1. Introduction
Probiotics are beneficial for the health of humans. The most popularly consumed probiotics are Lactobacillus acidophilus (Anjam et al., 2014). Research has demonstrated that the presence of L. acidophilus can produce some potential probiotic effects in humans, such as; performing as a barrier against pathogens, recovering lactose intolerance, boosting immune response, and decreasing cholesterol levels (Alberts, 1987). L. acidophilus can colonize the intestines of humans and inhibit pathogens, such as Escherichia coli, Salmonella typhimurium, Streptococcus aureus, and so on. These pathogens also inhabit the intestines (Anjam et al., 2014, Rolhion at al., 2015). These probiotics are applied to prevent diarrheal infections as well (Britton et al. 2009). According to the last Global Burden of Disease Study, approximately 2.39 billion people were infected by diarrheal cases globally and nearly 0.53 million under-five children died yearly. In lower and middle-income (LMI) countries, specifically, Bangladesh, incidence and case-fatality ratios are much higher as compared with that of developing countries (Sarker et al., 2018). In Bangladesh, the average cost per episode was US $ 67.18, while the moderate inpatient and outpatient charge were US $ 110.51 and US $ 23.62 respectively. The cost was substantially largest for impoverished households, 21.45% of household income, compared to 4.21% of the prosperous people (Sarker et al., 2018). L. acidophilus can be a cost-saving option for preventing diarrheal disease and pathogenic Escherichia coli-related other food-borne diseases of billions of people around the world.
Potential probiotic strains are vital for killing pathogens in the intestines. In Bangladesh most of all, marketed probiotics are purchased from foreign countries. There is very little research on isolated probiotics in Bangladesh (Anjuman et al., 2015). Moreover, assessment of the potential probiotic traits of a strain is essential for claiming it as a probiotic strain (Sarker et al., 2018; Chidre et al., 2017). Isolation of potential probiotics from Bangladeshi natural sources is significant to boost the immunity of the masses of this geographic territory. In this study, we focused on the isolation of a new probiotic strain of L. acidophilus from Bangladeshi natural sources.
Probiotic dahi is a good option for delivering probiotics to the intestines. Dahi is a customary yogurt or fermented milk product in the Indian subcontinent, usually produced from cow's milk, and also from buffalo milk, or goat milk. It is consumed all around the Indian subcontinent (Caballero et al., 2015). We can get two benefits from probiotic dahi: one is a test, and another one is probiotic culture. In the Bangladeshi market, there are no probiotic dahi having levels of specific probiotic bacteria (Anjuman et al., 2015).
The shelf life of dahi is another important factor for maintaining the quality of dahi up to the expiration date. There is a correlation between pH, temperature, probiotic growth, and contamination in dahi (Schmidt et al., 1996; Yang et al., 2018). The preparation of dahi in unhygienic environments and contaminated starter cultures can spoil dahi and cause food poison as well. Shelf life is dependent on this issue. Shelf life depends on another factor, that is, total live probiotic count Guesh et al., 2020; Fijan, 2016).
The main target of this topical investigation was to prepare a probiotic dahi having potential probiotic L. acidophilus in dosage form, that is, the number of probiotics would not be less than a certain number of colony forming units per gram of dahi. Secondly, we aimed to find the capability of dahi for inhibiting the growth of contaminations. Information on probiotic growth and inhibition of microbial contamination would be used for determining shelf life of dahi.
2. Materials and Methods
2.1. Probiotic isolation and identification
2.1.1. Probiotic isolation
Probiotic species were isolated from a number of samples of Dahi in Dhaka, Bangladesh. 1 mL of 10-9 decimal diluted sample (suspended in normal 0.9% (w/v) saline solution) was spread on 20-25mL MRS (de Man Rogosa Sharpe, Oxoid, UK) agar medium, and the plates were incubated for 24-72 h in 37°C. Based on morphology, the distinguished most common bacterial colonies were selected from MRS agar media. To purify colonies, the isolates were streaked on the same media and finally, the pure colonies were transferred to MRS broth with 15% glycerol for further research. The Leica MZ9.5 (Germany), a potent stereo microscopic instrument that features a fantastic 9.5:1 zoom ratio and magnification capabilities as high as 480x, was utilized to analyze the colony morphology.
2.1.2. Gram staining test, 16S gene sequencing, and phylogenetic tree
The selected colonies were examined using gram staining protocol according to Coica (2005) method, and then observed under light microscope by 100X resolution (Coico, 2005). The MRS-agar cultured colonies were used for genomic DNA isolation with Phenol chloroform chemical lysis method according to the manufacturer’s protocol. After DNA extraction, the concentration and purity of DNA was checked using Nanodrop® spectrophotometer ND2000 (Thermo Scientific, USA). The 260/280 ratio (absorbance at 260nm and 280nm) provided an indication of the purity of the DNA. The ratio of the sample obtained is above 1.8. In this study, 16S rDNA amplification and sequencing were performed based on the methodology described previously by Rahman et al. (2017). The universal 16S rRNA primer set for polymerase chain reaction (PCR) amplification was as follows: 27f (5'AGAGTTTGATCCTGGCTCAG-3'and 1492r (5'- GGTTACCTTGTTACGACTT-3') (Rahman et al., 2017).
The phylogenetic tree was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). This analysis involved 12 nucleotide sequences. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021).
2.1.3. Cellular morphology analysis under Transmission Electron Microscope
With a plasma cleaner (Power: Low, 12 sec), we hydrophilized the grid beforehand. After that, a tabletop centrifuge was put to work to spin the bacterial culture for two minutes at 2,000–4,000 rpm. Ten microliters of pure water were used for the re-suspension of the particle. After applying the specimen to the grid, it was left in place for one minute. The samples had been incubated for one minute, stained with a 1.8% uranyl acetate solution, and then allowed to air dry. The photos of bacterial samples were captured with a JEM 1010 electron transmission microscope (JEOL).
2.2. Probiotic potentiality and safety test
2.2.1. Antibiotic sensitivity test by agar well diffusion method
An antibiotic sensitivity test was conducted against the probiotic isolate JB2CON following the method of Chetan et al. (2017) with some modifications. 8 mL MRS agar (with log 8 CFU/mL of the isolate JB2CON) overplayed on previously solidified MRS agar (Chetan et al., 2017). Cefuroxim (100μl/30μg), Gentamycin (100μl/10μg), Chloramphenicol (100μl/30μg), Erythromycin (15 µg / 100 µL), Clindamycin (10 µg / 100 µL), Tylosin (30 µg / 100 µL), Amphicilin (10 µg / 100 µL), Vancomycin (30 µg / 100 µL), Kanamycin (30 µg / 100 µL), Streptomycin (10 µg / 100 µL), and Tetracycine (30 µg / 100 µL) were filled in a 7mm diameter well in MRS agar media with an upper layer of the probiotic bacterial strain. The test Petri dishes were incubated for 24 hours at 37°C. Antibiotic sensitivity for every antibiotic was tested twice.
2.2.2. Hemolysis test
To evaluate for hemolytic activity, blood agar (BD, USA) plates were streaked with MRS broth that contained JB2CON strain cultures and underwent incubation for 72 hours at 37 °C. Next, each of the plates were inspected to see if the Lactobacillus colonies were surrounded by any greenish (α-hemolysis) or clean (β-hemolysis) hemolytic zones, or if there was none at all (γ-hemolysis). The test was conducted for three times.
2.2.3. Acid tolerance
The acid tolerance of the isolate JB2CON was tested following the method of Ortakci et al. (2012) with some modifications (Ortakci et al., 2012). 0.2g NaCl plus 0.7 mL HCl (Stock solution), and sufficient water were used to make 100 mL simulated gastric juice (SGJ of pH 1.4). SGJ and sterilized dahi were mixed in a ratio of 1:4 for making a solution of pH 2.35 (pH <3) (Ortakci et al., 2012) This final solution was used for analyzing the capability of the isolate JB2CON in our study to tolerate acidity (pH <3). 1 mL probiotic isolate (1.28×108 CFU/mL) was added to 9 mL final solution of SGJ and dahi. Average CFU of the probiotic from two tests was counted after 24 hours of incubation at 37°C.
2.2.4. Bile salt tolerance
With minor adjustments, the bile tolerance of the isolate JB2CON was assessed using the methodology of Hassanzadazar et al. (2012) (Hassanzadazar et al., 2012). The isolate JB2CON was incubated for 24 hours at 35°C in MRS broth. Following this time, the cells were suspended using a slow vortex. After that, 1% cell suspension broth was mixed with the MRS broth containing bile salt (0.5% and 0.25% ox-gall). The isolate JB2CON's viability was assessed after 8 hours of incubation by culturing it in petri-dishes using the spread plate technique, which was then incubated for 5 days at 35°C. The control was the cell suspension broth that had been incubated at 35°C for 8 hours without any bile salt added. Two replicates were used for each test.
2.2.5. Antimicrobial activity test by agar well diffusion method
Antimicrobial activity test of the isolate JB2CON was conducted following the method of Chidre et al. (2017) with some modifications. Antagonistic effects against six pathogenic microorganisms were tested in TSA media (Chidre et al., 2017). 100μl supernatant of the cell suspension broth of the isolate JB2CON (final pH 3.98 after 48 hours incubation at 35°C) was placed in a 7mm diameter well in TSA media having a lawn of pathogenic microorganism. Two replicates of antagonistic tests were conducted against each pathogen. Salmonella typhimurium ATCC 14028, Staphylococcus aureus ATCC6538, Escherichia coli ATCC 8739, Bacillus subtitlis ATCC 6633, Pseudomonas aeruginosa ATCC 142, and Candida albicans ATCC 10231 were bought from a local supplier in Dhaka, Bangladesh. Antimicrobial activity against these pathogens was tested twice.
2.2.6. Antimicrobial activity test by time-kill assay against unknown contamination
In the agar diffusion assay we used specific pathogens of ATCC cultures, although in the time-kill assay we employed unknown contaminations. Antimicrobial activity test was performed with some modifications by time-kill assay described by Prabhurajeshwar and Kelmani (2019) (Prabhurajeshwar and Kelmani, 2019). For this assay, random contaminations and all ingredients for dahi (pasteurized milk, sugar, plus the isolate JB2CON) were added to each container before starting incubation. The contaminations were added from natural sources (water, glassware, sucrose, pasteurized milk, etc.) to each container. Three containers were prepared for three different temperatures: Ambient room temperature, 35°C, and 44°C. After 21 hours of incubation, microbial contaminants were observed for dahi samples in 100-fold dilution.
For the contamination test, we prepared media and conducted growth promotion tests according to the guideline of the nutrient and dietary supplements section in the United State Pharmacopeia (USP) (Radhakrishna, 2006). Total Non-Lactic Contamination (TNLC) was tested in Tryptic Soy Broth and Tryptic Soy Agar, and Total Yeast and Mold (TYMC) was tested in Tryptic Soy Broth and Sabouraud Chloramphenicol Agar. Moreover, specific pathogens, particularly, Salmonella tested on selective media, such as Rappaport Vassiliadis Salmonella Enrichment Broth, and Xylose Lysine Deoxycholate agar (XLD agar). The presence of E. coli was observed in MacConkey Broth and MacConkey Agar. Furthermore, the presence of S. aureus and P. aeruginosa were checked on Mannitol Salt Agar and Pseudomonas Cetrimide agar respectively (Radhakrishna, 2006).
2.3.1. Analysis of milk for preparation of dahi
Cow milk sample was collected from Dhaka Milk Industry, Dhaka, Bangladesh. Milk was analyzed for four properties, such as lactose, protein, fat, and minerals. Although these four properties were tested in Dhaka Milk Industry and the result was presented in their label, we re-tested these properties once by a milk analyzer (LACTOSCAN LW, Milkotronic Ltd.) following the method of AOAC (AOAC 13th edition, 2012).
2.3.2. Preparation of probiotic dahi
0.5 L cow milk with 30.0g sucrose was concentrated to 0.4L by heating. Then the temperature of the milk was lowered to about 45°C. 1.0g of the isolate JB2CON was added into each container. The containers were incubated at four different temperature conditions, such as ambient room temperature, 25°C, 35°C, and 44°C for 21 hours. For comparison, the pH of dahi was observed at four temperatures at the set time intervals.
2.4. Shelf-life study at storage condition
For selecting the suitability of the shelf life of probiotic dahi, antimicrobial activity test and shelf-life test were carried out simultaneously. An antimicrobial activity test was performed with some modifications by the time-kill assay described by Chidre et al. (2017) (Chidre et al., 2017). For this, a total of four containers (each one contained probiotic dahi) was incubated at 44°C for 5.50 hours and then stored at freeze temperature (2°C-8°C) for 14 days. Container-1 was filled with all ingredients of dahi (pasteurized milk, sugar, plus the isolate JB2CON) aseptically, and other containers contained the components of container-1 plus additional contaminations. Container-2 was allowed with unknown contamination (natural) and probiotic isolate, container-3 was filled with C. albicans ATCC 10231 plus probiotic isolate, and container-4 was inoculated with E. Coli ATCC 8739 plus probiotic isolate.
The shelf life of probiotic dahi was tested for 14 days in refrigerated (2°C- 8°C) conditions. L. acidophilus must be existed in concentrations of 105 - 106 CFU per mL in order to perform its probiotic actions (Alberts, 1987). As a result, we decided that the dosage of our probiotic dahi for the duration of its shelf life should be at least 106 CFU per mL. The total probiotic count after 14 days was compared with that on the first day at freezing temperature. This comparison was conducted for TNLC, TYMC, C. albicans ATCC 10231, and E. Coli ATCC 8739. The tests were performed twice.
2.5. Consumer acceptance of dahi
Probiotic dahi was prepared according to the process described at point 2.3.2 and incubated at 44°C for 5.50 hours. Consumer acceptance was assessed by a sensory test conducted by the method of Hashim et al. (2009) [
24]. 9-point hedonic scale was used, such as, like extremely=9, like very much=8, like moderately=7, like slightly=6, neither like nor dislike=5, dislike slightly=4, dislike moderately=3, dislike very much=2, and dislike extremely=1 (Hashim et al., 2009). 9-point hedonic scale was applied for four groups having four people in each group.
3. Results and Discussion
3.1. Probiotic isolation and identification
3.1.1. Probiotic isolation
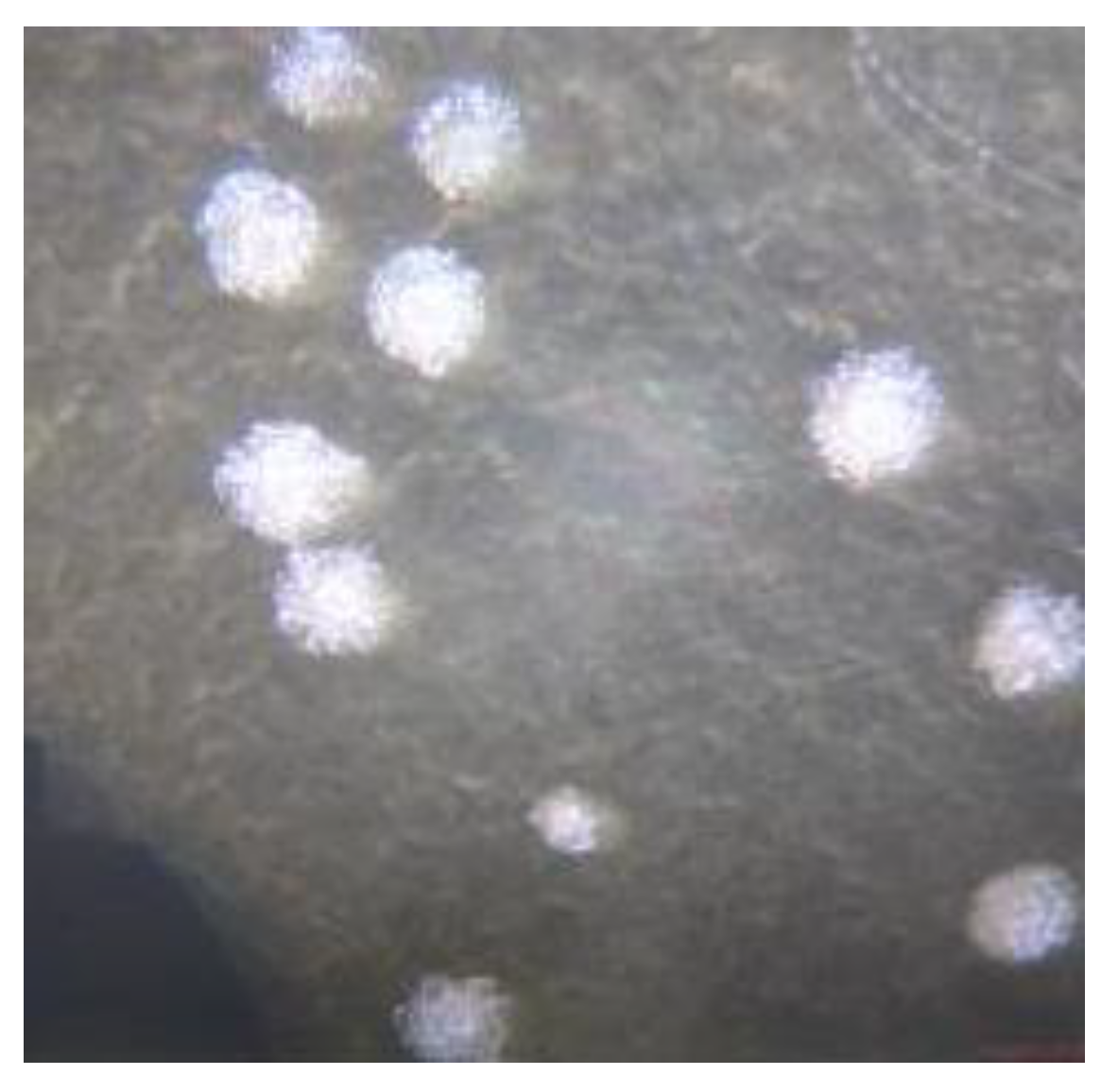
In MRS media, several colonies showed lactobacillus characteristics from morphological viewpoints (
Figure 1). The colonies were white, round, raised, and translucent (Kavitha et al., 2016).
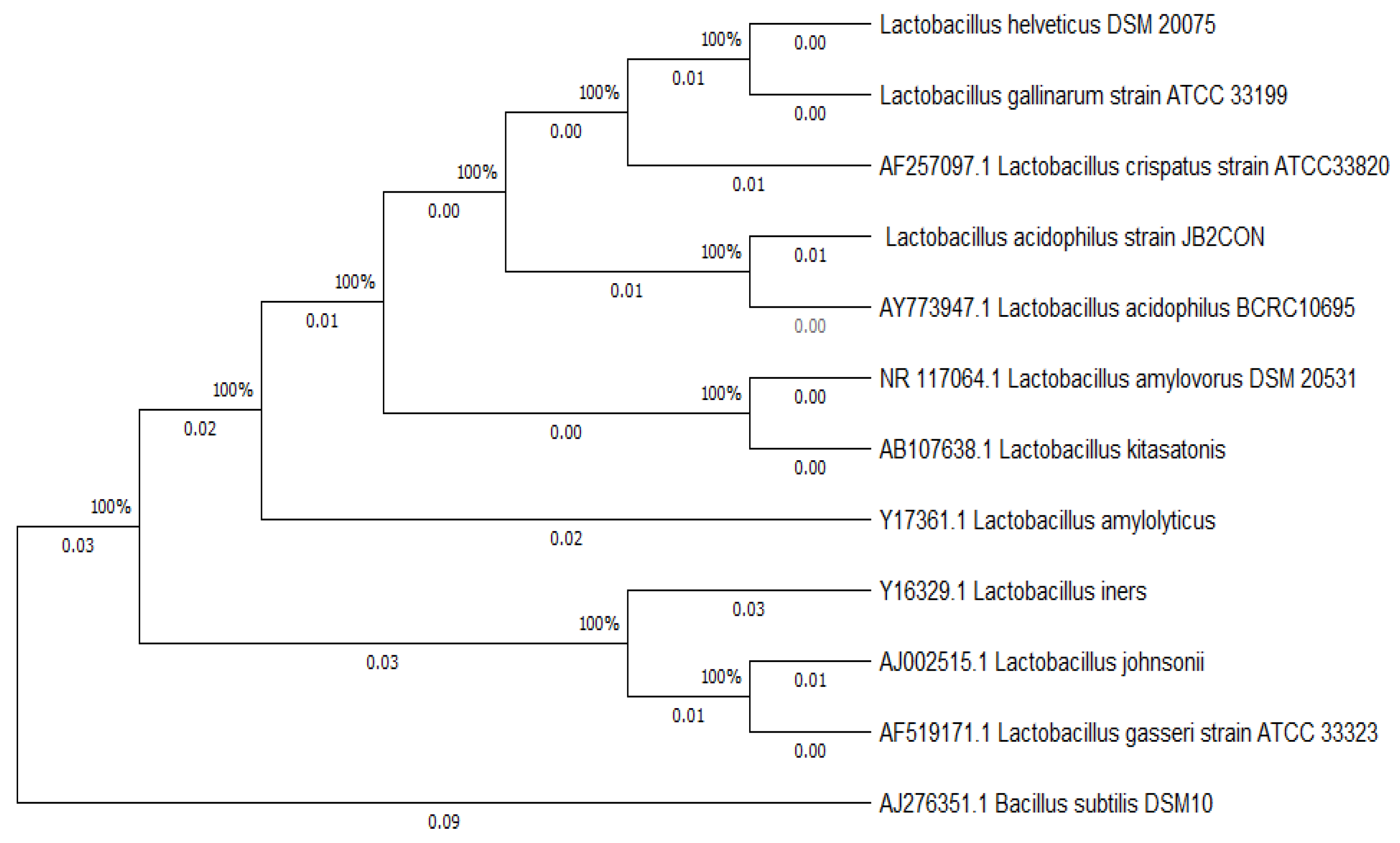
3.1.2. Gram staining test, 16S gene sequencing, and phylogenetic tree
The cells were gram-positive and long rod (Kavitha et al., 2016). According to the 16S gene sequence, the colony JB2CON was identified as
Lactobacillus acidophilus (accession no. OM909067, and strain JB2CON). The phylogenetic tree was constructed with the type strains of 10 species in
L. acidophilus subgroup (Matthew et al., 2013). In the tree of
L. acidophilus subgroup, our isolated strain JB2CON was placed with AY773947.1
L. acidophilus BCRC1069. Therefore, the strain JB2CON was identified as
L. acidophilus. The tree was rooted with the 16S rRNA gene from AJ276351.1 Bacillus subtilis DSM10 (
Figure 2).
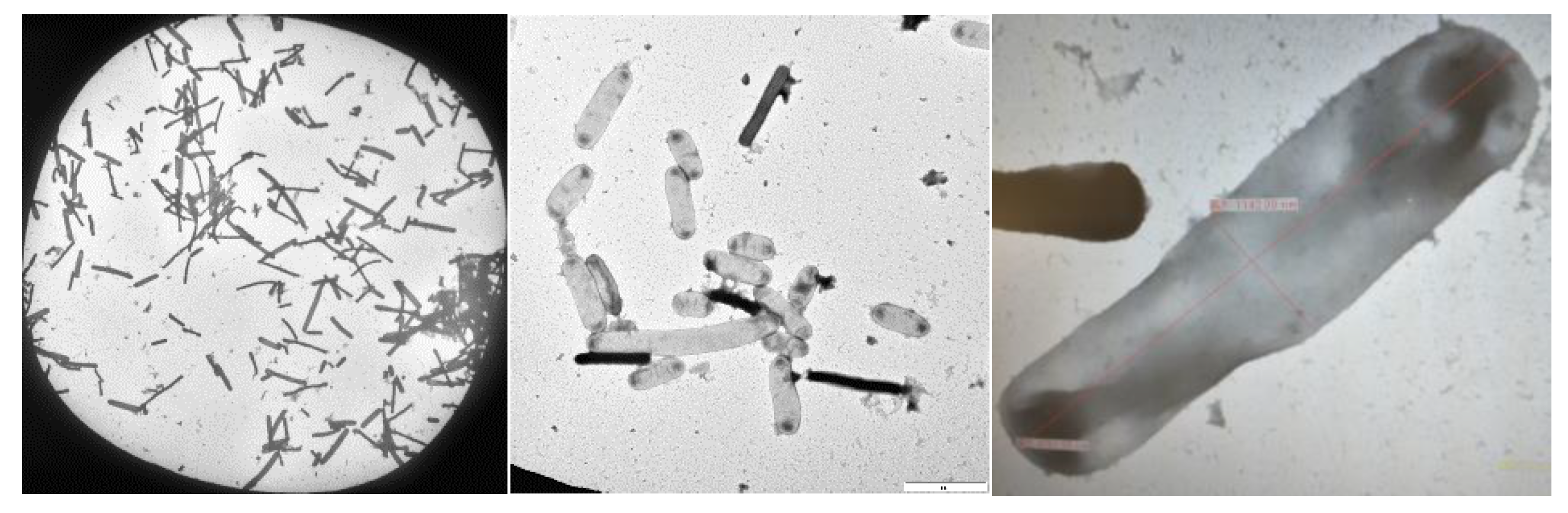
2.1.3. Cellular morphology analysis under Transmission Electron Microscope
Under a transmission electron microscope, microorganisms with rod shape and sizes between 2-5 μm were seen. In the course of our research, we identified rounded-end rods that had been found in pairs, short chains, or even as single cells (
Figure 3).
3.2. Probiotic potentiality and safety test
Lactobacillus acidophilus (LA) is the most common probiotic in the world. The probiotic potentiality of these bacteria was studied in previous studies (Kailasapathy and Chin, 2000). In the current study we tested three probiotic features of the isolate, L. acidophilus JB2CON (accession no. OM909067).
3.2.1. Antibiotic sensitivity test
L. acidophilus JB2CON (OM909067) in our study had a Zone of Inhibition (ZOI) larger than 20mm (diameter). According to the description of the Clinical and Laboratory Standards Institute (CLSI), they were susceptible to Cefuroxim (100μl/30μg), Gentamycin (100μl/10μg), Chloramphenicol (100μl/30μg), Erythromycin (15 µg / 100 µL), Clindamycin (10 µg / 100 µL), Tylosin (30 µg / 100 µL), Amphicilin (10 µg / 100 µL), Vancomycin (30 µg / 100 µL), Kanamycin (30 µg / 100 µL), Streptomycin (10 µg / 100 µL), and Tetracycine (30 µg / 100 µL) (
Table 1) (Prabhurajeshwar et al. 2019, CLSI, 2015). Every test for antibiotic sensitivity was performed thrice.
3.2.2. Hemolysis test
Hemolytic activity was not observed for the strain JB2CON. The isolate was identified as γ-hemolytic or non-hemolytic because there was no distinct transparency or greenish zone encircling their colonies on the blood agar Petri-dishes. A probiotic with these qualities is perfect (Halder et. al., 2017).
3.2.3. Acid tolerance
Acid tolerance test was conducted in a solution of pH 2.35, because pH of human stomach in natural state is between 1.5 and 3.5 (Helmenstine, 2020). Probiotic count in the tested solution plunged to (5.98±0) ×103 CFU/mL after 24 hours from (1.28±0) ×107 CFU/mL of initial count. Therefore, the probiotic L. acidophilus JB2CON (OM909067) was moderately tolerant to pH of gastric juice.
3.2.4. Bile salt tolerance
The good bacteria found in the human colon, known as probiotics, must adjust to bile salt. Since oxgall is typically used for this kind of test, we used it to assess the bile salt endurance of our isolate JB2CON (Hassanzadazar et al., 2012). After incubating the isolate for eight hours, we cultivated it on plates and saw growth on petridishes. When 0.025% and 0.5% oxgall were added, the CFU/mL decreased from (1.37±1.52) ×106 to (1.53±2.14) ×105 and (1.40±2.25) ×104, respectively, while it increased to (2.42±3.11) ×106 in the control broth. The outcomes supported the findings published by Ding et al. (2007), who noted that the probiotic strains in their investigation decreased after eight hours of incubation (Ding et al., 2007). For a bile sensitivity test in MRS broth, Hassanzadazar et al. (2012) selected a number of microbial cultures with bile concentrations as high as 0.3% as the test and 0% as the control. Despite the fact that our study contained a significantly higher concentration of live bacteria than theirs did, this experiment confirmed our findings (Hassanzadazar et al., 2012). As a result, the JB2CON strain is ideal for growing in the bile salt-surrounded human gut.
3.2.5. Antagonistic test
After 48 hours of incubation, the zone of inhibition was measured. The inhibition zones of more than 20mm, 10-20mm, and less than 10mm were accepted as strong, intermediate, and low inhibition respectively. In our study
L. acidophilus JB2CON (OM909067) had an intermediate inhibition against tested pathogens (Fijan et al., 2016). We might get a better result than this if we would carry out our experiment with simple spot-on lawn agar, because the Zone of Inhibition (ZOI) of a simple spot-on lawn agar is larger than that of the well diffusion method (Fijan et al., 2016) (
Table 2). We found the widest ZOI (14.5 mm) against
E. coli ATCC 8739, and no ZOI against
C. albicans ATCC 10231 by agar well diffusion method. That is why we conducted further anti-
E. coli, and anti-
C. albicans experiment in time-kill assay method during shelf-life test.
3.2.6. Antimicrobial activity test by time-kill assay for unknown contamination
Total Non-Lactic Contaminations were too numerous to count (TNTC) on TSA media. On the other hand, no CFU of TYMC (Total Yeast and Mold Count) was observed on SCA media at 35°C and 44°C, whereas TNTC was observed at ambient room temperature on this media (
Table 3). Further dilution was not performed for counting the exact CFU of unknown contamination, because the tested samples did not comply with the United State Pharmacopeia. In USP nutritional and dietary supplements guideline, microbial limit for TAMC is 1000 CFU/mL, and for TYMC it is 100 CFU/mL (Radhakrishna Tirumalai, 2006a). On the contrary, identical colonies of
E. coli, S. aureus, salmonella spp., and P. aeruginosa were not found on selective media (
Table 4). The samples complied with USP specification in terms of specific pathogenic microorganisms (Radhakrishna Tirumalai, 2006b). It indicates that probiotic dahi prepared by
L. acidophilus JB2CON (OM909067) can be a natural preservative for preventing the growth of bad bacteria (
E. coli, S. aureus, salmonella spp., and P. aeruginosa), albeit there are some resistant unknown bacteria and fungus can grow in dahi naturally. Thus, precautions should be taken for preparing dahi and selecting a starter culture.
3.3.1. Analysis of milk for preparation of dahi
Milk quality, such as protein and age of milk influence the titrable acidity of milk, and subsequently, titrable acidity and pH of dahi (Schmidt et al., 1996). Thus, we tested the milk properties of cow milk. Fat content was not less than 2%, and protein content was 3.5%. It was ideal full-fat milk (
Table 5) (Maryam et al., 2019).
3.3.2. Preparation of probiotic dahi
The Fastest solidification was found in the case of probiotic dahi at 44°C and it happened within 4.60±0.35 hours when pH reached 4.60±0.35 (
Table 6).
L. acidophilus is very well suited for living in dairy medium, as fermented milk is the ideal method of delivery for introducing
L. acidophilus into a gut microbiome (Meng et al., 2021).
3.4. Shelf-life study at storage condition
A total of four containers (containing probiotic dahi) were incubated at 44°C for 5.50 hours and then stored at refrigerator (2°C-8°C) for 14 days (Table: 16). In the previous study, it was confirmed that the viability of L. acidophilus cells stored at refrigerator (4 °C) is higher than that of cells stored at room temperature (25 °C) (Arepally et al., 2020). Thus, in the current study, we used refrigerator for storing our probiotic.
No E. Coli cell was found live on TSA media after 5.50 hours whilst C. albicans cells increased gradually and soared to 114 CFU/g after 14 days. This suggests that L. acidophilus JB2CON (OM909067) is strongly bactericidal against E. Coli. TNLC, and TYMC also increased steadily on TSA and SCA after 14 days. It indicates that probiotic dahi prepared by L. acidophilus JB2CON (OM909067) can be a natural preservative for inhibiting the growth of Escherichia Coli, although there are some resistant bacteria and fungi which can grow in dahi naturally. Thus, precautions should be taken for preparing dahi and selecting a starter culture. For setting shelf life, these factors should be considered carefully.
The container with pasteurized milk had no microbial contamination. Interestingly, in this container, the probiotic count of dahi increased 2.98-fold and soared to 170.0±0 ×109 CFU/30mL after day 14 from 60±4.24 ×109 CFU/30mL on day 1. We decided that our probiotic dahi should contain at least 106 CFU per milliliter for the duration of its shelf life, in accordance with Alberts' (1987) suggestion (Alberts, 1987). However, our yogurt with probiotics was far more concentrated—up to 14 days.
On the other hand, the pH of dahi went up slightly (0.06) from 3.65±0 to 3.59±0 (
Table 9). It indicates that CFU is increasing gradually for 14 days, but pH is not increasing noticeably by this time. This slight change in pH cannot affect the taste (sourness/acidity) of dahi. Therefore, the shelf life of 14 days is the ideal time duration for maintaining CFU and the taste of dahi According to the finding of our study, for maintaining a shelf life of 14 days, probiotic dahi should be prepared from pasteurized cow milk.
3.5. Consumer acceptance of dahi
The overall consumer acceptance of dahi was satisfactory (
Table 10). 9-point hedonic scale for sensory test of probiotic dahi was applied on total 16 people (four groups). The texture of dahi was neither liked nor disliked by the people of four groups. Basically, texture of dahi depends on various factors, such as, composition of raw milk, incubation time, additives, and so on (Yagmur et al., 2018). In the probiotic dahi, we did not add any extra additive, and fermentation time was reduced to decrease the sourness or acidity of the dahi. Therefore, our prepared probiotic dahi was soft dahi. However, the groups of people liked moderately the overall taste of the probiotic dahi, and also liked slightly or moderately the color, sweet, and sour.
4. Conclusions
The dahi in the extant study was prepared from a potential probiotic, L. acidophilus JB2CON (OM909067) isolated from dahi. As the probiotic strain had anti-Escherichia coli activity this probiotic, or pribiotic dahi can be an inexpensive option for recovering and preventing diarrheal disease and other Escherichia coli-related diseases in humans. This will save globally the cost and lives of billions of people who are infected with Escherichia coli. This is the greatest outcome of this research. Furthermore, according to our findings, contaminations in dahi originated from unhygienic practices and unidentified starter culture. These can reduce the shelf life of dahi and cause dahi spoilage. Therefore, precautions should be taken, and dahi can be prepared with pasteurized milk. Considering contamination, probiotic count, pH, and taste of dahi, the time duration of 14 days was an ideal shelf life for our probiotic dahi prepared from pasteurized milk.
Author Contributions
Conceptualization, Sk. Md. Jakaria Al-Mujahidy; Data curation, Sk. Md. Jakaria Al-Mujahidy and Shahidur Rahman; Formal analysis, Sk. Md. Jakaria Al-Mujahidy and Shahidur Rahman; Investigation, Md. Uddin; Methodology, Sk. Md. Jakaria Al-Mujahidy; Writing – review & editing, Md. Uddin.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data Availability Statement
Data will be provided upon request to the corresponding author.
Acknowledgments
This work was partially supported by Dairy Development Research Project, Bangladesh Livestock Research Institute, Regional Station, Baghabari, Shahjadpur, Sirajganj 6770.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alberts BM. Prokaryotic DNA Replication Mechanisms. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1987, 317, 395–420.
- Anjam, KCM., 2014. The Dynamic Interactions between Salmonella and the Microbiota, within the Challenging Niche of the Gastrointestinal Tract. Int. Sch. Res. Notices. [CrossRef]
- Anjuman, AB., Jakaria, DM., Sharif et al., 2015. Market Assessment and Product Evaluation of Probiotic Containing Dietary Supplements Available in Bangladesh Market. Int. J. Pharm. [CrossRef]
- AOAC (2012) Official methods of analysis. 13th Edition, 2012. The Association of Official Analytical Chemists, Inc; Washington, D.C 2012(25–78).
- Arepally D, Reddy RS, Goswami TK. Studies on survivability, storage stability of encapsulated spray dried probiotic powder. Curr. Res. Nutr. Food Sci. 2020, 3, 235–242.
- Britton, RA, and Versalovic, J., 2009. Probiotics and gastrointestinal infections. Interdiscip Perspect Infect Dis. 2008. [CrossRef]
- Caballero, B.; Finglas, P.; Toldra, F., 2015. Encyclopedia of Food and Health. Elsevier sci. 345–351. ISBN 978-0-12-384953-3.
- Chetan, S., Sachin, G., Nishchal, T et al., 2017. Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. Biotech. 7, 53. [CrossRef]
- Chidre, P., and Revanasiddappa, K.C., 2017. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 40, 270-283. [CrossRef]
- Chidre, P., and Revanasiddappa, K.C., 2017. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 40, 270-283. [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI), 2015. Performance standards for antimicrobial susceptibility testing: 25th informational supplement. CLSI document M100-S22. Clinical Laboratory Standard Institute, Wayne, PA.
- Coico, R., 2005. Gram Staining. Curr Protoc Microbiol. [CrossRef]
- Ding, W. K.; & Shah, N. P., 2007. Acid, Bile, and Heat Tolerance of Free and Microencapsulated Probiotic Bacteria. J. Food Sci. 72:M446–M450. [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Fijan, S., 2016. Antimicrobial effect of probiotics against common pathogens. In: Rao, V., & Rao, L. G. (Eds.), Probiotics and prebiotics in human nutrition and health. Intech Open 10.5772/63141.
- Fijan, S., 2016. Antimicrobial effect of probiotics against common pathogens. In: Rao, V., & Rao, L. G. (Eds.), Probiotics and prebiotics in human nutrition and health. Intech Open 10.5772/63141.
- Guesh, M., Diriba, M., Anteneh, T., and Tesfaye, S., 2020. Protective Effect of Potential Probiotic Strains from Fermented Ethiopian Food against Salmonella Typhimurium DT104 in Mice. Int. J. Microbiol. [CrossRef]
- Hashim, I.B., Khalil, A.H., and Afifi, H.S. Quality characteristics and consumer acceptance of yogurt fortified with date fiber. J. Dairy Sci. 2009, 92, 5403–5407. [CrossRef] [PubMed]
- Hassanzadazar H, Ehsani A, Mardani K, Hesari J. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. Vet Res Forum. 2012, 3, 181–5.
- Halder D, Mandal M, Chatterjee SS, Pal NK, Mandal S., 2017. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines. 5:31. [CrossRef]
- Helmenstine, A.M., 2020. What Is the pH of the Stomach? ThoughtCo. thoughtco.com/ph-of-the-stomach-608195.
- Kailasapathy, and Chin, J., 2000. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol. 78(1), 80-8. [CrossRef]
- Kavitha, P., Sindhuja, D., and Banumathi, M., 2016. Isolation and Biochemical Characterization of Lactobacillus species Isolated from Dahi. Int. J. Curr Microbiol App Sci. 5, 1042-1049. [CrossRef]
- Maryam, T., Mohammad, B., and Habibi, N. Mohebbat M., 2019. Effect of the milk fat content and starter culture selection on proteolysis and antioxidant activity of probiotic yogurt. Heliyon 5(2), e01204. [CrossRef]
- Matthew B., Sue P., Julian M., Eshwar M., 2013. The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success, FEMS Microbiology Letters. 349(2), 77–87. [CrossRef]
- Meng L, Li S, Liu G, Fan X, Qiao Y, Zhang A, et al., 2021. "The nutrient requirements of Lactobacillus acidophilus LA-5 and their application to fermented milk". Journal of Dairy Science. 104 (1), 138–150. [CrossRef]
- Molecular detection by analysis of the 16S rRNA gene of fecal coliform bacteria from the two Korean Apodemus species (Apodemusagrarius and A. peninsulae). Genet Mol Res. 16(1).
- Ortakci, F., Broadbent, J.R., McManus, W.R., and McMahon, D.J., 2012. Survival of microencapsulated probiotic Lactobacillus paracasei LBC-1e during manufacture of Mozzarella cheese and simulated gastric digestion. J. Dairy Sci. 95, 6274-6281. [CrossRef]
- Prabhurajeshwar, and Kelmani, C., 2019. Evaluation of antimicrobial properties and their substances against pathogenic bacteria in-vitro by probiotic Lactobacilli strains isolated from commercial yoghurt. Clin. Nutr. Exp. 23, 97-115.
- Radhakrishna Tirumalai, Ph.D., 2006. <2021>Microbiological enumeration tests-nutritional and dietary supplements. Pharmacopeial Forum, 29, 268.
- Radhakrishna Tirumalai, Ph.D., 2006. <2022>Microbiological procedure for absence of specified microorganism- nutritional and dietary supplements. Pharmacopeial Forum, 29, 287.
- Radhakrishna, T., 2006. <2023>Microbiological attributes of non-sterile nutritional and dietary supplements. Pharmacopeial Forum. 30, 1818.
- Rahman, M.M., Yoon, K.B., Lim, S.J., Jeon, M.G., Kim, H.J., Kim, H.Y., Cho, J.Y., Chae H.M., and Park, Y.C., 2017.
- Rolhion, N, and Chassaing, B., 2016. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci. 371, 20150504. [CrossRef]
- Saitou N. and Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425.
- Sarker, A.R., Sultana, M., and Mahumud, R.A., 2018. Economic costs of hospitalized diarrheal disease in Bangladesh: a societal perspective. Glob Health Res Policy. 3. [CrossRef]
- Schmidt, K., Stupar, J., Shirley, J., Adapa, S., and Sukup. Factors affecting titratable acidity in raw milk. Kansas Agricultural Experiment Station Research Reports. Dairy Day 1996, 2, 60–62.
- Tamura K., Stecher G., and Kumar S., 2021. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. [CrossRef]
- Yagmur, E.K., Ibrahim, A., and Senol, K. Determination of Texture Profile Analysis of Yogurt Produced By Industrial and Traditional Method. Int J Sci Technol Res. 2018, 4, 66–70.
- Yang, E., Fan, L., and Yan, J., 2018. Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Expr. 8. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).