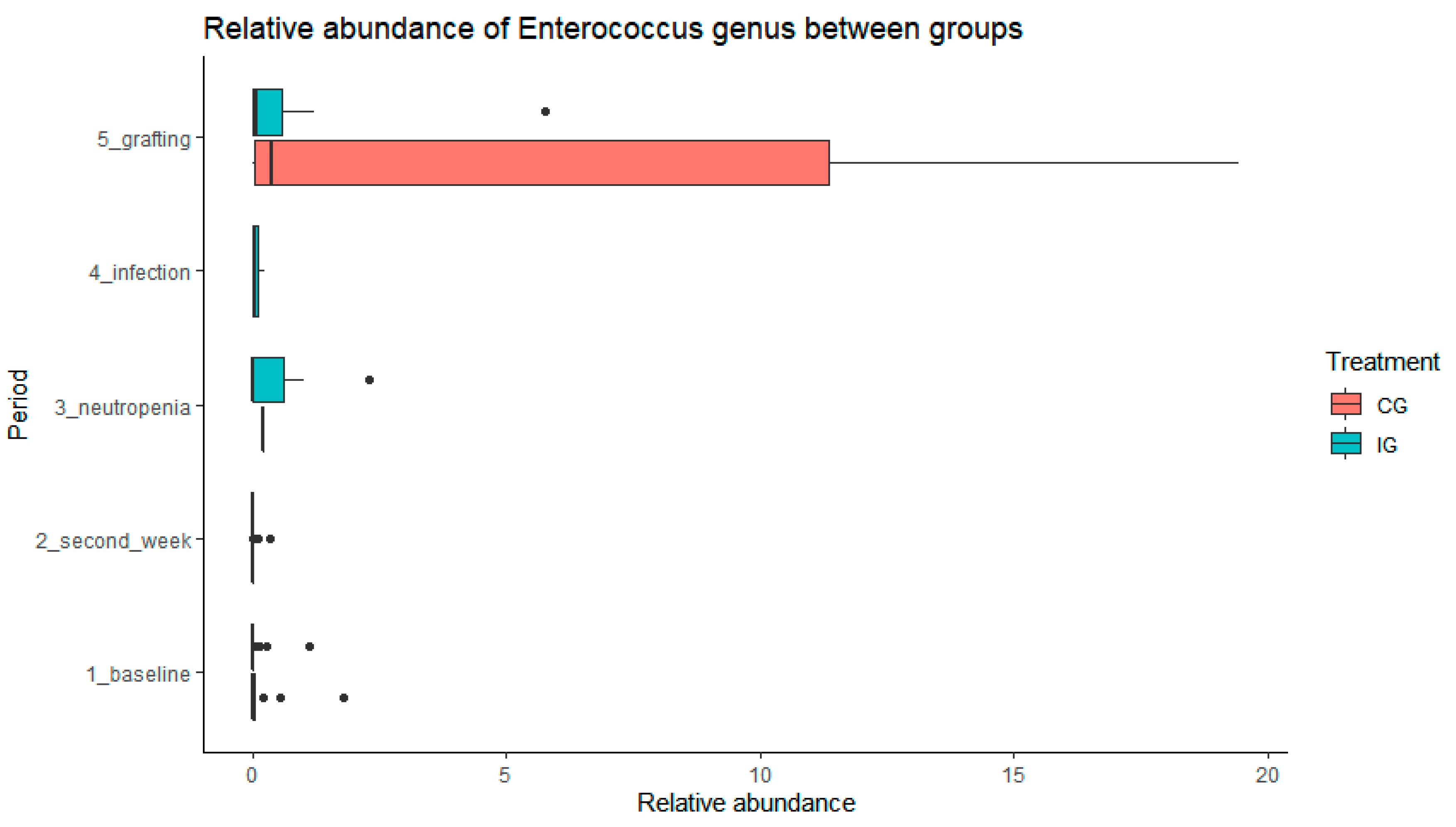
We observed an increase of abundance of
Enterococcus in the CG with trend to significance compared with the IG (p=0,07) in paired sample analysis, considering patients with stool collected on both periods (baseline and grafting)
Figure 6.
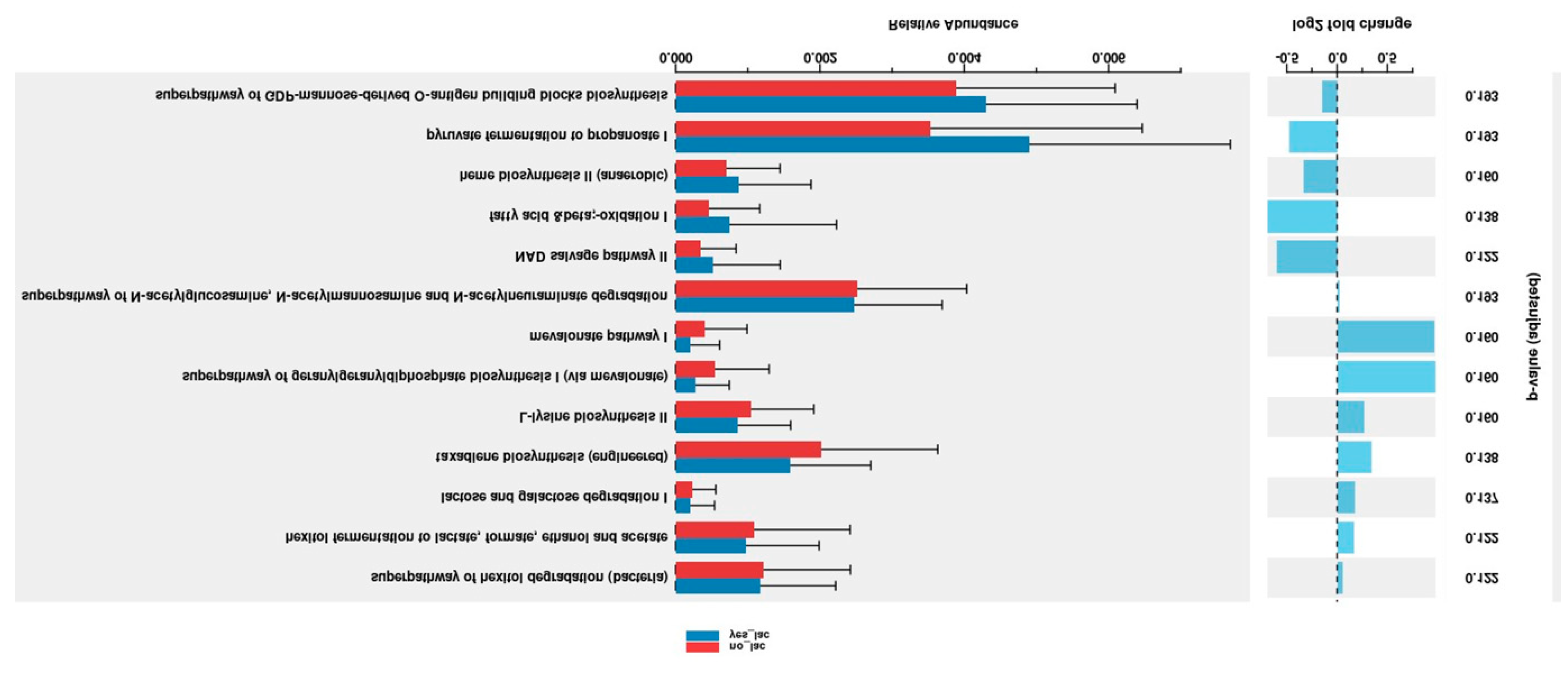
The PICRUSt2 program was used for prediction of metagenome functions of gut microbiome of HSCT patients. We observed greater non-significant pyruvate fermentation to propanoate I (p=0.193) relative abundance in the IG comparing with CG (
Figure 7).
yes-Lac: L. plantarum group (intervention group); no-Lac; control group.
3. Discussion
In the present study we did not observe a significant reduction in MDRO colonization or infection following L. plantarum ingestion compared to the control group. However, all infections in the intervention group were MBI-LCBI, with most of them occurring during neutropenia and after cessation of L. plantarum use. Of note, L. plantarum was safe and tolerable before the onset of neutropenia in all individuals undergoing HSCT and it may potentially reduce fecal levels of Enterococcus genus in HSCT candidates. There was a decrease in the presence of Roseburia and Coprococcus in both groups in grafting period. On the other hand, the consumption of L. plantarum was associated with significantly increased presence of the Lactobacillales and decrease of Turicibacter on gut microbiome of HSCT patients in second-week period. None of the patients in the intervention group had BSI caused by L. plantarum or severe adverse events.
Throughout the HSCT period, there was a significant reduction in alpha diversity in the gut microbiome especially between baseline and second-week versus neutropenia and grafting. Additionally, we observed intense intraindividual variability in the alpha diversity between the time points as shown previously by Taur et al [
11]. Interestingly, there was a reduction of uniformity of the species observed between time baseline and grafting in both groups slightly smaller in CG then IG. At baseline, at least 50.9% of bacteria were predominantly from the phyla Bacteroidetes, followed by Firmicutes(38.3%) and Proteobacteria(4.5%). These phyla were still predominant at grafting for both IG and CG.
Although
Enterococcus abundance had a greater rise in the CG, there were no significant differences between groups probably because of
L. plantarum cessation in neutropenia and the small size of the sample studied. Previous studies have observed that the increase of
Enterococcus in the microbiome at the engraftment period is associated with BSI during the HSCT[
11,
12,
13]. Thus, the smaller
Enteroccus abundance found in the IG might be advantageous. No differently abundant taxa were identified between the time points evaluated, except for changes in
Lactococcus and
Turicibacter on the second week of
L. plantarum use. These findings can be explained perhaps by intraindividual variability of the microbiome. Nonetheless, the increase of
Lactococcus may be worthy of note because this genus expresses an antimicrobial peptide involved in human gut homeostasis and may reduce VRE[
11,
12,
13].
Turicibacter might contribute to host metabolic and homeostatic mechanisms as well [
14].
The
Enterobacteriaceae family, mainly
K. pneumonia, was responsible for the highest number of infections in patients in the IG. We observed intestinal domination by
Enterobacteriaceae in only one patient who developed BSI. Interestingly, the IG developed MBI-LCBI in the neutropenia period after
L. plantarum was discontinued and in samples from these individuals we did not find members of the
Lactobacillales order. These changes may be due to interactions between
L. plantarum and other microorganisms and metabolites[
12]. Several methods have been developed to predict functions from 16S rRNA sequence data, including PICRUSt2 that we applied in the present study [
15]. We observed a non-significant increased on metabolites such as pyruvate fermentation to propanoate I (p=0.193) relative abundance in the IG . Bacteria’s from
Firmicutis phylum as f_
Lactobacillaceae,
g_Lactobacillus have been associated with pyruvate fermentation to propanoate I in Parkison diseases [
16]. However, the role of
L. plantarum on metagenomics functions needs to be clarified.
Of note,
L. plantarum had an average 86%(±11%) drug-target engagement at 43(±29) days of consumption and it was safe, tolerable and associated with an significant increase in the abundance of the Lactobacillales in the present study. None of the patients in the IG had BSI caused by
L. plantarum or serious adverse events. Similar with two recent studies in HSCT patients that support the safety of probiotics in theses patients [
9,
17], pointing out that the organisms included in over-the-counter probiotics are a rare cause of BSI during pre-engraftment period [
17], indicating the feasibility of probiotic
L. plantarum in children undergone HSCT without associated BSI or serious adverse events [
9].
This study has limitations as the duration of L. plantarum consumption by patients varied according to the patient's availability and clinical condition for admission to HSCT. The sample size of individuals was small and not controlled by clinical features that potentially could influence the results.
4. Materials and Methods
This study was conducted at the Hospital das Clinicas, Faculdade de Medicina, Universidade de Sao Paulo, Brazil. The Cell Therapy Clinical Unit (CTCU) is a 10-room ward for adult patients, with double bedrooms for autologous transplants and single bed rooms for allogeneic transplants.
This exploratory clinical investigation included patients seen from November 2017 to May 2020 who were colonized by MDROs prior to their HSCT. We utilized a convenience sample of patients treated with L. plantarum (IG) and an untreated control group (CG). The inclusion criteria were: oncohematological disease and indication for HSCT, being over 18 years old with no history of previous gastrointestinal tract surgery and to be colonized by MDROs identified by a positive surveillance culture (SC) prior to HSCT. The study protocol followed Helsinki’s declaration and was approved by the Institute’s ethical committee (CAPPesq approval 2.126.478), it was registered on the Registro Brasileiro de Ensaios Clínicos (ReBEC number RBR-2ztbwgr) and all participants signed voluntarily the written informed consent form.
Demographic and clinical variables were assessed, such as age, gender, underlying diseases, HSCT, mucositis, weight, height, body mass index (BMI) and classification of nutritional status by WHO or PAHO criteria at the time of admission to the study. Feces consistency was determined using the Bristol fecal scale prior to and following the consumption of
L. plantarum. Gastrointestinal toxicities (mucositis and diarrhea) were measured by Commun Terminology Criteria for Adverse Events 5.0 version (CTCAE) [
18]. Neutrophil recovery was defined as the third day of an absolute neutrophil count >500 neutrophils mm
-3.
L. plantarum, G18 lineage, was administrated as a probiotic to the treatment group at a dose of 5 x 10
9 Colony Forming Units (CFU) twice daily in the form of gastric release capsules produced in a compounding pharmacy. The HSCT pharmacy supervised capsule dispensing and assessed quantifying drug-target engagement. Consumption was terminated in cases of HSCT neutropenia. All symptoms in those taking
L. plantarum were assessed according to CTCAE protocol[
18]. The
L. plantarum capsule content was analyzed by 16S rRNA next generation sequencing (NGS), by growth in
Lactobacillus culture medium (MRS Agar) and by mass spectrometry (Maldi-TOF-Buker-Germany).
MDROs: Vancomycin resistant
Enterococcus; Enterobacteria resistant to carbapenems and/or colistin, producing ESBL (enzymes produced by certain bacteria that are able to hydrolyze extended spectrum cephalosporin); Gram-negative non-fermenting bacteria (
Pseudomonas aeruginosa and
Acinetobacter baumannii) resistant to carbapenems and/or colistin[
19].
Colonization: Colonization was evaluated by SC using rectal swab samples and qPCR analysis for the main resistance genes
blaVIM - Metallo-beta-lactamase VIM,
blaSPM - Metallo-beta-lactamase SPM,
blaKP C- Carbapenem-hydrolyzing beta-lactamase KPC,
mcr-1- Phosphoethanolamine-lipid A transferase MCR-1,
blaOXA-23-Carbapenem-hydrolyzing beta-lactamase OXA-23,
blaOXA-48-Carbapenem-hydrolyzing beta-lactamase OXA-48,
blaOXA-143-Carbapenem-hydrolyzing beta-lactamase OXA-143,
vanA-Vancomycin resistance protein VanA,
vanB-Vancomycin resistance protein VanB. The DNA sequences of oligonucleotides used in qPCR are shown in Table 1-supplementary material [
20,
21,
22]
. Infections were defined using the criteria of the Centres for Disease Control and Prevention (CDC)[
23]: - Central Line-Associated Bloodstream Infections (CLABSI) - as a laboratory confirmed bloodstream infection where an eligible BSI organism is identified on the day of the event or the preceding day.
- Mucosal Barrier Injury Laboratory-Confirmed Bloodstream Infection (MBI-LCBI) - characterizes the LCBI in patients immunosuppressed by microbiological translocation of the gastrointestinal tract due to persistent neutropenia (neutrophils <500 cells / mm3) or diarrheal episodes (1 liter or more of diarrhea in 24 hours) or graft-versus-host disease (GVHD) in allogeneic HSCT patients, within seven days of a positive blood culture. Decolonization was defined as the SC changing from positive to negative after the intervention period.
Stool samples were collected in an outpatient setting according to hospital protocol. The SC was performed weekly in inpatient settings for HSCT patients. We initiated bacterial cultures in selective media followed by qPCR analysis for the main resistance genes
i.e. a multiplex PCR for detecting genes related to
Metallo-β-lactamase resistance in
P. aeruginosa:
IMP, VIM and
SPM; for detecting genes related to oxacillinase resistance in
A. baumanii:
OXA-23 and
OXA-143; for detecting genes related to
Van A and
Van B resistance; and for detecting genes related to
blaKPC antimicrobial resistance (table 1 on supplementary material). The stool samples were obtained prospectively in a sterile bottle containing guanidine and stored at -20°C until DNA extraction, as previously described[
24]. The first collection was carried out before the consumption of the probiotic and was classified as ‘baseline’. Patients were instructed to date and store the samples in a freezer until the material was delivered to the researcher or collected on the day of delivery. The collection was performed weekly after initiating the consumption of
L. plantarum and continued until the grafting procedure.
The composition of the prokaryotic communities was determined based on partial 16S rRNA (V4 region) sequences directly amplified using a bacterial/archaeal primer set 515F/806R[
25] from each DNA sample. PCR amplification, library preparation, and sequencing followed the procedures described by Ribeiro et al
. [
24]. Briefly, approximately 0.25 g of feces was used for DNA isolation using the DNeasy PowerSoil Kit (Qiagen, Germantown, MD, USA), following the manufacturer’s instructions. The template preparation was performed by the Ion Chef System(Thermo Fisher Scientific, MA, USA), using the Ion PGM Hi-Q View Chef Kit. Sequencing was performed in the Ion Personal Genome Machine(PGM), using the Ion PGM Hi-Q Sequencing Kit and the Ion 318 Chip v2, following the instructions of the manufacturer(Thermo Fisher Scientific, MA, USA). Samples beneath 85.000 reads were re-sequenced.
The 16S rRNA gene data pre-processing and diversity estimates were performed using Quantitative Insights Into Microbial Ecology (QIIME2) version 2019.10[(26]. Demultiplexed sequence data were denoised with DADA2 (via q2-dada2) with the default parameters: 260 bp in length and an average quality Phred score of ≥30 to generate the amplicon sequence variants (ASVs)[
27,
28]
. A phylogenetic tree was built by inserting the sequences into the Greengenes 13_8 reference tree, using q2-fragment-insertion plugin[
18], which uses the SATé-enabled phylogenetic placement insertion method[
29]. Alpha-diversity metrics (number of observed ASV, Pielou’s evenness, Shannon diversity, and Faith’s phylogenetic diversity), and Beta-diversity metric (Bray Curtis and unweighted and weighted UniFrac) were estimated using q2-diversity after samples were rarefied to 32.000 sequences per sample(30–32). The rarefaction curves for the samples reached the plateau, indicating that there was good representation of the microbial community. The Principal Coordinates Analysis (PCoA) plot for each of the beta-diversity metrics was generated using the phyloseq package as implemented in R (33). The ASVs were taxonomically classified using the q2-feature-classifier(34) (naive Bayes classifier) against Greengenes 13_8 99% OTUs reference sequences[
35,
36]. Metagenomic functions was analyzed using Package: ggpicrust2, Type: Package, Title: Make 'PICRUSt2' Output Analysis and Visualization Easier, Version: 1.7.2, R version 4.3.1 (2023-06-16) [
37] .
A database containing all demographic and clinical variables was built using the Collaborative software Airtable(San Francisco, California, US). Categorical variables were described as counts and proportions. Variables with a normal and asymmetric distribution were described as mean (range). Normality was evaluated with visual inspection of histograms and the Shapiro-Wilk test. The Student t test was used for comparison of normal variables, and the Wilcoxon test was used for non-normal variables. Categorical variables were compared by the chi square and Fisher exact test. Compositions of microbiota communities were summarized by proportion at different taxonomy levels, including specie, genus, family, order, class, and phylum ranks. For all analyses, significance was determined as p< 0.05. The Kruskal-Wallis test was performed to explore differences in alpha-diversity metrics. Differences in community composition (beta-diversity) were assessed using non-parametric Analysis of Similarities (ANOSIM) tests. To determine the features most likely to explain differences between periods and groups, we employed the algorithm Linear discriminant analysis Effect Size (LEfSe)(36). We used a Wilcoxon rank-sum test and Wilcoxon signed rank test, adjusted by the Bonferroni correction, to compare continuous microbiota features among groups and time periods, and Fisher’s exact test to compare categorical decolonization data. All these analyzes were performed using R, version3.6.
5. Conclusions
In conclusion, ingestion of L. plantarum capsules was safe and tolerable. Although it may have impact on Enterococcus abundance and reduction of Turicibacter, it has not showed a significant impact on Gram-Negative MDRO abundance in HSCT patients. Further studies are necessary to confirm our findings.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org., Table S1:. Table 1. DNA sequences of oligonucleotides used in polymerase chain reactions.
Author Contributions
Conceptualization, BDGCM, ECS, TG, SFC.; methodology, BDGCM, LAMF, ECS, SFC ; software, JVS, LAMF. TFB, ; validation, BDGCM, SFC; formal analysis, BDGCM, RCM, JVS, TFB, CFS ; resources, ECS and SFC.; data curation, BDGCM, RCM, JVS, TFB, CFS , SFC.; writing—original draft preparation, BDGCM; SFC.; writing—review and editing, BDGCM, JVS, SW, SFC .; supervision, LAMF, SFC.; funding acquisition, SFC. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by São Paulo Research Foundation (FAPESP) [18/13147-3 to S.F.C.]. The funders had no role in study design, methods, data collection and analysis, decision to publish or preparation of this manuscript. All authors had final responsibility for the decision to submit the manuscript for publication.
Institutional Review Board Statement
The present study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving human subjects were approved by the Ethics Committee of Hospital das Clinicas da Faculdade de Medicina da Universidade de São Pualo, São Paulo, Brasil for analysis of research projects (CAPPesq approval 2.126.478)
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Data availability: Microbiome sequencing data are available from Bioproject (NCBI - National Center for Biotechnology Information, U.S. National Library of Medicine) with the accession number PRJNA724885.
Acknowledgments
We would like to acknowledge Kelly Dias for guiding the participants and the nurses who collected biological samples, especially Juliana Turca and Liliane Pinheiro. We would also like to acknowledge the pharmacy team, especially Tamires Fernandes and Anna Bortulucci.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ferreira AM, Moreira F, Guimaraes T, Spadão F, Ramos JF, Batista MV, et al. Epidemiology, risk factors and outcomes of multi-drug-resistant bloodstream infections in haematopoietic stem cell transplant recipients: importance of previous gut colonization. J Hosp Infect,100 (2018), pp. 83–91. Available from: https://doi.org/10.1016/j.jhin.2018.03.004. [CrossRef]
- Bilinski J, Robak K, Peric Z, Marchel H, Karakulska-Prystupiuk E, Halaburda K, et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol Blood Marrow Transplant. 22 (2016),pp.1087–1093. [CrossRef]
- Tacconelli E, Mazzaferri F, Smet AM De, Bragantini D, Eggimann P, Huttner BD, et al. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin Microbiol Infect, 25(2019), pp. 807–817. [CrossRef]
- Aira A, Fehér C, Rubio E, Soriano A. The Intestinal Microbiota as a Reservoir and a Therapeutic Target to Fight Multi-Drug-Resistant Bacteria : A Narrative Review of the Literature. Infect Dis Ther, 8 (2019), pp. 469–482. [CrossRef]
- Wu G, Zhang C, Wang J, Zhang F, Wang R, Shen J, et al. Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children. Sci Rep [Internet]. 6 (2016), p. 24030. Available from: http://dx.doi.org/10.1038/srep24030. [CrossRef]
- Esaiassen E, Hjerde E, Cavanagh JP, Pedersen T. Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Front Pediatr, 6 (2018),p. 347. [CrossRef]
- Salomão MCC, Heluany-filho MA, Menegueti MG, Marlieke EA de K, Martinez R, Bellissimo-Rodrigues F. A randomized clinical trial on the effectiveness of a symbiotic product to decolonize patients harboring multidrug-resistant Gram-negative bacilli. Rev Soc Bras Med Trop, 49 (2016), pp. 559–566. [CrossRef]
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol, 11 (2014), pp. 506–514. [CrossRef]
- Ladas EJ, Bhatia M, Chen L, Sandler E, Petrovic a, Berman DM, et al. The safety and feasibility of probiotics in children and adolescents undergoing hematopoietic cell transplantation. Bone Marrow Transplant [Internet]. 51 (2015), pp.:1–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26569091. [CrossRef]
- Araos R, Tai AK, Snyder GM, Blaser MJ, Agata EMCD. Predominance of Lactobacillus spp. Among Patients Who Do Not Acquire Multidrug-Resistant Organisms. Clin Infect Dis, 7 (2016), pp.:937–943. [CrossRef]
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 55 (2012), pp. 905–914. [CrossRef]
- Carvalho R, Vaz A, Pereira FL, Dorella F, Aguiar E, Chatel JM, et al. Gut microbiome modulation during treatment of mucositis with the dairy bacterium Lactococcus lactis and recombinant strain secreting human antimicrobial PAP. Sci Rep, 8 (2018),pp. 1–10. [CrossRef]
- Kim SG, Becattini S, Moody TU, Shliaha P V, Littmann ER, Seok R, et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature, 572 (2019), pp. 665–669. [CrossRef]
- Heidenreich D, Kreil S, Jawhar M, Müller N, Nolte F, Becker KP, et al. Course of colonization by multidrug-resistant organisms after allogeneic hematopoietic cell transplantation. Ann Hematol, 97 (2018),pp. 2501–2508. [CrossRef]
- Cohen SA, Woodfield MC, Boyle N, Stednick Z, Boeckh M, Pergam SA. Incidence and outcomes of bloodstream infections among hematopoietic cell transplant recipients from species commonly reported to be in over-the-counter probiotic formulations. Transpl Infect Dis. 2016;18(5):699–705. [CrossRef]
- Hoffman JM and Margolis KG.,* Building community in the gut: a role for mucosal serotonin. Nat Rev Gastroenterol Hepatol, 1 (2020), pp. 6–8. [CrossRef]
- Douglas, G.M., Maffei, V.J., Zaneveld, J.R. et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38, 685–688 (2020). [CrossRef]
- 17-Li Z, Lu G, Li Z, Wu B, Luo E, Qiu X, Guo J, Xia Z, Zheng C, Su Q, Zeng Y, Chan WY, Su X, Cai Q, Xu Y, Chen Y, Wang M, Poon WS, Luo X.Altered Actinobacteria and Firmicutes Phylum Associated Epitopes in Patients With Parkinson's Disease. Front Immunol. 2021 Jul 2;12:632482. [CrossRef]
- National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE) verson 5.0. In: US Department of Health and Human Services, National Institutes of Health, National Cancer Institute, 2020.
- Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multi-drug resistant organisms in healthcare settings. J Korean Med Assoc, 61 (2018), pp. 26–35. [CrossRef]
- Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents, 27 (2006),pp. 351–353. [CrossRef]
- Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother, 12 (2009),pp. 5035–5038. [CrossRef]
- Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol, 45 (2007),pp. 544–547. [CrossRef]
- CDC - Center for Disease Control and Prevention. Bloodstream Infection Event ( Central Line-Associated Bloodstream Infection and Non-central line-associated Bloodstream Infection ). Publ on-line http//www.cdc.gov/nhsn/ [Internet]. 2016;(January):1–32. Available from: Centers for Disease Control (CDC)/National Healthcare Safety Network (NHSN). Bloodstream Infection Event (Central Line-Associated Bloodstream Infection and Non-central line-associated Bloodstream Infection.. Available at: http://www.cdc.gov/nhsn/PDFs/pscM .
- Ribeiro RM, de Souza-Basqueira M, de Oliveira LC, Salles FC, Pereira NB, Sabino EC. An alternative storage method for characterization of the intestinal microbiota through next generation sequencing. Rev Inst Med Trop Sao Paulo, 60 (2018), pp.1–11. [CrossRef]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 108 (2011),pp.4516–4522. [CrossRef]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol, 37 (2019),pp.852–857. [CrossRef]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods,13(2016), pp.581–853. [CrossRef]
- Janssen S, Mcdonald D, Gonzalez A, Navas-molina JA, Jiang L, Xu Z. Phylogenetic Placement of Exact Amplicon Sequences. mSystems. 3 (2018), pp.1–14. [CrossRef]
- Mirarab S, Nguyen N, Warnow T. SEPP: SATé-enabled phylogenetic placement. In: Pacific Symposium on Biocomputing. 2012. pp. 247–258. [CrossRef]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 61(1992),pp. 1–10. [CrossRef]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and Qualitative Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities, 73 (2007), pp.1576–1585. [CrossRef]
- Lozupone C, Knight R. UniFrac : a New Phylogenetic Method for Comparing Microbial Communities. 71 (2005),pp. 8228–8235. [CrossRef]
- McMurdie PJ, Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One, 8 (2013), p.4. [CrossRef]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2 ’ s q2-feature-classifier plugin. 2018, pp. 1–17. [CrossRef]
- Mandal S, Treuren W Van, White RA, Eggesbø M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. 1 (2015), pp. 1–7. [CrossRef]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol [Internet]. 2011;12(6):R60. Available from: http://genomebiology.com/2011/11/6/R60. [CrossRef]
- Chen Yang, Jiahao Mai, Xuan Cao, Aaron Burberry, Fabio Cominelli, Liangliang Zhang, ggpicrust2: an R package for PICRUSt2 predicted functional profile analysis and visualization, Bioinformatics, Volume 39, Issue 8, August 2023, btad470. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).