1. Introduction
Fragrant rice is favored by consumers due to its excellent cooking qualities, aroma, and high economic value (Mo et al., 2015; Ashraf et al., 2023). The cultivation of fragrant rice in our country has been experiencing the phenomenon of “losing fragrance as it is cultivated more”. The rice cannot meet the market demand for high-quality fragrant rice, resulting in long-term reliance on imports. The annual import volume has reached 3 million tons (Bao et al., 2021). The development of high-quality germplasm resources and the establishment of high-yield cultivation technology system of fragrant rice are two important steps for improving fragrant rice production (Shan et al., 2015; Wang et al., 2023).
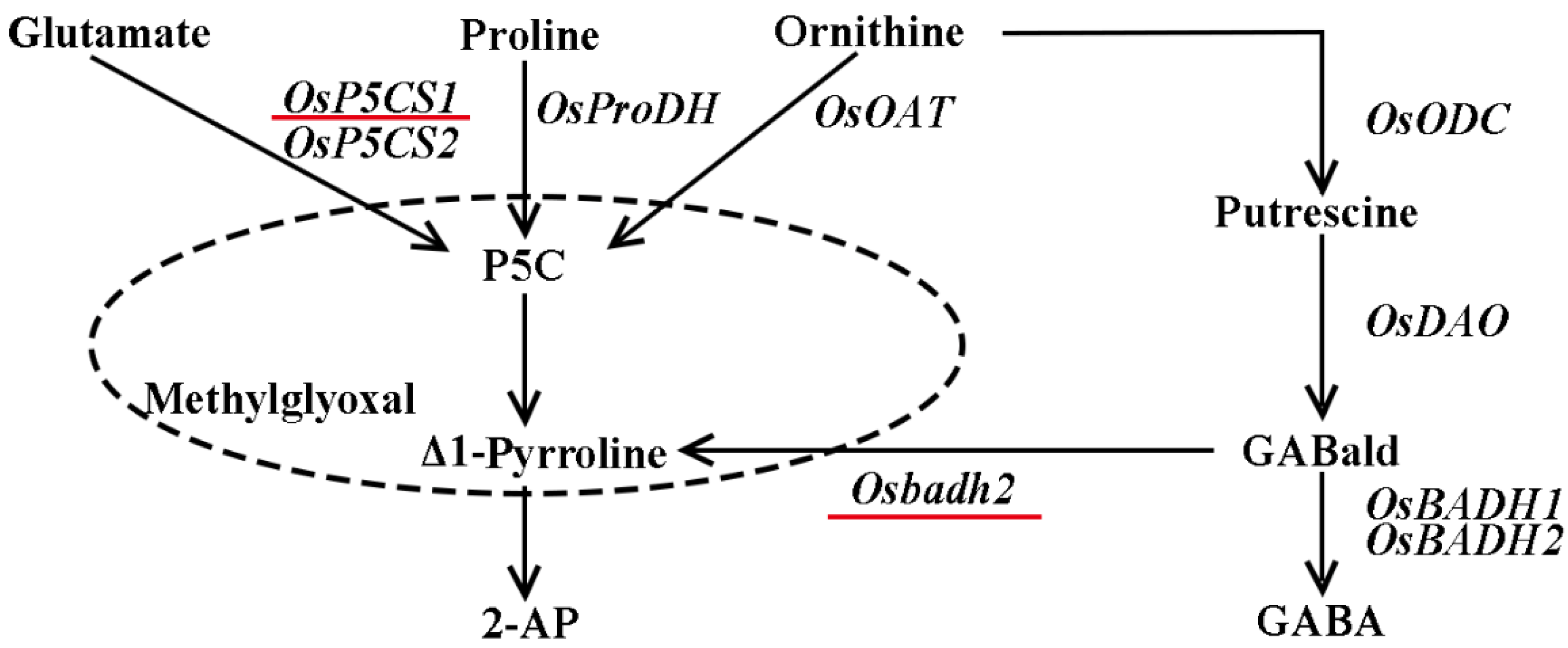
Fragrant rice is a special type of rice where the stems, leaves, panicles, flowers, and grains emit fragrance, apart from the roots (Mo et al., 2015). The fragrant aroma of cooked fragrant rice and its rich nutritional content, including various amino acids, have made it highly favored by people in rice-consuming regions around the world (Ashraf et al., 2023). Rice aroma contains about 200 volatile compounds, whereas the 2-acetyl-1-pyrroline (2-AP) is considered as the most important for aroma production in fragrant rice (Jezssek et al., 2002; Poonlaphdecha et al., 2016; Imran et al., 2023). There are two biosynthetic pathways for 2-AP: firstly, proline, ornithine and glutamate are catalyzed by proline dehydrogenase (PRODH), ornithine aminotransferase (OAT) and pyrroline-5-carboxylic acid synthase (P5CS), respectively to form pyrroline-5-carboxylic acid (P5C), which directly reacts with methylglyoxal (MG) to form 2-AP in a non-enzymatic reaction (Huang et al., 2008), and/or P5C is decarboxylated to form Δ1-Pyrroline catalyzed by pyrroline-5-carboxylate decarboxylase (P5CR), and Δ1-Pyrroline directly forms 2-AP with MG (Wakte et al., 2016; Li et al., 2016). Secondly, ornithine is catalyzed by ornithine decarboxylase (ODC) to form putrescine, and then diamine oxidase (DAO) catalyzes putrescine to form γ-aminobutyraldehyde (GABald). The GABald is converted to γ-aminobutyric acid (GABA) catalyzed by a functional betaine aldehyde dehydrogenase (BADH), which inhibits the biosynthesis of 2-AP in non-aromatic rice. Conversely, GABald cannot be converted to GABA under the action of a nonfunctional betaine aldehyde dehydrogenase (badh), resulting in the accumulation of 2-AP in aromatic rice (Bradbury et al., 2008; Bao et al., 2018).
With the establishment of the genetic map of rice, important QTLs and potential candidate genes for explaining the molecular characteristics of fragrance in fragrant rice have been identified (Prodhan and Shu, 2020). The OsBADH2 gene encoded a functional betaine aldehyde dehydrogenase, and inhibited 2-AP biosynthesis in rice (Huang et al., 2008). Down-regulation of OsBADH2 gene expression by RNAi interference techniques and TALEN technology resulted in the elevation of 2-AP content in non-aromatic rice (Niu et al., 2008; Shan et al., 2015). The CRISPR/Cas9-mediated editing of OsBADH2 gene induced 2-AP production in fragrant rice (Tang et al., 2020). Moreover, the proline is an important precursor for 2-AP biosynthesis (Huang et al., 2008). The P5C synthase encoded by the OsP5CS gene had dual functions as glutamate kinase and γ-glutamyl phosphate reductase, and plays its role as a rate-limiting enzyme in proline biosynthesis (Szabados and Savouré, 2010). The rice genome contains two homologous OsP5CS genes i.e., OsP5CS1 and OsP5CS2. The OsP5CS1 is located on chromosome 1 and often induced by salt, dehydration and cold stress whereas the OsP5CS2 is located on chromosome 5 and induced by salt and mannitol stress (Rai and Penna, 2013; Forlani et al., 2015). Over-expression of the OsP5CS1 gene in aromatic rice increased 2-AP content (Kaikavoosi et al., 2015; Hinge et al., 2016). Compared with conventional rice, the expression of OsP5CS1 gene in aromatic rice was positively correlated with 2-AP accumulation (Ghosh and Roychoudhury, 2018). There have been many studies on the regulation of P5CS expression in different plant tissues in response to different environmental signals, but little is known about the transcriptional regulation of P5CS gene. Cis-acting element analysis of promoters of OsP5CS1 and OsP5CS2 genes in rice showed that binding sites of 24 different types of transcription factors were detected in the promoters of the two genes. This includes transcription factor families such as AP2, bZIP, MYB, and NAC (Bao et al, 2021). So, it has not yet been studied to determine the transcription factors that bind to the promoter of OsP5CS1 gene and how the expression of OsP5CS1 gene is regulated to affect the content of 2-AP.
The long-term non-degradation of aroma in the origin of aromatic rice was closely related to the high zinc (Zn) content in soil (Hu et al., 2001). Previously, Foliar application of Zn fertilizer promoted the accumulation of fragrance in fragrant rice. Mo et al. (2016) pointed out that exogenous Zn fertilizer application promoted the accumulation of 2-AP in detached panicles of fragrant rice by increasing proline content. Luo et al. (2019) found that foliar application of Zn fertilizer enhanced P5CS enzyme activity, leading to increased accumulation of 2-AP in fragrant rice. Bao et al. (2021) reported that foliar Zn application up-regulated the expression of OsP5CS1 gene and promoted the accumulation of 2-AP in aromatic rice.
Zn, as a cofactor in enzymes, transcription factors, and protein-protein interaction domains, plays structural and catalytic roles in many proteins, making it an essential micronutrient for plants (Maret and Li, 2009). Zinc-binding proteins constitute approximately 10% of the proteome in eukaryotes (Choi and Bird, 2014). Plants rely on a tightly regulated Zn homeostasis network to maintain sufficient intracellular Zn levels and prevent toxicity caused by Zn deficiency or excess accumulation (Colvin et al., 2010). The bZIPs is a large family of transcription factors in plants, and contain a bZIP domain that consists of basic domain and leucine zipper (Joo et al., 2021). These residues were located on adjacent helices, with a basic region consisting of approximately 16 amino acid residues containing a nuclear localization signal, followed by a conserved N-XT-R/K motif that directly bound to DNA. In the leucine zipper region, leucine residues were repeated every 7 amino acids and were sometimes substituted by isoleucine, phenylalanine, valine, or methionine. The leucine residues determined the dimerization specificity of bZIP proteins. Therefore, these variant residues represent the specificity for determining homodimerization and heterodimerization (Nijhawan et al., 2008). bZIP proteins were the most diverse family in plants and have been identified in whole-genome sequences of various species, including Arabidopsis, rice, maize (Zea mays L), sorghum (Sorghum bicolor (L.) Moench), grape (Vitis vinifera L.), tomato (Lycopersicon esculentum Mill.), and apple (Malus pumila Mill.) (Joo et al., 2021). Seventy-five putative genes encoding bZIP proteins have been identified in Arabidopsis. These bZIP proteins were classified into 10 groups based on sequence domain similarity and biological function (Jakoby et al., 2002). The F-bZIP proteins were the central mediator of Zn deficiency reaction (Lilay et al., 2020; 2021) whereas the F-bZIP transcription factors AtbZIP19 and AtbZIP23 maintained Zn levels in cells through N-terminal Cys-his rich motif and single Cys or His residue binding Zn2+ in Arabidopsis thaliana (Assuncao et al., 2010; Lilay et al., 2021). Plants rely on a strict network of Zn homeostasis to maintain Zn levels in the cells to avoid damages caused by excess and deficiency of Zn (Colvin et al., 2010), however, the regulation of 2-AP by bZIP transcription factor needs investigations. The targeted editing of 2-AP biosynthetic gene OsBADH2 by CRISPR/Cas9 technology and the development of high-quality aromatic rice resources are becoming important. The present study was therefore conducted to develop the Osp5cs1 knockout mutant and OsP5CS1 overexpression lines by genetic transformation in the perspective of OsBADH2 knockout in indica rice.
2. Materials and Methods
2.1. Plant Materials and Experimental Details
The experiment was conducted at the Guangzhou Key Laboratory for Research and Development of Crop Germplasm Resources, Zhongkai University of Agriculture and Engineering, Guangzhou, China (23104 N, 113281 E) from January 2021 to June 2022. The Osp5cs1 knockout mutant and OsP5CS1 over-expression lines were constructed by genetic transformation of indica rice cultivar i.e., ‘Zhonghua11’, which knocked out OsBADH2 to produce fragrance. Three Osbzip60-like knockout mutant lines and OsbZIP60-like over-expression lines were also constructed with ‘Zhonghua11’ producing aroma as background material. The wild-type, the Osbzip60-like mutants, and OsbZIP60-like over-expression plants at two weeks of seedling age were cultured in normal nutrient solution and Zn-deficient medium for two weeks and used to determine the 2-AP concentrations.
2.2. Real-Time PCR Quantification of Gene Expression
The RNA was extracted with MegBio Plant Total RNA Mini-Extraction Kit, then reverse transcribed into cDNA by Prime Script® RT reagent kit with gDNA Eraser kit. The relative expression of OsP5CS1 gene was analyzed by Thermo Fisher real-time fluorescence quantitative PCR kit. Using the rice actin gene as the endogenous reference gene, three statistical replicates, the primer sequences of OsP5CS1 and actin were F 5′-TTTTGAGTCCCGACCTG-3′, R 5′-TTCACCAACATTACGAGGA-3′, F 5′-GATCACTGCCTTGGCTCCTA-3′ and R 5′-GTACTCAGCCTTGGCAATCC-3′, respectively (Bao et al., 2022a).
2.3. Determination of the 2-AP Contents
The leaves were cut into pieces, and then the 2-AP was extracted by distillation and extraction using a distillation extractor, and a Shimadzu GC-MS QP 2010 plus gas mass spectrometer was used. The 2,4,6-Trimethylpyrimidine (TMP) was used as the internal standard, and the column was RESTEK Rxi-5ms with a length of 30 m, an inner diameter of 0.32 mm, and a film thickness of 0.25 μm. The column temperature heating program was as follows: 40 °C for 1 min, then heated to 65°C at 2°C min-1, held for 1 min; then heated to 220°C at 10°C min-1, the carrier gas was high-purity helium (purity > 99.999 %), constant pressure splitless injection mode with 10 μL injection volume (Bao et al., 2021).
2.4. Promoter Cis-Acting Element Analysis
2.5. Yeast One-Hybrid Assays
The Yeast ‘Y187’ was inoculated in YPDA liquid medium. Three single colonies were randomly selected from the plate, diluted, coated on the corresponding defective plate without adding histidine with different concentrations (0, 10, 20, 30, 40, 50, 75 mM 3AT) of 3AT, and incubated at 30℃ for 3 days. The plasmid containing pHIS2::OsP5CS1 promoter was transformed into Y187 yeast as receptive cells, and then plasmid pGADT7::bZIP60 was transferred into receptive cells and coated on the selected 3AT plate. These positive clones were amplified from yeast cells for DNA sequencing and BLAST analysis against sequences in GenBank database (Lilay et al., 2020).
2.6. Electrophoretic Mobility Shift Assay (EMSA)
The 5'-biotinylated oligonucleotide (5'-ATG CAT GCA TGC ATG CAT GCA TGC-3') was used as probes and incubated with the nuclear extract at room temperature for 30 min. The entire reaction mixture was run on a non-denaturing 0.5×TBE 6% polyacrylamide gel for 1h at 60 V at 4°C and then transferred onto Biodyne® B nylon membranes (Pall Corporation). Signals were visualized with reagents included in the kit and ChemiDoc XRS (Bio-Rad Laboratories, UAS) (Hellman and Fried, 2007).
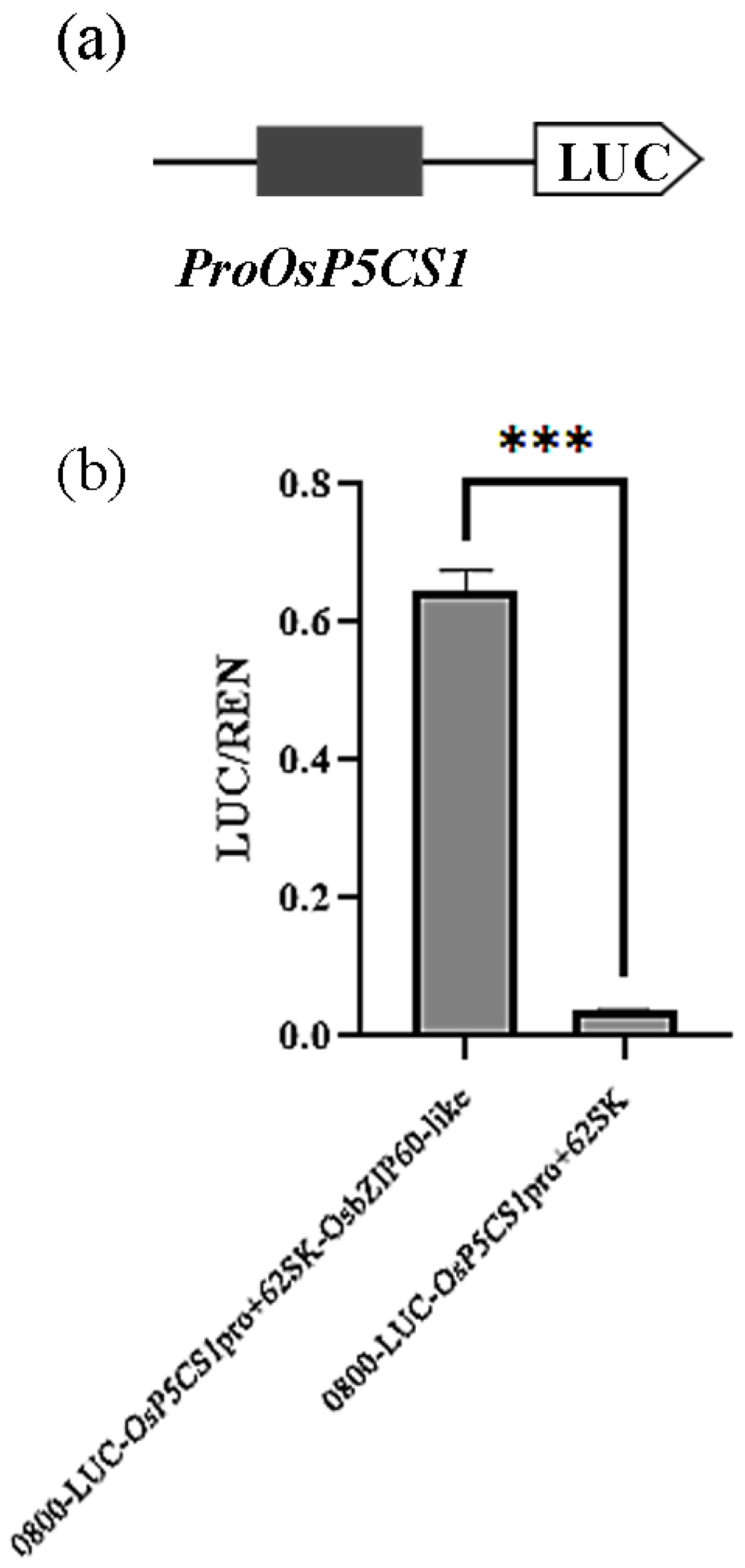
2.7. Dualluciferaseassaysystem
The OsP5CS1 promoter (2000bp) was ligated into the pGL3::LUC (containing Firefly luciferase) vector by homologous recombination to construct a reporter gene vector. The OsbZIP60-like was connected to Ubi promoter expression vector. The reporter gene vector, expression vector and pRT::TK vector (containing Renilla luciferase) included in the experimental group whereas the reporter gene vector, blank control expression vector and pRT::TK vector included in the control group were transformed into Arabidopsis protoplasts by agrobacterium transformation method, respectively. Luciferase and Renilla substrates were added, and the activity of reporter genes in Firefly luciferase and Renilla luciferase was detected by enzyme marker. The LUC/REN ratio showed transcriptional regulatory activity of OsbZIP60-like on OsP5CS1 promoters (Fang et al., 2021).
2.8. Statistical Analyses
Data were analyzed by SPSS 25 (Analytical Software, Chicago) whereas the means were separated by the least significant difference (LSD) test at P < 0.05 level.
3. Results
3.1. The OsP5CS1 Gene and 2-AP Biosynthesis
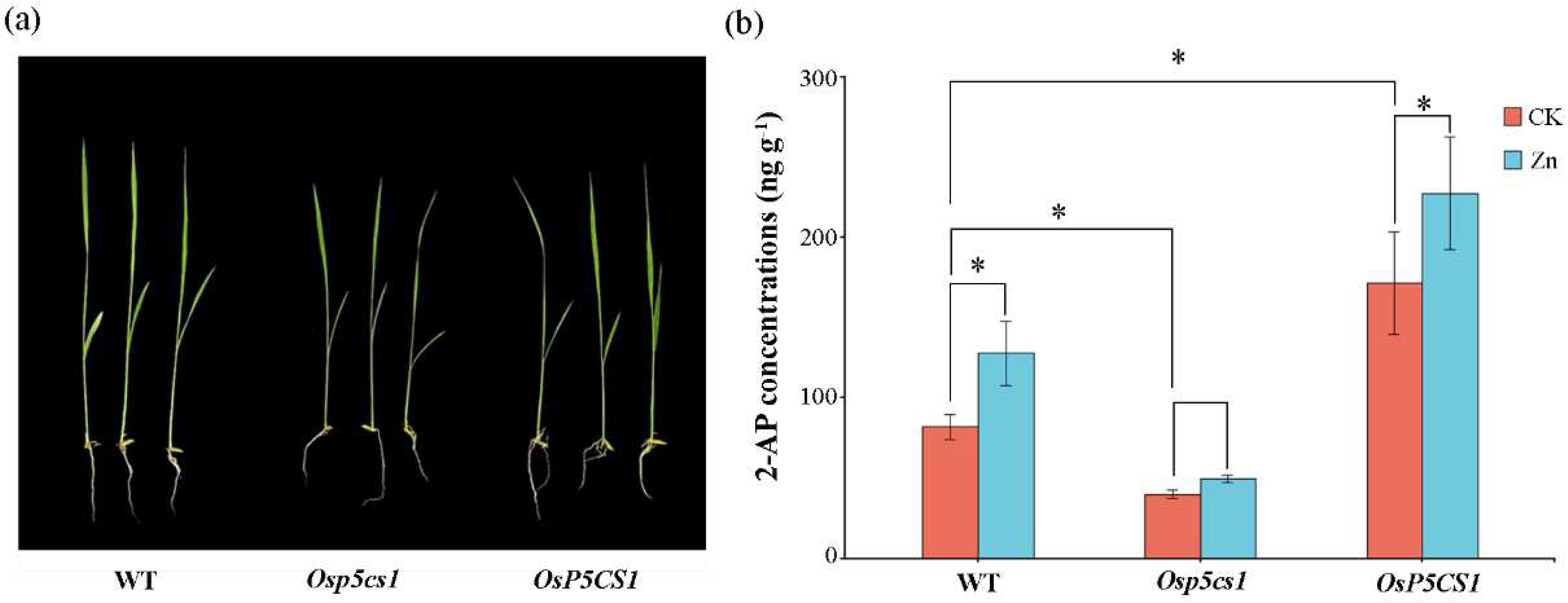
Compared with wild type (WT), the growth of
Osp5cs1 plants were inhibited, whereas the
OsP5CS1 plants were similar to the WT (
Fig. 2a). Compared with WT, the 2-AP concentrations in the
Osp5cs1 plants were decreased by 51.11 %, and the 2-AP concentrations in the
OsP5CS1 plants were increased by 52.26 %. In the WT, the concentrations of 2-AP were increased by 35.98 % under Zn treatment compared with CK. In the
OsP5CS1 plants, the 2-AP concentrations were increased by 24.59 % under Zn treatment compared with CK (
Fig. 2b). This result proves that
OsP5CS1 gene was a key gene in the 2-AP biosynthesis pathway of fragrant rice.
Figure 1.
2-AP biosynthetic pathway.
Figure 1.
2-AP biosynthetic pathway.
3.2. Analysis of Cis-Acting Elements in the OsP5CS1 Promoter
Plant CARE was used to analyze the type, quantity and position of the cis-acting elements of the
OsP5CS1 promoter to predict the function of the promoter. It was found that there are many cis-acting elements with different functions in the promoter. There were multiple cis-acting elements that may bind to bZIP60, such as ABRE, ABRE3a, ABRE4, G-Box, and ERE (
Table 1)
3.3. OsbZIP60-Like Transcription Factor
The
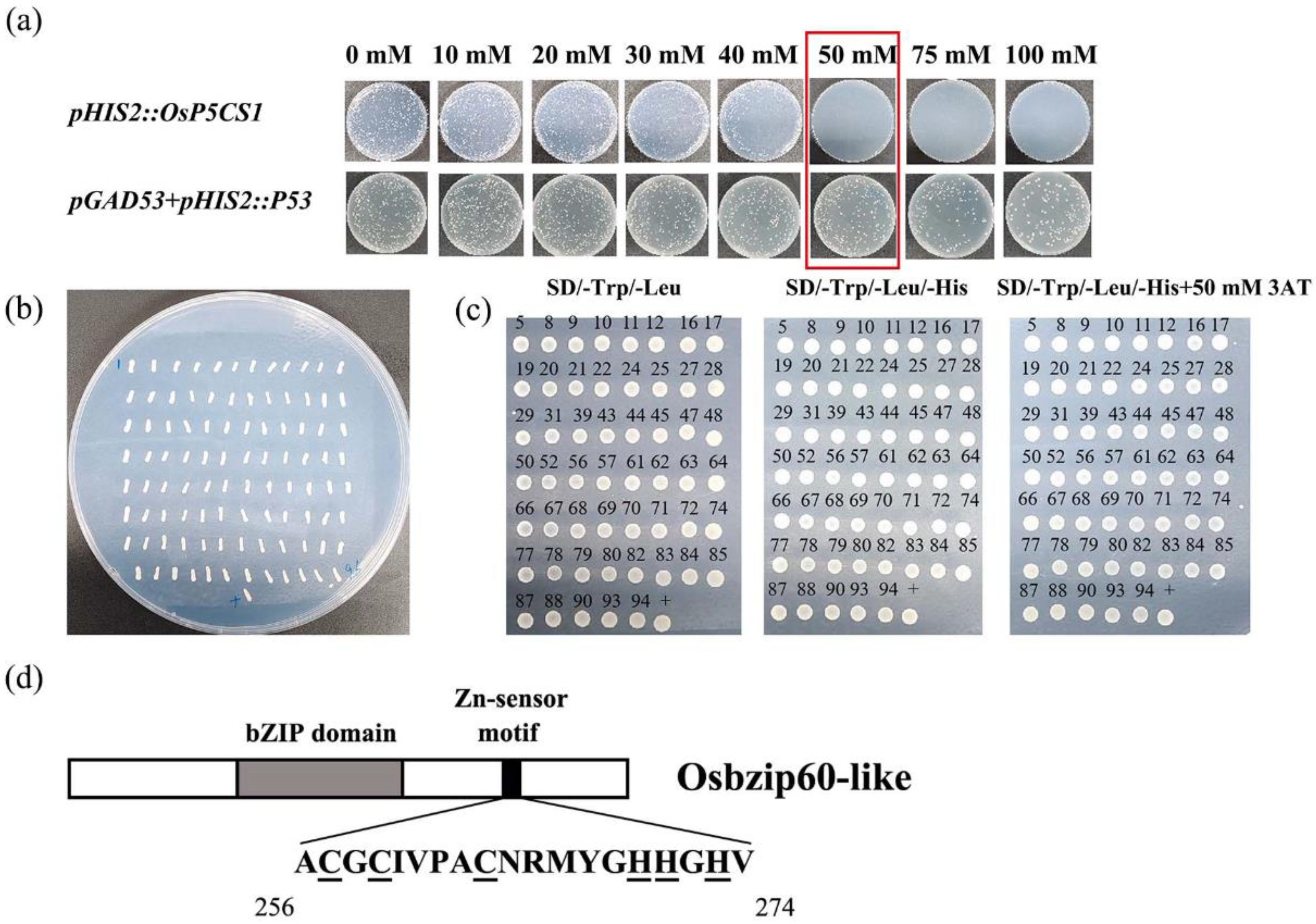
OsP5CS1 gene promoter sequence was ligated into the bait vector and transformed into yeast. No colony growth of transformants on the medium supplemented with 50 mM 3AT was established indicating that the
HIS3 reporter gene was not activated, so yeast one-hybrid was performed on 50 mM 3AT (
Fig. 3a). Through the library of rice nuclear gene, 96 positive clones were screened (
Fig. 3b). After PCR amplification, Seqman and BLAST comparison, the empty and repeated sequences were removed, and the 53 different sequences were obtained. Fifty-three positive clones were inoculated on SD-TLH+50 mM 3AT-deficient plates, diluted with sterile water, and then placed on SD-TL, SD-TLH, and SD-TLH+50 mM 3AT-deficient plates for rotary verification (
Fig. 3c). The protein LOC4344086 appeared several times in the sequencing of positive clones and showed strong positive signals. The sequence alignment showed that the fragment sequence was 100% similar to the OsbZIP60-like gene sequence of rice. According to the amino acid structure analysis, the OsbZIP60-like was identified as a group F-bZIP transcription factor, in addition to the BRLZ basic domain and leucine zipping dimer domain typical of bZIP family, the amino acid composition was rich in Cys and His residues binding to Zn
2+ (
Fig. 3d). The results preliminarily indicate that the transcription factor OsbZIP60-like specifically binds to the
OsP5CS1 promoter.
3.4. The OsP5CS1 Required the OsbZIP60-Like for Transactivation
Eleven presumed OsbZIP60-like cis-elements were identified in the promoter region of
OsP5CS1 (2000 bp). The
OsP5CS1 promoter was connected to the firefly lucifin reporter (
Fig. 4a), and the
OsP5CS1 promoter was strongly activated by OsbZIP60-like (
Fig. 4b).
3.5. The Transcription Factor OsbZIP60-Like Specifically Bound to the OsP5CS1 Promoter
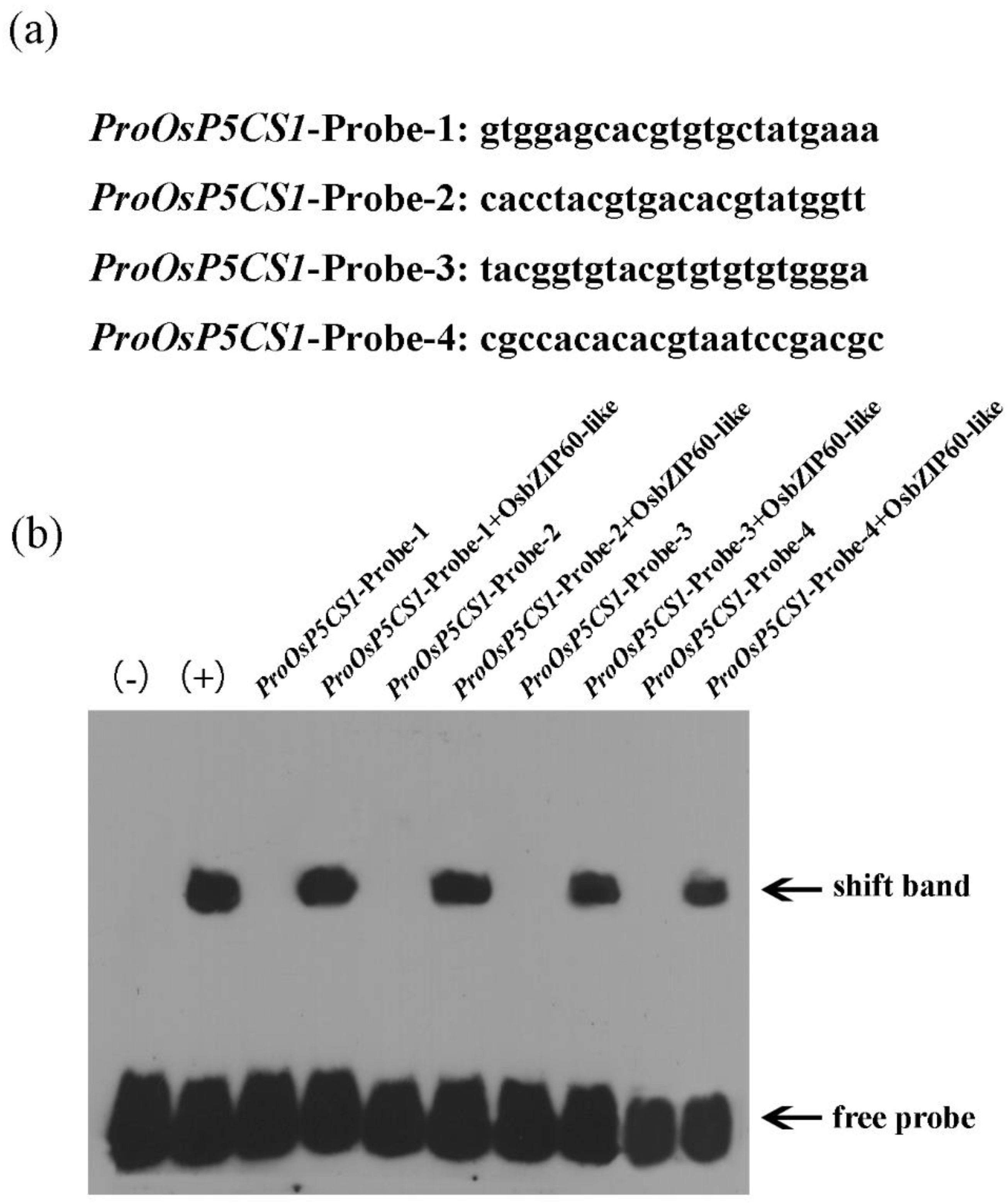
The purified OsbZIP60-like protein was incubated with four selected labeled promoter fragments (
Fig. 5a). Specific DNA complexes were detected in four probes (
Fig. 5b). The results confirmed that the transcription factor OsbZIP60-like specifically bound to the
OsP5CS1 promoter.
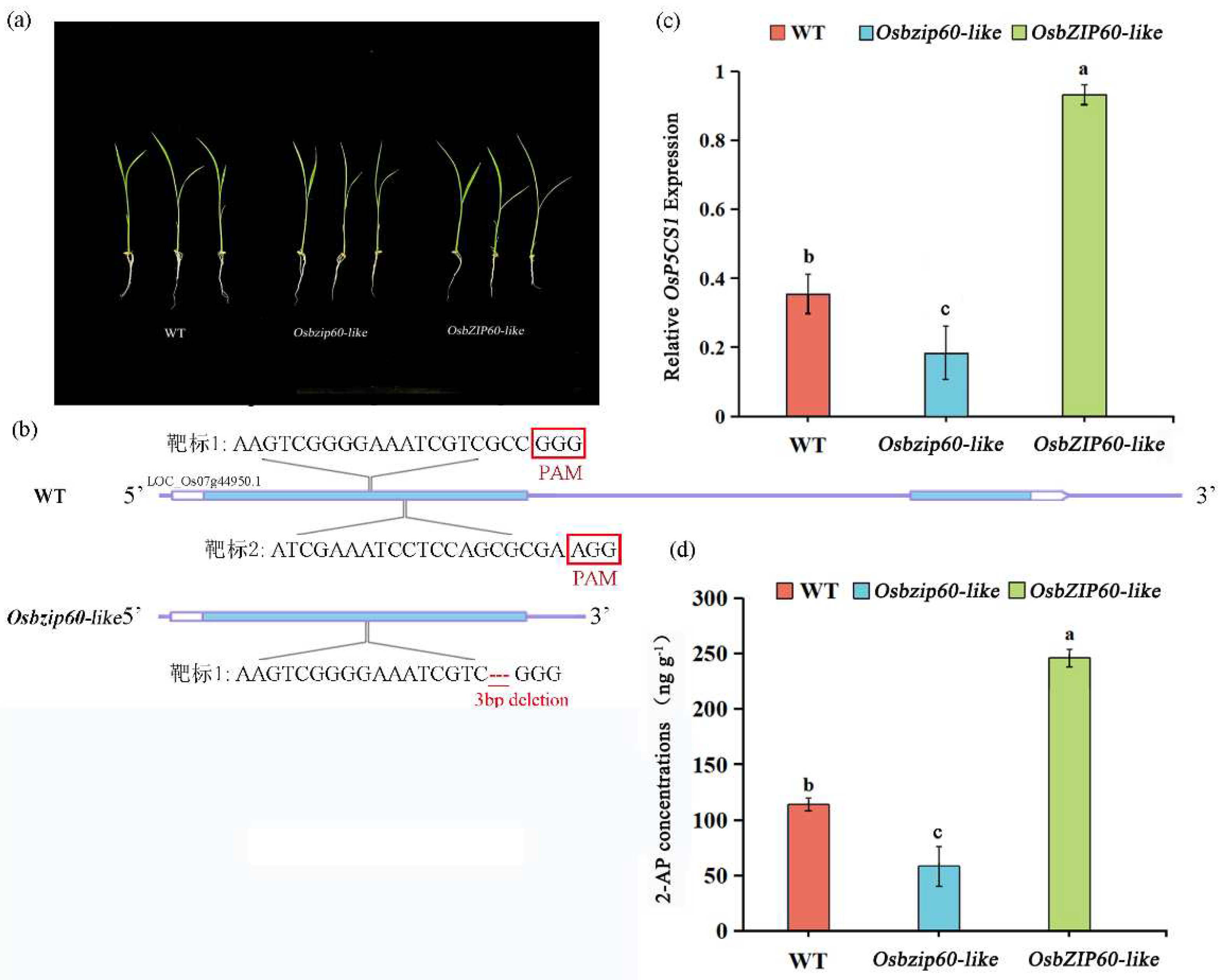
3.6. Transcription Factor OsbZIP60-Like and 2-AP Biosynthesis
There was no significant difference in the growth of the WT, the
Osbzip60-like mutants and the
OsbZIP60-like over-expressing plants (
Fig. 6a). The 3bp deletion of the target site in the mutant affected the function of the protein (
Fig. 6b). Compared with the WT, the expression of
OsP5CS1 gene was down-regulated by 48.65% in
Osbzip60-like mutants and up-regulated by 124.32% in
OsbZIP60-like over-expressing plants (
Fig. 6c). Compared with the WT, the 2-AP concentrations were decreased by 45.45% in
Osbzip60-like mutants and increased by 100% in
OsbZIP60-like over-expressing plants (
Fig. 6d). Overall, transcription factor OsbZIP60-like was a positive regulator of 2-AP accumulation.
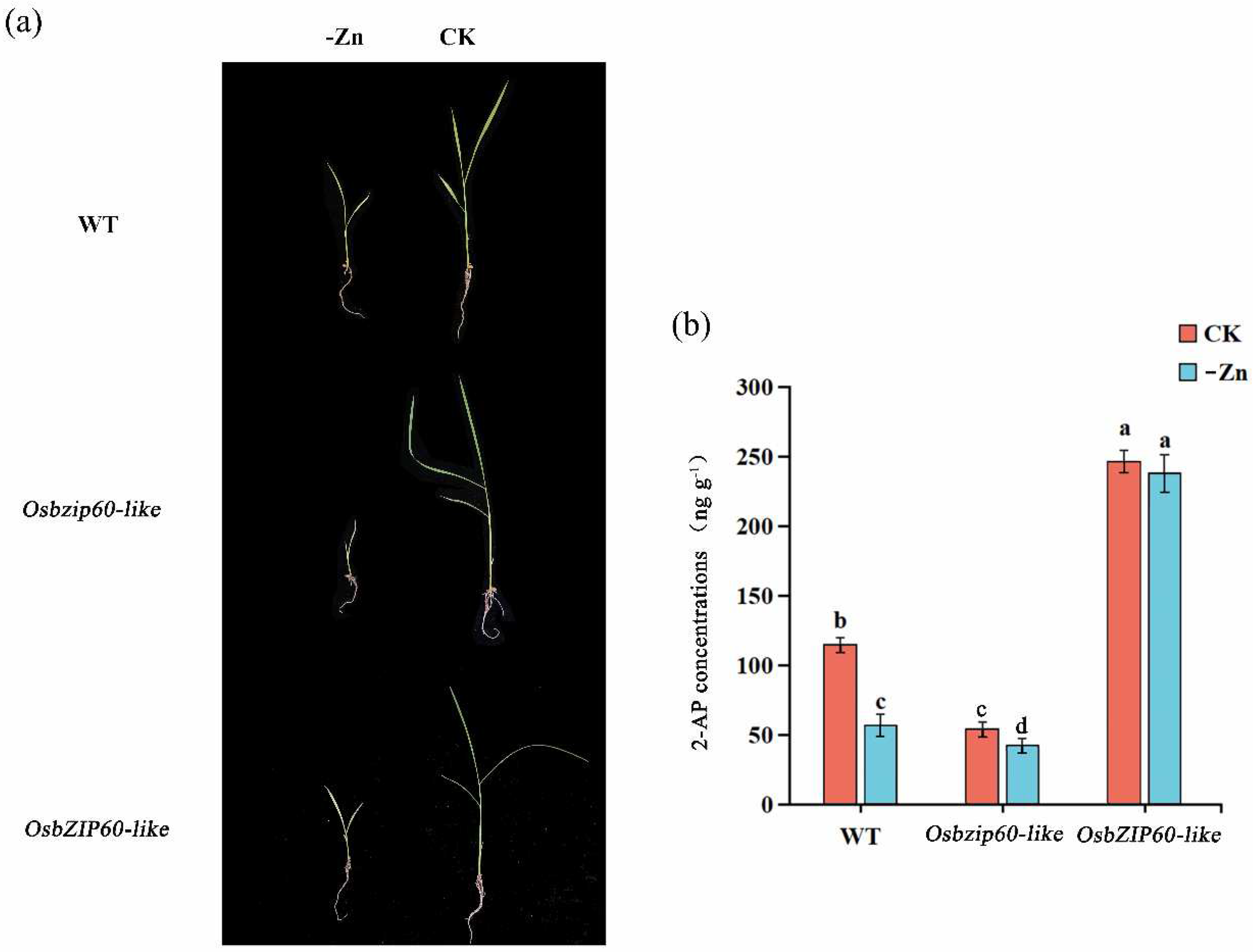
3.7. OsbZIP60-Like Mutants Were Sensitive to Zn Deficiency
The WT, the
Osbzip60-like mutants and the
OsbZIP60-like over-expressing plants were cultured in normal and Zn-deficient medium for two weeks, respectively (
Fig. 7a). Compared with the controls, the 2-AP concentrations were decreased by 47.82% and 27.27% in the WT and
Osbzip60-like mutants in Zn deficient conditions. Moreover, the concentrations of 2-AP in the
OsbZIP60-like over-expressing plants were remained stable after Zn deficiency treatment (
Fig. 7b). Overall, the results showed that the
Osbzip60-like mutant was sensitive to Zn deficiency.
4. Discussion
The expression pattern of
OsP5CS1 gene was consistent with the accumulation pattern of 2-AP content by foliar Zn application of fragrant rice at heading stage (Bao et al., 2021). In order to further verify that
OsP5CS1 gene is the node gene in 2-AP biosynthesis of fragrant rice, CRISPR/Cas9 technology was used to construct the knockout mutant of
Osp5cs1 gene, and
OsP5CS1 over-expression vector was constructed to obtain over-expression plants. Compared with the WT, the 2-AP concentrations were significantly decreased in
Osp5cs1 mutants and increased in
OsP5CS1-over-expressing plants (
Fig. 2b). The
OsP5CS1 gene was proved to be a key gene in the 2-AP biosynthesis pathway of fragrant rice. There are many studies on the regulation of
P5CS expression in different plant tissues in response to different environmental signals, but little is known about the transcriptional regulation mechanism of the
P5CS gene. Cis-acting element analysis was performed on the promoters of rice
OsP5CS1 and
OsP5CS2 genes, respectively. Twenty-four different types of transcription factors binding sites were predicted in the promoters of these two genes, including AP2, bZIP, MYB, and NAC families of transcription factors (Zarattini and Forlani, 2017). The transcription factors binding to the promoter of the
OsP5CS1 gene has not been confirmed, and how the expression of the O
sP5CS1 gene is regulated to affect the concentration of 2-AP has not been reported yet. In present study, the Yeast one-hybrid assays and EMSA techniques determined that the transcription factor OsbZIP60-like specifically bound to the promoter of the
OsP5CS1 gene (
Fig. 3 and
Fig. 4) whereas the OsbZIP60-like was required for transcriptional activation of
OsP5CS1 gene expression (
Fig. 5).
Zn-binding proteins account for about 10% of the proteome in eukaryotes (Choi and Bird, 2014). The F-bZIP transcription factors AtbZIP19 and AtbZIP23 were central regulators of Zn deficiency response in Arabidopsis, and the double mutants were sensitive to Zn deficiency (Assunção et al., 2010; Lilay et al., 2021). Under Zn deficient conditions, the AtbZIP19 and AtbZIP23 could bind to the promoter of Zn transport-related genes, activate their transcription, and maintain Zn homeostasis in cells (Ishimaru et al., 2005). Under Zn-sufficient conditions, he Zn
2+ bound to the Cys-His-rich motif of AtbZIP19 and AtbZIP23 inhibited their own activity and failed to activate transcription of downstream Zn transport genes (
ZIP or
NAS) (Jamsheer and Kumar, 2021). The OsbZIP48 had the highest sequence similarity with AtbZIP19 and AtbZIP23 in rice, and was considered to be a Zn receptor in rice, maintaining Zn homeostasis (Lilay et al., 2020). However, F-bZIP transcription factors as a Zn receptor regulating aroma of fragrant rice has not been reported yet. We used the
OsP5CS1 promoter as bait and screened OsbZIP60-like through the yeast one-hybrid assays (
Fig. 3). Protein domain analysis revealed that OsbZIP60-like belongs to F-bZIP, and amino acids contain Cys and His residues bound to Zn
2+ (
Fig. 3d). The EMSA demonstrated that OsbZIP60-like specifically bound to
OsP5CS1 gene in vitro (
Fig. 4). The dual luciferase reporting system found that OsbZIP60-like promoted transcriptional activation of
OsP5CS1 (
Fig. 5b). Compared with the WT, the
OsP5CS1 gene expression was significantly down-regulated in
Osbzip60-like mutant, and the 2-AP concentrations were significantly decreased. The
OsP5CS1 gene expression was significantly up-regulated in
OsbZIP60-like over-expressed plants, and the 2-AP concentrations were significantly increased (
Fig. 6cd). And the
Osbzip60-like mutants were sensitive to Zn deficient conditions (
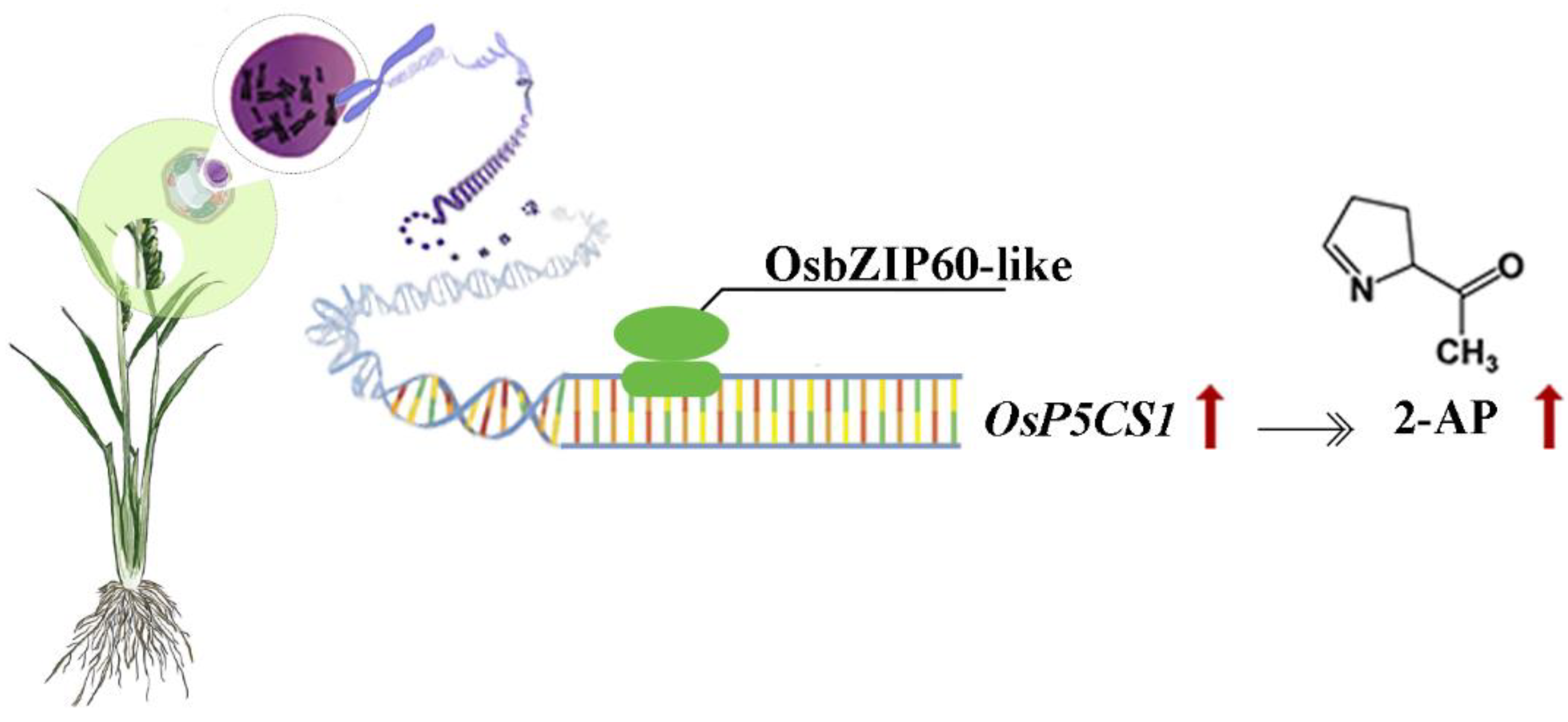
Fig. 7). In this study, OsbZIP60-like acted as a positive regulator to increase 2-AP content by increasing the expression of
OsP5CS1 gene (
Fig. 8). However, whether the OsbZIP60-like is a Zn receptor and how OsbZIP60-like as a zinc receptor regulates the mechanism of 2-AP in fragrant rice still needs to be explored.
Author Contributions
GB designed the experiment. GB, UA, LL and JQ performed the experiment, data collection, lab analysis, and data analysis. GB and LL contributed in providing chemicals, reagent, analyses, and tools. GB, UA, CW and YZprepare the initial draft. GB and UA finalized the initial draft. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the Natural Science Foundation of China (32001115), and the open competition program of top ten critical priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2022SDZG05), and the grant from the Basic and Applied Basic Research Foundation of Guangdong Province (2022A1515111059), the grant from the Guangzhou Science and Technology Plan Project (2023A04J1432).
References
- Ashraf U, Hussain S, Shahid M, Anjum S, Kondo M, Mo Z, Tang X. (2022) Alternate wetting and drying modulated physio-biochemical attributes, grain yield, quality, and aroma volatile in fragrant rice. Physiologia Plantarum, 174(6): e13833. [CrossRef]
- Assunção A, Herrero E, Lin Y, Huettel B, Talukdar S, Smaczniak C, Immink R, van Eldik M, Fiers M, Schat H, Aarts M. (2010) Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proceedings of the National Academy of Sciences of the United States of America, 107: 10296-10301. [CrossRef]
- Bao G, Ashraf U, Wan X, Zhou Q, Li S, Wang C, He L, Tang X. (2021) Transcriptomic analysis provides insights into foliar zinc application induced up-regulation in 2-Acetyl-1-pyrroline and related transcriptional regulatory mechanism in fragrant rice. Journal of Agricultural and Food Chemistry, 69: 11350–11360. [CrossRef]
- Bao G, Ashraf U, Wang C, He L, Wei X, Zheng A, Mo Z, Tang X. (2018) Molecular basis for increased 2-acetyl-1-pyrroline contents under alternate wetting and drying (AWD) conditions in fragrant rice. Plant Physiology and Biochemistry, 133: 149-157. [CrossRef]
- Bao G, Huang S, Ashraf U, Qiao J, Zheng A, Zhou Q, Li L, Wan X. (2022a) Insights of improved aroma under additional nitrogen application at booting stage in fragrant rice. Genes, 13, 2092. [CrossRef]
- Bao G, Zhou Q, Li S, Ashraf U, Huang S, Miao A, Cheng Z, Wan X, Zheng Y. (2022b) Transcriptome analysis revealed the mechanisms involved in ultrasonic seed treatment-induced aluminum tolerance in peanut. Froniters in Plant Science, 12: 807021. [CrossRef]
- Bradbury L, Gillies S, Brushett D, Waters D, Henry R. (2008) Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Molecular Biology, 68: 439-449. [CrossRef]
- Choi S, Bird A J. (2014) Zinc’ing sensibly: controlling zinc homeostasis at the transcriptional level. Metallomics, 6: 1198-1215. [CrossRef]
- Colvin R A, Holmes W R, Fontaine C P, Maret W. (2010) Cytosolic zinc bufering and mufing: their role in intracellular zinc homeostasis. Metallomics, 2: 297-356. [CrossRef]
- Fang Q, Zhou F, Zhang Y, Singh S, Huang C. (2021) Degradation of STOP1 mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant Journal, 106(2): 493-506. [CrossRef]
- Forlani G, Bertazzini M, Zarattini M, Funck D. (2015) Functional characterization and expression analysis of rice δ1-pyrroline-5-carboxylate dehydrogenase provide new insight into the regulation of proline and arginine catabolism. Frontiers in Plant Science, 6: 591-608. [CrossRef]
- Ghosh P, Roychoudhury A. (2018) Differential levels of metabolites and enzymes related to aroma formation in aromatic indica rice varieties: comparison ith non-aromatic varieties. 3 Biotech, 8: 25-38. [CrossRef]
- Hellman L, Fried M. (2007) Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nature protocols, 2: 1849-1861. [CrossRef]
- Hinge V, Patil H, Nadaf A. (2016) Comparative characterization of aroma volatiles and related gene expression analysis at vegetative and mature stages in basmati and non-basmati rice (Oryza Sativa L.) cultivars. Applied Biochemistry and Biotechnology, 178: 619-639. [CrossRef]
- Huang T, Teng C, Chang J, Chuang H, Ho C, Wu M. (2008) Biosynthetic mechanism of 2-acetyl-1-pyrroline and its relationship with delta1-pyrroline-5-carboxylic acid and methylglyoxal in aromatic rice (Oryza sativa L.) callus. Journal of Agricultural and Food Chemistry, 56(16): 7399-7404. [CrossRef]
- Hu S, Huang Q, Xu Q. (2001) Study on the relationship between aromatic rice quality and trace element content. Crop science, 15(4): 12-15. [CrossRef]
- Imran M, Shafiq S, Ashraf U, Qi J, Mo Z, Tang X. (2023) Biosynthesis of 2-Acetyl-1-pyrroline in fragrant rice: recent insights into agro-management, environmental factors, and functional genomics. Journal of Agricultural and Food Chemistry, 71(10): 4201-4215. [CrossRef]
- Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa N. (2005) OsZIP4, a novel zinc-regulated zinc transporter in rice. Journal of Experimental Botany, 56: 3207-3214. [CrossRef]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, Group B. (2002) bZIP transcription factors in Arabidopsis. Trends in Plant Science, 7: 106-111. [CrossRef]
- Jamsheer K M, Kumar M. (2021) Transcription factors as zinc sensors in plants. Trends in Plant Science, 26(8): 761-763. [CrossRef]
- Jezussek M, Juliano B, Schieberle P. (2002) Comparison of key aroma compounds in cooked brown rice varieties based on aroma extract dilution analyses. Journal of Agricultural and Food Chemistry, 50: 1101-1105. [CrossRef]
- Joo H, Baek W, Lim C, Lee S. (2021) Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses. Current Genomics, 22: 4-15. [CrossRef]
- Kaikavoosi K, Kad T, Zanan R, Nadaf A. (2015) 2-Acetyl-1-Pyrroline augmentation in scented indica Rice (Oryza sativa L.) varieties through Δ1-Pyrroline-5-Carboxylate Synthetase (P5CS) gene transformation. Applied Biochemistry Biotechnology, 177: 1466-1479. [CrossRef]
- Li M, Ashraf U, Tian H, Mo Z, Pan S, Anjum S, Duan M, Tang X. (2016) Manganese-induced regulations in growth, yield formation, quality characters, rice aroma and enzyme involved in 2-acetyl-1-pyrroline biosynthesis in fragrant rice. Plant Physiology and Biochemistry, 103:167-75. [CrossRef]
- Lilay G, Castro P, Guedes J, Almeida D, Campilho A, Azevedo H, Aarts M, Saibo N, Assunção A. (2020) Rice F-bZIP transcription factors regulate the zinc deficiency response. Journal of Experimental Botany, 71: 3664-3677. [CrossRef]
- Lilay G, Persson D, Castro P, Liao F, Alexander R, Aarts M, Assunção A. (2021) Arabidopsis bZIP19 and bZIP23 act as zinc sensors to control plant zinc status. Nature Plants, 7: 137-143. [CrossRef]
- Luo H, Du B, He L, He J, Hu L, Pan S, Tang X. (2019) Exogenous application of zinc (Zn) at theheading stage regulates 2-acetyl-1-pyrroline (2-AP) biosynthesis in different fragrant ricegenotypes. Scientific Reports, 9: 19513-19523. [CrossRef]
- Maret W, and Li Y. (2009) Coordination dynamics of zinc in proteins. Chemical Reviews, 109:4682-4707. [CrossRef]
- Mo Z, Li W, Pan S, Fitzgerald T, Xiao F, Tang Y, Wang Y, Duan M, Tian H, Tang X. (2015) Shading during the grain filling period increases 2-acetyl-1-pyrroline content in fragrant rice. Rice, 8: 9-19. [CrossRef]
- Nijhawan A, Jain M, Tyagi A, Khurana J. (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiology, 146(2): 333-350. [CrossRef]
- Niu X, Tang W, Huang W, Ren G, Wang Q, Luo D, Xiao Y, Yang S, Wang F, Lu B, Gao F, Lu T, Liu Y. (2008) RNAi-directed downregulation of OsBADH2 results in aroma (2-acetyl-1-pyrroline) production in rice (Oryza sativa L.). BMC Plant Biology, 8: 100-110. [CrossRef]
- Poonlaphdecha J, Gantet P, Maraval I, Sauvage F, Menut C, Morère A, Boulanger R, Wüst M, Gunata Z. (2016) Biosynthesis of 2-acetyl-1-pyrroline in rice calli cultures: demonstration of 1-pyrroline as a limiting substrate. Food Chemistry, 197: 965-971. [CrossRef]
- Prodhan Z, Shu Q. (2020) Rice aroma: A natural gift comes with price and the way forward. Rice Science. 27: 86-100. [CrossRef]
- Rai A, Penna S. (2013) Molecular evolution of plant P5CS gene involved in proline biosynthesis. Molecular Biology Reports, 40: 6429-6435. [CrossRef]
- Shan Q, Zhang Y, Chen K, Zhang K, Gao C. (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnology Journal, 13(6): 791-800. [CrossRef]
- Szabados L, Savouré A. (2010) Proline: a multifunctional amino acid. Trends in Plant Science. 15: 89-97. [CrossRef]
- Tang Y, Abdelrahman M, Li J, Wang F, Ji Z, Qi H, Wang C, Zhao K. (2020) CRISPR/Cas9 induces exon skipping that facilitates development of fragrant rice. Plant Biotechnology Journal, 19: 642-644. [CrossRef]
- Wang X, Ren Y, Ashraf U, Gui R, Deng H, Dai L, Tang X, Wang Z, Mo Z. (2023) Optimized splitting and nitrogen rates of liquid fertilizer management improve grain yield, biomass accumulation, and nutrient uptake of late-season indica fragrant rice. Journal of the Science of Food and Agricultural, 103(14): 6800-6813. https://doi.org/10.1002/jsfa.12767.Wakte K, Zanan R, Hinge V, Khandagale K, Nadaf A, Henry R. (2016) Thirty-three years of 2-acetyl-1-pyrroline, a principal basmati aroma compound in scented rice (Oryza sativa L.): a status review. Journal of the Science of Food and Agriculture, 97: 384-395. https://doi.org/10.1002/jsfa.7875.
- Zarattini M, Forlani G. (2017) Toward unveiling the mechanisms for transcriptional regulation of proline biosynthesis in the plant cell response to biotic and abiotic stress conditions. Frontiers in Plant Science, 8: 927-935. [CrossRef]
Figure 2.
Comparison of 2-AP concentrations in wild type (WT), Osp5cs1 mutant and OsP5CS1 over-expressing plants. (a) Phenotype of seedlings after the second week of growth (b) 2-AP concentrations; * means there was a difference at P<0.05 by LSD test.
Figure 2.
Comparison of 2-AP concentrations in wild type (WT), Osp5cs1 mutant and OsP5CS1 over-expressing plants. (a) Phenotype of seedlings after the second week of growth (b) 2-AP concentrations; * means there was a difference at P<0.05 by LSD test.
Figure 3.
OsbZIP60-like transcription factor screened by yeast single hybrid. (a) Background screening of defective plates with different concentrations (0, 10, 20, 30, 40, 50, 75 mM 3AT) of 3AT, (b) 96 monoclonal colonies grew on SD-TLH+50 mM 3AT plates of the screening librar, (c) Rotational validation of monoclonal; (+) positive control pGAD53m+pHIS2::p53, (d) OsbZIP60-like protein structure analysis.
Figure 3.
OsbZIP60-like transcription factor screened by yeast single hybrid. (a) Background screening of defective plates with different concentrations (0, 10, 20, 30, 40, 50, 75 mM 3AT) of 3AT, (b) 96 monoclonal colonies grew on SD-TLH+50 mM 3AT plates of the screening librar, (c) Rotational validation of monoclonal; (+) positive control pGAD53m+pHIS2::p53, (d) OsbZIP60-like protein structure analysis.
Figure 4.
The transcription factor OsbZIP60-like specifically bound to the OsP5CS1 promoter. (a) Oligonucleotide sequences used in EMSA, (b) OsbZIP60-like binds to ProOsP5CS1-Probe-1, ProOsP5CS1-Probe-2, ProOsP5CS1-Probe-3 and ProOsP5CS1-Probe-4. "-" and "+" denote negative and positive controls, respectively.
Figure 4.
The transcription factor OsbZIP60-like specifically bound to the OsP5CS1 promoter. (a) Oligonucleotide sequences used in EMSA, (b) OsbZIP60-like binds to ProOsP5CS1-Probe-1, ProOsP5CS1-Probe-2, ProOsP5CS1-Probe-3 and ProOsP5CS1-Probe-4. "-" and "+" denote negative and positive controls, respectively.
Figure 5.
The OsP5CS1 required the OsbZIP60-like for transactivation. (a) The OsP5CS1 promoter was connected to the firefly lucifin reporter, (b) The corresponding relative ratio of LUC/REN was shown on the right. Error bars indicate SE from six replicates. Asterisks indicate significant differences as determined by t-test analysis (P< 0.05).
Figure 5.
The OsP5CS1 required the OsbZIP60-like for transactivation. (a) The OsP5CS1 promoter was connected to the firefly lucifin reporter, (b) The corresponding relative ratio of LUC/REN was shown on the right. Error bars indicate SE from six replicates. Asterisks indicate significant differences as determined by t-test analysis (P< 0.05).
Figure 6.
Comparison of 2-AP concentrations in wild type (WT), OsbZIP60-like mutant and OsbZIP60-like-1-CP complementing lines. (a) The growth phenotypes of wild-type (WT), Osbzip60-like mutant and OsbZIP60-like over-expressed plants at the second-week seedling stage, (b) Osbzip60-like mutant constructed by CRISPR/Cas9 system, (c) wild-type (WT), The expression of OsP5CS1 gene in Osbzip60-like mutants and OsbZIP60-like over-expressed plants, (d) The determination of 2-AP concentrations in wild type (WT), OsbZIP60-like mutants and OsbZIP60-like over-expressed plants. Different letters above the bar graphs indicate differences at P < 0.05 by the LSD test.
Figure 6.
Comparison of 2-AP concentrations in wild type (WT), OsbZIP60-like mutant and OsbZIP60-like-1-CP complementing lines. (a) The growth phenotypes of wild-type (WT), Osbzip60-like mutant and OsbZIP60-like over-expressed plants at the second-week seedling stage, (b) Osbzip60-like mutant constructed by CRISPR/Cas9 system, (c) wild-type (WT), The expression of OsP5CS1 gene in Osbzip60-like mutants and OsbZIP60-like over-expressed plants, (d) The determination of 2-AP concentrations in wild type (WT), OsbZIP60-like mutants and OsbZIP60-like over-expressed plants. Different letters above the bar graphs indicate differences at P < 0.05 by the LSD test.
Figure 7.
The effect of zinc deficiency on 2-AP accumulation in WT, Osbzip60-like mutanst and OsbZIP60-like over-expressed plants. (a) WT, Osbzip60-like mutants and OsbZIP60-like over-expressed plants were cultured with zinc deficiency for 2 weeks. (b) CK: controlled water spray treatment; -Zn: zinc deficiency treatment. Different letters above the bar graphs indicate differences at P < 0.05 by the LSD test.
Figure 7.
The effect of zinc deficiency on 2-AP accumulation in WT, Osbzip60-like mutanst and OsbZIP60-like over-expressed plants. (a) WT, Osbzip60-like mutants and OsbZIP60-like over-expressed plants were cultured with zinc deficiency for 2 weeks. (b) CK: controlled water spray treatment; -Zn: zinc deficiency treatment. Different letters above the bar graphs indicate differences at P < 0.05 by the LSD test.
Figure 8.
Hypothesized model of OsbZIP60-like regulation of 2-AP content. OsbZIP60-like, as a positive regulator, increased 2-AP content by increasing the expression of OsP5CS1 gene.
Figure 8.
Hypothesized model of OsbZIP60-like regulation of 2-AP content. OsbZIP60-like, as a positive regulator, increased 2-AP content by increasing the expression of OsP5CS1 gene.
Table 1.
Prediction of cis-acting elements in the OsP5CS1 promoter.
Table 1.
Prediction of cis-acting elements in the OsP5CS1 promoter.
| Response element type |
Number of cis-acting elements |
| Abscisic acid (ABA) responsiveness |
|
| ABRE |
11 |
| ABRE3a |
4 |
| ABRE4 |
4 |
| Salicylic acid (SA) responsiveness |
|
| ERE |
1 |
| TCA-element |
1 |
| Auxin responsive element |
|
| TGA-element |
1 |
| Defense and stress responsiveness |
|
| TC-rich repeats |
1 |
| Light responsiveness |
|
| ATCT-motif |
4 |
| G-box |
10 |
| GATA-motif |
1 |
| TCC-motif |
2 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).