1. Background
Chronic cough is a common clinical manifestation especially among a cohort of patients suspected for pulmonary tuberculosis (PTB) (Siddharthan et al., 2019, van Gemert et al., 2015). However, where diagnosis is not definitive, there is often a lapse in suspicion of other aetiological agents like fungi and non-tuberculous (TB) bacteria. The challenge is that the burden of invasive pulmonary fungal infections is underestimated and increasing, yet pulmonary fungal infections are often misdiagnosed as pulmonary tuberculosis mostly in developing countries like Uganda. The prevalence of hematological malignancies, pulmonary tuberculosis, HIV/AIDs, diabetes, Covid 19 and extensive immunosuppressive drugs are all incriminated for such an increase, representing a major unmet need (Amiri et al., 2018, Zhao et al., 2021, Chong et al., 2021, Fekkar et al., 2021). Like in some previous global studies, we have recently highlighted how common fungal co-infections are in this patient cohort at Mbarara Regional Referral Hospital (Njovu et al., 2021, Hanson et al., 2019, Smith and Kauffman, 2012, Silveira and Paterson, 2005, Yamamura and Xu, 2021, Tobón and Gómez, 2021) and here we continue to highlight their medical importance. Recently the WHO released a priority pathogen list highlighting the need for improved surveillance and diagnostic capacity for fungal pathogens of critical, neglected importance (Tacconelli et al., 2018, Burki, 2023, Parums, 2022). Furthermore, little is known on the role of fungal-bacterial di-kingdom aetiology in chronic pulmonary infection or co-colonization Fortunately, the recent outbreak of Covid 19 has disclosed a number of such events especially the co-infection reports by black fungus and further compounds what we are seeing with TB (Chong et al., 2021, Rothe et al., 2021, Rawson et al., 2020, Hughes et al., 2020, Sreenath et al., 2021, Reece et al., 2017). In this regard, we have initiated investigations into fungal-bacterial co-infections in association with TB (Itabangi et al., Amiri et al., 2018, Njovu et al., 2021, Njovu et al., 2023). So far, our preliminary data suggest that fungal-bacterial di-kingdom synergistic aetiology is of major importance that needs critical attention clinically. For instance, although the etiology of invasive chronic pulmonary infections is often attributed to Mycobacterium tuberculosis, our data suggests otherwise and reveals that numerous unappreciated invasive pulmonary fungal opportunists and other non-TB bacterial commensals can also co-infect or be the primary cause of chronic pulmonary disease (Njovu et al., 2023, Njovu et al., 2021). Our preliminary hospital data show that up to 92% of patients suspected of pulmonary tuberculosis are diagnosed with pulmonary fungal infections (mycoses) (Njovu et al., 2021). However, the challenge is that current technical expertise and diagnostic approaches are not designed to routinely assess poly-microbial synergistic infections, and may therefore miss a possible fungal-bacterial co-infection. We therefore, hypothesized here that the co-colonization of a host by multiple microbial communities can be shaped by the environmental frequency, antimicrobial resistance and interaction profiles of both commensal fungal and bacterial opportunists; and that understanding the nature, diversity, and environmental origin of these communities will provide important insight into their pathological potential. A major unanswered question however is: how widespread are these types of interactions in patient samples, and how do they influence virulence and antimicrobial profiles in the context of patient co-colonization and co-infection. Fungi and bacteria are found together in a myriad of environments and particularly in biofilms, where adherent species interact through diverse signaling mechanisms (Reece et al., 2017, Du et al., 2022, Shirtliff et al., 2009). Of course, this co-existence still remains elusive to us, but recent studies suggest that such interactions may influence virulence and antimicrobial resistance (Rawson et al., 2020, Itabangi et al., 2022, Jabbari Amiri et al., 2016, Njovu et al., 2021, Njovu et al., 2023)). For instance, we have demonstrated that a bacterial endosymbiont (Ralstonia pickettii) of the fungus Rhizopus microsporus influences its resistance against antifungals, oxidative and nitrosative stresses(Itabangi et al., 2022). Moreover, disrupting these microbial communities pharmacologically can have unanticipated results. For example, in instances where co-infection results in antagonistic or mutual inhibitory interactions, treatment with antifungals or antibacterial agents may promote or unleash the growth of the off-target microbiota, with implications for morbidity and mortality in the at-risk individuals (Schönherr et al., 2017). Addressing these challenges at the level of patient populations requires a step-change in the awareness of fungal-bacterial co- infections and the capacity of our medical staff to diagnose and treat these poly microbial infections as equally important. Thus, in this study we explore a phenotypic outlook of some of the fungal-bacterial poly microbial communities associated with chronic pulmonary disease and their association with Mycobacterium tuberculosis.
2. Methods and Materials
2.1. Study Area and Population
The study was conducted at Mbarara Regional Referral hospital (MRRH)which is a tertiary care facility and Teaching Hospital in South Western Uganda among presumptive pulmonary tuberculosis patients
2.2. Study Design
This was a laboratory-based cross-sectional study, with a sample size of 151 participants. The study was a diagnostic-based observational approach with a primary outcome of improving diagnostic profile.
2.3. Patients and Samples
2.3.1. Selection Criteria
The target population under study were patients suspected for chronic pulmonary disease at MRRH. Participants were included if they were 18 years of age and above, consented and with at least one of the following qualities; present with PTB like symptoms, no prior history of TB, are smear and Gene Xpert negative or positive, Chest X ray positive or negative, LAM negative or positive with persistent pulmonary symptoms; and or on an anti-TBs, severely ill but can produce sputum. However, patients that were HIV/AIDs positive or negative with TB like symptoms but on anti-TB treatment with good prognosis, on Anti-TB severely ill but cannot produce sputum, critically ill and pregnant women were excluded from the study.
2.3.2. Recruitment Strategy
Patients were recruited by convenient sampling as they reported for medical services at the HIV/TB clinic of Mbarara Regional referral hospital (MRRH). The recruitment of study participants closely followed the existing diagnosis and point-of-care protocols.
2.3.3. Sample Collection
From the 151 recruited patients, a total of 302 sputa ((an on-spot and early morning) samples were collected. The collection of two samples was mainly a means of quality control of the collected samples. Baseline data including demographic and clinical history was also collected using a patient reported questionnaire. The samples were collected according to established guidelines for collection of sputum samples for TB diagnosis. Several precautions and check were taken to endeavor collected samples were the appropriate quality, quantity and in the right containers without any leakages.
2.3.4. Biographical Data Collection
Biographical data was collected through the administration of an open ended questionnaire by a research assistant
2.4. Sample Processing
2.4.1. Direct Smear Microscopy
The criteria for fungal and non-TB bacterial diagnosis was based on the presence of pus cells with bacterial cells, budding yeast cells, Pseudo hyphae, filamentous fungal structures and other fungal fruiting bodies in a direct Gram stained smear or wet mount preparation. Accordingly, Gram smears were prepared fixed with alcohol or heat, stained with crystal violet, decolorized with 70% ethanol, counterstained with nigrosine red, dried and examined using X100 objective. Wet mount smears were mad e by transferring a small portion of the sputa on a microscopic slide with a drop of 20% w/v KOH solution, a coverslip added and examined using X10 and X40 objectives. For culture reading of filamentous fungi, colonies on basic (e.g. Sabouraud) or sporulation (e.g. Potato dextrose) agar plates were prepared with Lacto phenol cotton blue (LPCB) stain and examined microscopically using X10 and 40 Objectives.
2.4.2. Germ Tube Test
For yeast isolates, germ tube test was performed to differentiate between C. albicans and non albicans candida (NAC). Accordingly, a small portion of an 18-72 hrs old culture of the yeast was suspended in 0.5ml of human serum in a test-tube. Positive and negative control was set. All the test tubes were incubated at (370C) for 2-3 hrs. A drop of the yeast suspension was placed on a clean glass slide, a cover slip added and examined microscopically for presence or Absence of germ tubes.
2.4.3. Cultural Methods
2.4.3.1. Non-TB Bacterial Cultivation and Identification
This included streaking specimens on both basic and identification media. Accordingly, sputa were streaked on Nutrient, Blood, Chocolate or MacConkey agars and incubated at (370C). For anaerobic enhancement, streaked plates were also incubated at (370C) in aerobic jar under 5% CO2. The identification of bacterial isolates was also enhanced by Bacterial Analytical profile index (API). This is based on the biochemical assimilation and fermentation of specific sugars by bacteria. The API were set according the manufacturer’s instructions.
2.4.3.2. Fungal Cultivation and Identification
This included streaking specimens on both basic and identification media. Accordingly, sputa were streaked on Sabouraud dextrose agar (SDA, and incubated at (270C) and (370C) respectively for primary isolation of fungi. For sporulation and conidiation, fungal isolates were sub cultured on Potato dextrose agar (PDA) (for filamentous fungi), Rice agar (for yeasts). Following, sporulation and conidiation enhancement for fruiting bodies by filamentous fungi, fungal fruiting structures were identified using LPCB wet mounting. Yeasts, were identified through sub culturing on Chromogenic Agar medium and incubated at (370C) for 48 hrs. identification of the colonies was based on the colour produced by specific yeast species for instance, C. albicans produced light green, C. glabrata produce small, light-pink colonies and C. famata white to light pink colonies (Nadeem et al., 2010) etc. This identification was further reinforced and controlled with API 20 Candida kits. The API was set according to the manufacturer’s instructions based on the principle of biochemical assimilation and fermentation of specific sugars by yeasts.
2.4.3.3. Molecular detection of MTB Bacteria
2.4.3.4. Gene Xpert (Molecular Assay)
This was preformed according to shear and Perween’ protocol, 2004. Accordingly, Sputa samples were collected from targeted patients. Sputum was liquefied and inactivated with 2:1 sample reagent ratio and 2 ml of material was transferred into the test cartridge. Cartridge was inserted into MTB/RIF test platform. Sample automatically filtered and washed. Ultrasonic lysis of filter captured organisms to release DNA. The DNA molecules mixed with dry PCR reagents. Semi-nested real time amplification and detection in integrated reaction tube printable results reported as “MTB detected; or MTB not detected.
2.4.4. Ethical Consideration
Ethical approvals were sought and obtained from Mbarara university of science and Technology (Ref: MUREC1/7-0709/20), European and developing countries Trail partnership (EDCTP) (Ref: TMA2019CDF-2789), Uganda National council of science and Technology (Ref: HS1233ES) and the study was also registered in the International Standard Randomized Controlled Trail Number (ISRCTN) registry (Ref: ISRCTN 33572982). The study has been conducted according to Uganda National and European Union legislation.
2.4.5. Informed Consent
Target participants were fully informed of the purpose and procedures of the study, their obligations, potential benefits and risks. To this effect an informed consent form was prepared and approved by the institutional review board for use and this was signed voluntarily during implementation stage by each of the study participants prior to enrolment.
2.4.6. Data Analysis
Raw data was collected into Epi-collect version 5. For statistical analyses data were imported into Stata version 11 and graph pad version 6, where analyses were performed using percentages, odds ratios, multivariate and univariate functions; and t-student test.
3. Results
3.1. Demographic Characteristics
A total of 151 patients suspected for
TB and presenting with chronic pulmonary disease symptoms were recruited in a study at MRRH. These patients were examined for Pulmonary tuberculosis (PTB) using chest X-ray, sputum Xpert MTB/RIF and Lateral flow urine Lipoarabinomannan (LAM) assay. According to aetiology patients were divided into mono-kingdom fungal (Yeast (n=92) or mold (n=81)) infection (Total (n=173)),
Mycobacterium tuberculosis infection (n=58) and non-TB bacterial infection (n=140); and di-kingdom fungal-bacterial co-infections (n=129) group. The average age for the patients recruited was 46.05263 (STD 16.31) with a range of 18-97 years. In respect to gender, males accounted for 63.6% and 36.4% for females. Out of 151 study participants, 59.6% (n=90) were HIV positive, 38.4% (n=58) were HIV negative and 3 % (n=3) had unknown HIV status. The average duration for cough among participants was 80.73 (STD 13.56) days with a range of 18-90 days. For treatment history, 21.1 % (n=32) had been treated for TB and completed treatment whilst, 39.1% (n=59) were undergoing TB treatment as shown in
Table 1.
3.2. Distribution of Fungal and Non-TB Bacterial Species
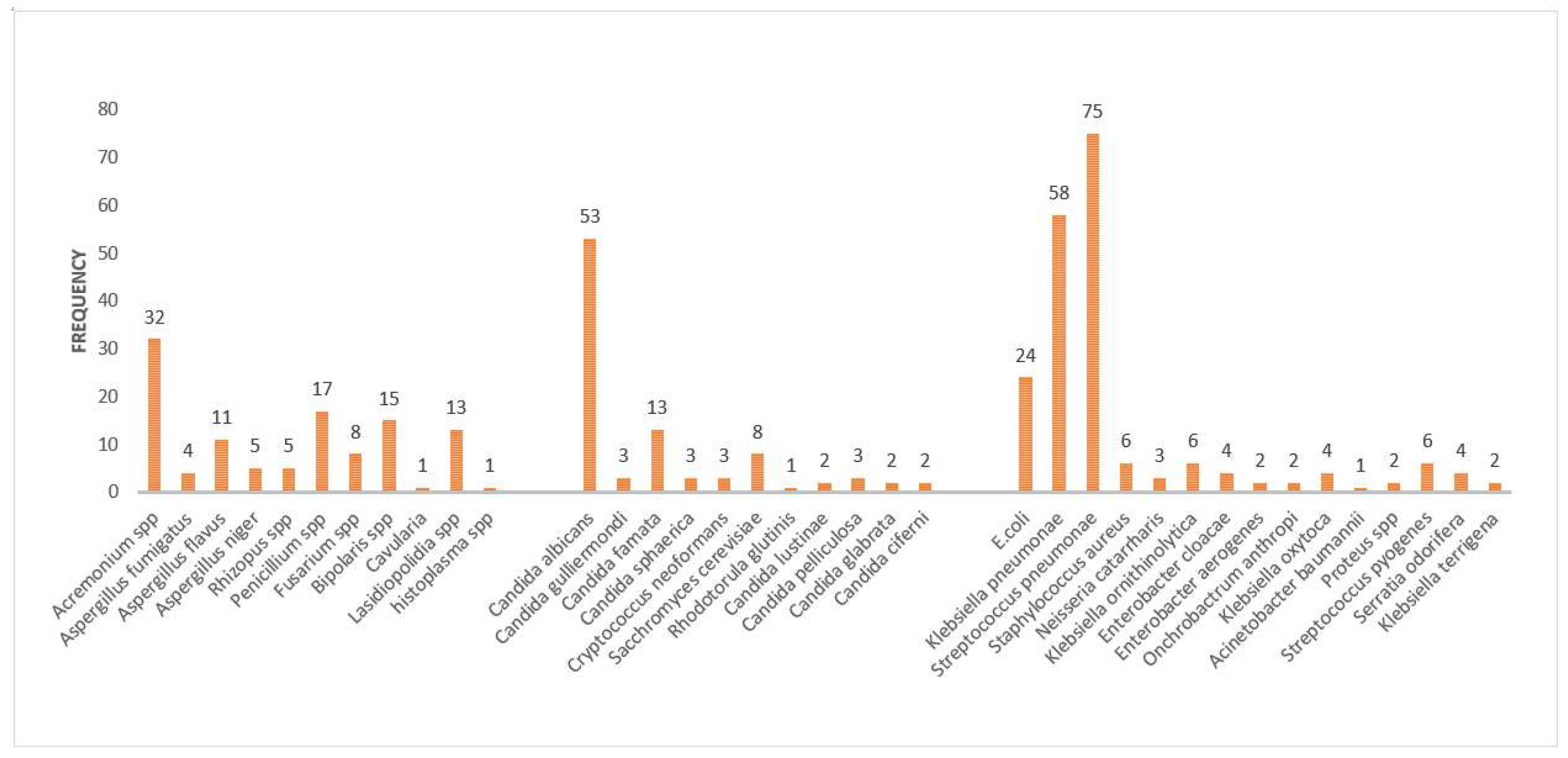
Patients presenting with chronic pulmonary disease seem to present a richly diversified aetiological profile, which mostly is ignored during clinical and laboratory diagnosis. Here we present a panel of aetiological profiles of 151 cases. From the 151 patients, 23 species of fungus, 16 species of non-TB bacterial isolates were detected by conventional laboratory based diagnostic testing. Among the fungal aetiological agents, the most commonly observed yeast species included:
Candida albicans (n=53) followed by
Candida famata (n=13), Saccharomyces cerevisiae (n=8), Cryptococcus neoformans (n=3), Candida pelliculosa (n=3), Candida sphaerica (n=3), Candida guillermondi (n=3), Candida lustinae (n=2), Candida glabrata (n=2), Candida ciferii (n=2) and Rhodotorula glutins (n=1).Among the molds
, Acremonium spp (n=32)was the most commonly observed, followed by
Bipolaris spp (n=15)., Lasiodiopolidia spp (n=13),Aspergillus flavus (n=11), Penicillium spp (n=8), Fusarium spp (n=8), Aspergillus niger (n=5), Rhizopus spp (n=5), Aspergillus fumigatus (n=4), and
Curvularia spp (n=1) and
Histoplasma capsulatum (n=1). On the other hand, non-TB bacterial aetiology was observed in this patient cohort. Accordingly; the aetiology was dominated by
Streptococcus pneumoniae (n=75) Klebsiella pneumoniae (n=58), these were then followed by
E. coli (n=24), Streptococcus pyogenes (n=6), Staphylococcus aureus (n=6), Proteus spp (n=2), Neisseria catarrharis (n=3),, Enterobacter cloacae (n=2), Klebsiella ornithinolytica (n=6), Acinetobacter baumannii (n=1), Enterobacter aerogenes (n=2), Ochrobactrum anthropi (n=2), Klebsiella terrigena (n=2)., Serritia liquefaciens (n=2), Klebsiella Oxytoca (n=4)., and Serritia odorifera (n=4) (
Figure 1 )
3.3. Association between Having Fungal, Non TB Bacterial and TB Pathogens
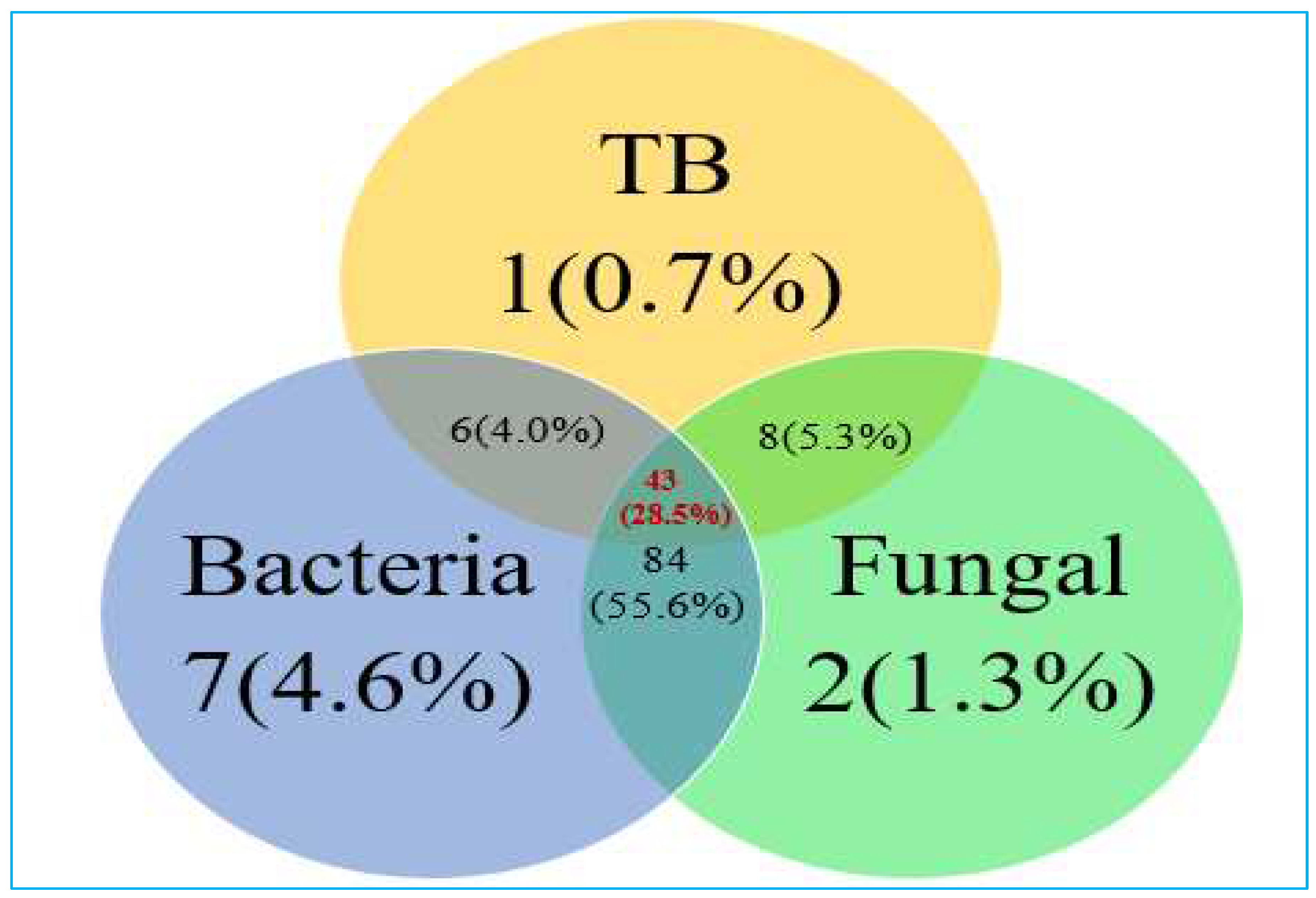
The study focused on patients with chronic pulmonary disease who were under suspicion for Tuberculosis (TB), a total cohort of 151 individuals were meticulously examined and assessed. Our study reveals intriguing findings with regard to fungal co-infections in this patient population. Among the enrolled patients, a notable proportion, precisely 28.5%, demonstrated concurrent infections involving TB, fungal, and bacterial pathogens, showcasing a complex interplay of these microbial agents. Notably, over half of the patients, accounting for 55.6%, manifested infections attributed exclusively to bacterial and fungal agents while remaining free from TB. Within this cohort, it was observed that a minority, less than 10%, exhibited singular infections. Specifically, 0.7% of patients were diagnosed with TB infections, 1.3% with fungal infections as the solitary pathogenic agent, and 4.6% with exclusive non-TB bacterial infections (
Figure 2). These findings underscore the significance of considering fungal–bacterial co-infections in the diagnostic and treatment strategies for patients with chronic pulmonary diseases who are suspected of Tuberculosis. The intricate relationship between these pathogens necessitates a nuanced approach in clinical management and highlights the potential impact of fungal pathogens in this clinical context
3.4. Distribution of Fungal-Bacterial Di-Kingdom Co-Existence Categories
-
1.
Distribution of di-kingdom (Fungal (yeast/mold) and non-TB bacterial) aetiological categories in relation to mono TB infection
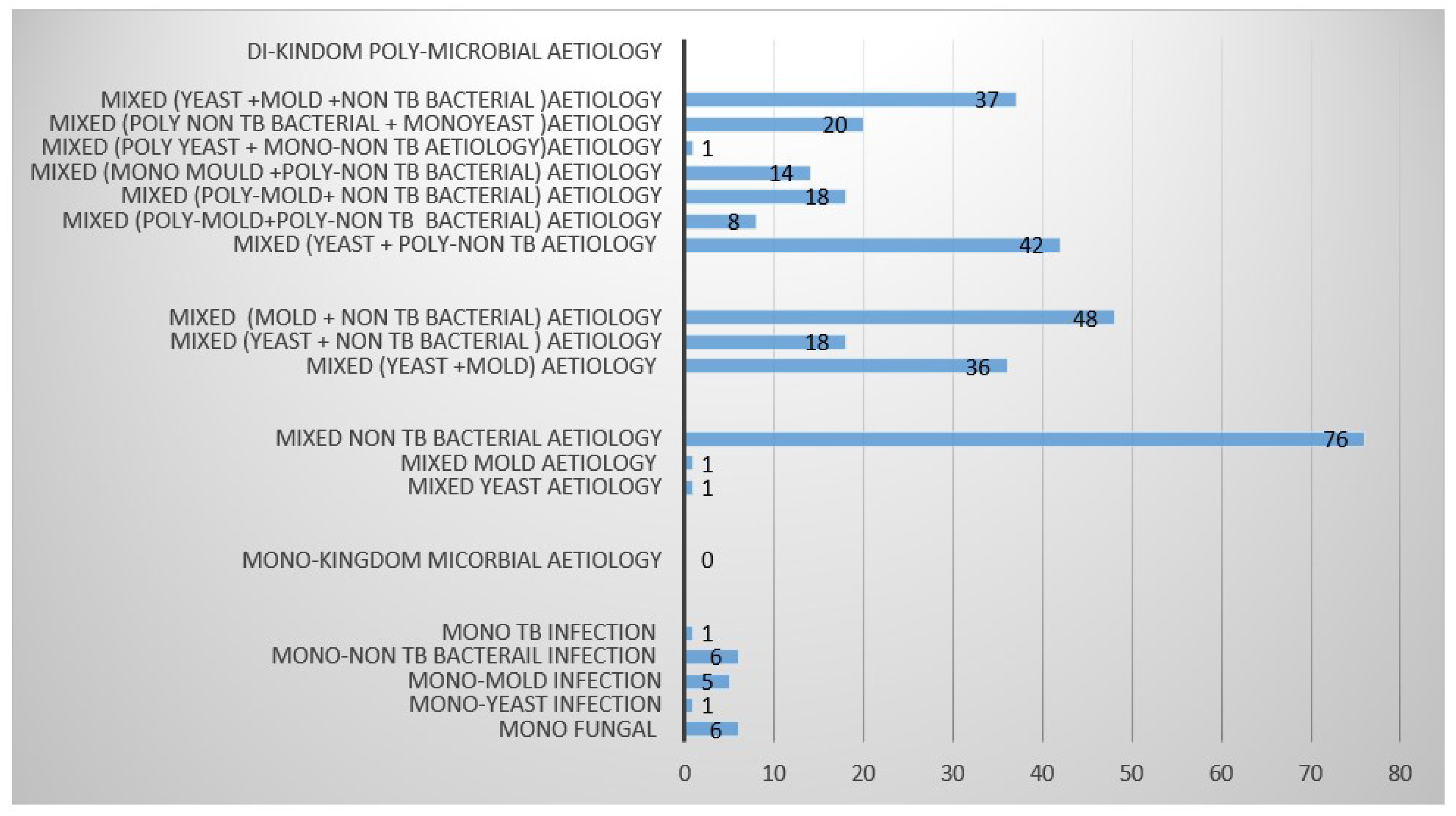
We have observed four major aetiological categories in this targeted patient cohort. Specifically, they included yeasts, molds,
Mycobacterium tuberculosis (MTB) and non-TB bacterial aetiology. Mono kingdom aetiology was mostly due to fungal (molds, yeasts) (n=173), followed non-TB bacteria (n=140), and MTB (n=58) refer to
Table 1. However, we further sub categorized these into mixed di-kingdom aetiology based on the three-mono kingdom aetiologies. Accordingly, di-kingdom aetiology was mostly due to mixed (yeast +poly –non TB bacterial aetiology (n=42), followed by Mixed (yeast +Mold +non TB bacterial) (n=32) co-infections, mixed (poly Yeast + mono-non TB bacterial aetiology) (n=20), Mixed (poly Mold + non-TB bacterial aetiology) (n=18), (molds + non-TB bacterial) co-infections followed by mixed (yeast + non TB bacterial) co-infections. However, mono kingdom mixed (yeast + mold) co-infections were also observed. From the di-kingdom category, we also observed poly-fungal or non-TB bacterial involvement as well. Accordingly, mixed (mono yeast +poly non-TB bacterial) aetiology was the most observed, followed by mixed (yeast + mold + non-TB bacteria), mixed (poly non-TB + mono Yeast), mixed (poly mold + non TB bacterial), mixed (mono mold + non TB bacteria), mixed (poly mold + poly non Tb bacteria) and mixed (poly yeast + mono non TB bacteria) aetiology. When compared with MTB aetiology, only a few cases presented with mono MTB aetiology.
Figure 3
3.5. Distribution Profile Panel of Fungal - Bacterial Di-Kingdom Co-Existence Aetiology in Association with Pulmonary TB
Di-Kingdom aetiological pairings
In this patient category, a wide range of aetiological agents were identified. Specifically, di-kingdom aetiological pairings associated with pulmonary TB are highlighted here but for the picture, consult Table1. Among these Acremonium spp +Streptococcus pneumonia e (n=4), Acremonium spp + K. pneumoniae (n=5), Acremonium spp + C. albicans + K. pneumoniae (n=3), Acremonium spp + Lasiodiopolidia spp + S. pneumoniae (n=2), Acremonium spp + Streptococcus pneumoniae + K. pneumoniae (n=2), Aspergillus niger + C. albicans + K. pneumoniae + E. coli (n=1), Bipolaris spp + Lasiodiopolidia spp + K. pneumoniae + S. pneumoniae (n=2), Candida albicans + S. pneumoniae, Candida fermata + K. pneumoniae + S. pneumoniae, C. albicans + K. pneumoniae + S. pneumoniae + E.coli, C. fermata +E. coli + K. pneumoniae + S. pneumoniae, Cryptococcus neoformans + E. coli, C. albicans + Serritia odorifera, Bipolaris spp + Lasiodiopolidia spp + K. pneumoniae + S. pneumoniae and Penicillium Spp + S. pneumoniae were the most prevalent Table 2 . We further investigated their occurrence with TB treatment and post treatment trends as describe below.
Mono-Kingdom Fungal and non-TB bacterial aetiological pairings
The most prevalent mono-kingdom fungal aetiological pairings included Acremonium spp + Penicillium spp and Acremonium spp + C. albicans while K. pneumoniae + S. pneumoniae was the most prevalent mono kingdom non-TB bacterial aetiology.
Association with MTB
We also investigated the association of these aetiological pairings with MTB. Accordingly, we noted the association between; Acremonium spp + Klebsiella pneumoniae, Acremonium spp + Streptococcus pneumoniae + K. pneumoniae, Aspergillus flavus + Penicillium spp + E. cloacae, Candida albicans + S. pneumoniae, C. albicans + K. pneumoniae + S. pneumoniae + E. coli, C. fermata +E. coli + K. pneumoniae + S. pneumonia, C. albicans + E. coli, C. albicans + Klebsiella ornithinolytica + K. pneumoniae, Penicillium Spp + Rhizopus spp + S. pneumoniae and Acremonium spp + Penicillium spp.
Table 3.
Fungal- bacterial di-kingdom aetiological pairings in association with Pulmonary TB.
Table 3.
Fungal- bacterial di-kingdom aetiological pairings in association with Pulmonary TB.
| |
Assn with TB |
| |
|
|
|
|
|
|
|
|
|
| Fungal-Bacterial Di-Kingdom poly Microbial aetiological pairings |
1 |
2 |
3 |
4 |
5 |
1 |
2 |
3 |
|
| |
|
|
|
|
|
|
|
|
|
| Acremonium spp + E. coli |
|
|
|
|
|
|
|
|
|
| Acremonium spp+ C. albicans + Klebsiella pneumoniae +Streptococcus pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Streptococcus pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + C. albicans + Klebsiella pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp+ Fusarium Spp + Streptococcus pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Candida sphaerica + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Lasiodiopolidia spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + K. orithrolytica + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Aspergillus fumigatus + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Fusarium spp + Bipolaris spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Bipolaris spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Lasiodiopolidia spp + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Rhizopus spp +Saccharomyces cerevisiae + Acinetobacter baumannii |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Klebsiella pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Penicillium spp + E.coli +S. pneumoniae +K. Pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + A. fumigatus + Penicillium spp + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Streptococcus pneumoniae + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + S. pneumoniae + Staphylococcus aureus |
|
|
|
|
|
|
|
|
|
| Acremonium spp + C. albicans + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Aspergillus flavus + Bipolaris spp + K. pneumoniae + S. pneumoniae + E.coli |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Candida glabrata + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| Aspergillus Spp + C. glabrata + K. pneumoniae + S. pneumoniae + Streptococcus pyogenes |
|
|
|
|
|
|
|
|
|
|
Aspergillus flavus + Fusarium spp + S. pneumoniae + S. pyogenes + S.aureus
|
|
|
|
|
|
|
|
|
|
| Aspergillus niger + C. albicans + Lasiodiopolidia spp + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus niger + Penicillium spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus niger + C. fermata + E. coli |
|
|
|
|
|
|
|
|
|
| Aspergillus spp + Lasiodiopolidia spp + C. glabrata + S. pneumoniae + S. pyogenes + S. aureus |
|
|
|
|
|
|
|
|
|
| Aspergillus fumigatus + Proteus spp |
|
|
|
|
|
|
|
|
|
| Aspergillus niger + C. albicans + K. pneumoniae + E. coli |
|
|
|
|
|
|
|
|
|
| Aspergillus fumigatus + S. pneumoniae + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus spp + C. albicans + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus spp + Fusarium spp + C. guillermondi + S. pneumoniae +S. aureus + Neisseria catarrharis |
|
|
|
|
|
|
|
|
|
| Aspergillus flavus + Candida fermata + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus flavus + Penicillium spp + E. cloacae |
|
|
|
|
|
|
|
|
|
| Aspergillus spp + Curvularia spp + C. fermata + Enterobacter aerogenes |
|
|
|
|
|
|
|
|
|
| Aspergillus spp + Penicillium spp + Fusarium spp + Klebsiella ornithinolytica |
|
|
|
|
|
|
|
|
|
| Aspergillus flavus + Penicillium spp + S. pneumoniae + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Aspergillus Spp + Penicillium spp + C. albicans + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + Penicillium spp + Lasiodiopolidia spp + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + C. albicans + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + Lasiodiopolidia spp + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + S. pneumoniae + N. catarrharis |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + Candida lustinae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + C. albicans + E.coli |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + C. albicans +K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + Lasiodiopolidia spp + S. pneumoniae + E. coli |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| Candida ciferii + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Candida albicans + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Candida albicans + Klebsiella terrigena |
|
|
|
|
|
|
|
|
|
| Cryptococcus neoformans + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Candida fermata + E.coli |
|
|
|
|
|
|
|
|
|
| Candida fermata + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + K. pneumoniae + S. pneumoniae + E.coli |
|
|
|
|
|
|
|
|
|
| C. fermata +E. coli + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Candida sphaerica + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Candida guillermondi + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. pelliculosa + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + S. aureus + S. pyogenes |
|
|
|
|
|
|
|
|
|
| C. albicans + N. catarrharis |
|
|
|
|
|
|
|
|
|
| C. albicans + Histoplasma capsulatum + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + Rhizopus spp + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + C. fermata +K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Cryptococcus neoformans + E. coli |
|
|
|
|
|
|
|
|
|
| C. albicans + E. coli |
|
|
|
|
|
|
|
|
|
| C. fermata + E. cloacae |
|
|
|
|
|
|
|
|
|
| C. albicans + Klebsiella ornithinolytica + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. ciferii + E. cloacae |
|
|
|
|
|
|
|
|
|
| Candida tropicalis + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. pelliculosa + Penicillium spp + Enterobacter aerogenes + S. pyogenes |
|
|
|
|
|
|
|
|
|
| C. pelliculosa + Penicillium spp + Ochrobactrum anthropi + streptococcus pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + Penicillium spp + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. guillermondi + Ochrobactrum anthropi |
|
|
|
|
|
|
|
|
|
| C. albicans + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + K. terrigena |
|
|
|
|
|
|
|
|
|
| C. albicans + Serritia odorifera |
|
|
|
|
|
|
|
|
|
| C. lustinae +Klebsiella oxytoca |
|
|
|
|
|
|
|
|
|
| C. albicans + Lasiodiopolidia spp + Klebsiella ornithinolytica |
|
|
|
|
|
|
|
|
|
| C. albicans + K. oxytoca + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. tropicalis + Klebsiella ornithinolytica + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| C. albicans + K. oxytoca |
|
|
|
|
|
|
|
|
|
| Saccharomyces cerevisiae + S. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| S. cerevisiae + E. coli + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| S. cerevisiae + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| S. cerevisiae + Penicillium spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| Penicillium Spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Penicillium Spp + Rhizopus spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Fusarium spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| Fusarium spp + K. pneumoniae |
|
|
|
|
|
|
|
|
|
| Lasiodiopolidia spp + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| Poly Fungal aetiological pairings |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Penicillium spp |
|
|
|
|
|
|
|
|
|
| Acremonium spp + C. albicans + Penicillium Spp |
|
|
|
|
|
|
|
|
|
| Acremonium spp + C. albicans |
|
|
|
|
|
|
|
|
|
| Acremonium spp + Aspergillus |
|
|
|
|
|
|
|
|
|
| Bipolaris spp + C. albicans |
|
|
|
|
|
|
|
|
|
| C. spaerica + Fusarium spp |
|
|
|
|
|
|
|
|
|
| S. cerevisiae + Rhizopus spp |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| POLY non TB Bacterial pairings |
|
|
|
|
|
|
|
|
|
| K. pneumoniae + S. pneumoniae + S. pyogenes |
|
|
|
|
|
|
|
|
|
| K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
|
|
|
| K. pneumoniae + S. pneumoniae + E.coli |
|
|
|
|
|
|
|
|
|
| S. pneumoniae + S. aureus |
|
|
|
|
|
|
|
|
|
| S. pneumoniae + S. pyogenes + S. aureus |
|
|
|
|
|
|
|
|
|
| K. pneumoniae + S. pneumoniae + E.coli + Proteus spp |
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
3.5.1. Distribution Profile Panel of Fungal Bacterial Di-Kingdom Co-Infections during TB Treatment
When we looked at the aetiological prevalence of aetiological pairings during TB treatment, we noted that Acremonium spp + K. pneumoniae 4(80%) p=0.0378, and Bipolaris spp + Penicillium spp + E. coli 2(40%) p=0.0234 were the most prevalent pairings noted during TB treatment. Other pairings occurred in equal measures including Acremonium spp + K. pneumoniae, Acremonium spp + Lasiodiopolidia spp + S. pneumoniae, Bipolaris spp + C. albicans + S. pneumoniae 1(20%).
Table 4.
Fungal Bacterial aetiological pairings during TB treatment.
Table 4.
Fungal Bacterial aetiological pairings during TB treatment.
| Aetiology |
1 |
2 |
3 |
4 |
5 |
|
| |
|
|
|
|
|
|
| Acremonium spp + K. pneumoniae |
|
|
|
|
|
|
| Acremonium spp + Penicillium spp + C. albicans |
|
|
|
|
|
|
| Acremonium spp + Lasiodiopolidia spp + S. pneumoniae |
|
|
|
|
|
|
| C. albicans + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
| Aspergillus fumigatus + Penicillium + E. cloacae |
|
|
|
|
|
|
| Bipolaris spp + Penicillium spp + E.coli |
|
|
|
|
|
|
| Penicillium spp + S. pneumoniae |
|
|
|
|
|
|
| Aspergillus fumigatus + Penicillium + E. cloacae |
|
|
|
|
|
|
| Aspergillus spp + Curvularia spp + C. fermata + Enterobacter aerogenes |
|
|
|
|
|
|
| C. albicans + Klebsiella ornithinolytica + K. pneumoniae |
|
|
|
|
|
|
| C. guillermondi + Ochrobactrum anthropi |
|
|
|
|
|
|
| E. cloacae |
|
|
|
|
|
|
| Cryptococcus neoformans + E.coli |
|
|
|
|
|
|
| C. lustinae + Klebsiella oxytoca |
|
|
|
|
|
|
| Bipolaris spp + C. albicans + S. pneumoniae |
|
|
|
|
|
|
| Fusarium spp + S. pneumoniae |
|
|
|
|
|
|
| C. albicans + K. terrigena |
|
|
|
|
|
|
| S. pneumoniae + S. aureus |
|
|
|
|
|
|
| S. pneumoniae + S. pyogenes + S. aureus |
|
|
|
|
|
|
| C. fermata + K. pneumoniae + E.coli |
|
|
|
|
|
|
| Bipolaris spp + Penicillium spp + Lasiodiopolidia spp + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
| Bipolaris spp + C. albicans + E.coli |
|
|
|
|
|
|
3.5.2. Distribution Profile Panel of Fungal Bacterial Di-Kingdom Co-Infections Post TB Treatment
Some of the recruited patients had a history of post treatment symptoms of MTB, these are often attributed to treatment failure by anti TB drugs suggestive of anti TB drug resistance. However, here we noted some significant di-kingdom aetiological pairings which included Acremonium spp + Klebsiella pneumoniae 5(100%) p=0.0016, Acremonium spp + C. albicans + Klebsiella pneumoniae 2(40%); p=0.0385, Candida albicans + S. pneumoniae, C. albicans + E. coli, C. fermata + K. pneumoniae + S. pneumoniae p=0.0321, C. ciferii + K. pneumoniae (p=0.0054).
Table 5.
Fungal Bacterial aetiological pairings during post TB treatment.
Table 5.
Fungal Bacterial aetiological pairings during post TB treatment.
| Aetiology |
1 |
2 |
3 |
4 |
5 |
|
| |
|
|
|
|
|
|
| Acremonium spp + E. coli |
|
|
|
|
|
|
| Acremonium spp + Penicillium spp |
|
|
|
|
|
|
| Acremonium spp + Aspergillus flavus + S. pneumoniae |
|
|
|
|
|
|
| Acremonium spp + Rhizopus spp +Saccharomyces cerevisiae + Acinetobacter baumannii |
|
|
|
|
|
|
| Acremonium spp + Klebsiella pneumoniae |
|
|
|
|
|
|
| Acremonium spp + Bipolaris spp + S. pneumoniae |
|
|
|
|
|
|
| Acremonium spp + C. albicans + Klebsiella pneumoniae |
|
|
|
|
|
|
| Penicillium Spp + Rhizopus spp + S. pneumoniae |
|
|
|
|
|
|
| Aspergillus flavus + Candida fermata + S. pneumoniae |
|
|
|
|
|
|
| Fusarium spp + K. pneumoniae |
|
|
|
|
|
|
| C. fermata + E. cloacae |
|
|
|
|
|
|
| C. ciferii + E. cloacae |
|
|
|
|
|
|
| S. cerevisiae + Rhizopus spp+ S. pneumoniae |
|
|
|
|
|
|
| C. pelliculosa + Penicillium spp + Ochrobactrum anthropi + streptococcus pneumoniae |
|
|
|
|
|
|
| Candida albicans + S. pneumoniae |
|
|
|
|
|
|
| C. albicans + E. coli |
|
|
|
|
|
|
| C. albicans + K. oxytoca + S. pneumoniae |
|
|
|
|
|
|
| C. fermata + K. pneumoniae |
|
|
|
|
|
|
| C. fermata + K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
| C. ciferii + K. pneumoniae |
|
|
|
|
|
|
| C. albicans + Bipolaris spp + S. pneumoniae |
|
|
|
|
|
|
| C. tropicalis + Klebsiella ornithinolytica + K. pneumoniae |
|
|
|
|
|
|
| K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
| Candida sphaerica +K. pneumoniae + S. pneumoniae |
|
|
|
|
|
|
4. Discussion
Aetiological co-infection is a simultaneous infection of a host infection niche by more than one disease causing agent (Hanson et al., 2019). This trend in humans has been ignored or unknown and seems to be increasing every passing day. In fact, it has been hypothesized that poly microbial infections might be more common and aggressive than single infections (Hughes et al., 2020, Naseef et al., 2022, Zhao et al., 2021). Indeed, co-infections can be of a particular significance since the disease causing agents can interact with co-infecting agents and the human host at a much subtler level than can be detected. What we are so far seeing is that the level of communication can either be of a positive or negative nature (MacAlpine et al., 2023, Kapitan et al., 2019, Itabangi et al., 2022). Either way the effects seem impactful depending on the involved agents, underlying co-morbidities and micro-environments of interactions (Kapitan et al., 2019). Of course, where the molecular and phenotypic communications are overly positive, the strength of a disease progression is enhanced as opposed to a negative interaction (Kapitan et al., 2019, Peleg et al., 2010, Todd et al., 2019, Eichelberger and Cassat, 2021). However, negative interactions may also present adverse effects depending on the signaling capacity, amount and potency of bi-products produced (Hanson et al., 2019, Casadevall and Pirofski, 2003, Matsuo et al., 2019).
While we recognize tuberculosis and HIV as a major and longstanding co-infection scenario, it’s prudent that other possible co-infections are also investigated and brought to light. Specifically, we also note di-kingdom synergistic co-infections between bacteria and fungi. These cross-kingdom interactions further complicate our superficial view of disease outcomes and may require a much deeper insight of into the molecular and metabolic aspects. Indeed, fungi and bacteria are known to formulate all sorts of relationships ranging from mutualistic, commensalitic, antagonistic and endosymbiotic modes. All these can impact disease progression differently when established in a common infection niche (Nogueira et al., 2019, Allison et al., 2016, Kapitan et al., 2019).
Undeniably, di-kingdom aetiological synergy is becoming a common phenomenon among an array of patient populations (Allison et al., 2016, Hughes et al., 2020). In a patient cohort like those presenting with chronic pulmonary diseases suspected for pulmonary TB, it’s still far-sighted especially in yet to develop settings, and so there is need that definitive aetiology is established in suspected cases. With often a gap in suspicion of poly microbial aetiology among this patient populations, the burden and aetiology of invasive pulmonary infections due to non-TB aetiology is underestimated and thus worth investigating. Here we provide an insight into the di-kingdom poly microbial aetiological synergy associated with chronic pulmonary disease. We highlight that fungal-bacterial co-infections can be a common feature among this patient population. Specifically, we note that non-TB bacterial aetiology is as common yet the suspicion index for these infections is low in our routine practices. For instance, in the Ugandan clinical guidelines (UCG 2016), fungal pathogens were never given enough attention apart from the common infections like candidiasis. Here we show that the diversity of fungal aetiological agents is expanding and deserves enough attention. Similarly, so, the ability of fungi and bacteria to associatively co-infect the same host is now common and may deserve more attention than before.
Though globally pulmonary TB infections remain a major threat, it’s important to note that other aetiologies such as pulmonary fungal and non-TB bacterial aetiological agents are common. Further still, it’s also important to note that there is an interesting twist to this profile though not often investigated. The twist here is a co-colonization or co-existence of fungi and bacteria in the same infection niche. This cross-kingdom aetiological pattern seems to affect either microbial consortia’s ability to adapt with a subsequent influence on the virulence attributes of the microbes involved (Nogueira et al., 2019). A few studies so far highlight, the possibility of a cross talk between di-kingdom microbes (Dixon and Hall, 2015). Indeed, Fungal bacterial interactions can display a wide range of effects which may not be identical in vivo and in vitro. For instance, there has been an in vitro di-kingdom cross talk demonstration between Candida albicans and Pseudomonas aeruginosa that displays mostly an antagonistic relationship whilst the interaction in the human host displayed synergistic effects on their virulence that resulted in higher mortality (Bergeron et al., 2017, Neely et al., 1986, Nogueira et al., 2019, Roux et al., 2009). Indeed, this co-infection revealed significant upregulation of the proinflammatory cytokine IL-6 and a less prominent increase of IL-8 which is a potent chemo-attractant of neutrophils. This is significant evidence that indeed, microbial co-existence might shape phenotypic and genomic signatures of involved microorganisms
The question thus remains, what is the significance of mixed both mono and di-kingdom aetiological pairings observed in this patient population. Here, we note diversity in both mono and di-kingdom aetiological pairings which proves hard to pin point which of these would be a perfect fit to study detail in order to get a much clearer insight into what could be happening. However, just our knowledge of their prevalence is good enough to enhance clinical management. When compared to their association with TB, there seems to be a correlation between the most common aetiological pairings and TB prevalence. Whether this has an immunological aspect to it remains to be explored but could clearly mean that cross kingdom aetiology might prove more significant than we currently rate it.
Thus, our findings here demonstrate some rare aetiological pairings from what have been shown before. Accordingly, Acremonium spp + K. pneumoniae parings were the most prevalent. This was followed by Candida albicans + S. pneumonia pairings, C. albicans + K. pneumoniae + S. pneumoniae + E. coli pairings and C. fermata +E. coli + K. pneumoniae + S. pneumoniae pairings as the next commonly prevalent. This profile shows that the taxonomical range is also expanding to now encompass agents that have for a very long time been regarded environmental opportunists like Acremonium spp. However, we also note in the literature that this is not necessarily out of range for instance, Itabangi et al., described the influence of a bacterial endosymbiont Ralstonia pickettii on the virulence of an opportunistic fungus Rhizopus microsporus (Itabangi et al., 2022). The study describes an addictive mutualistic relationship between the two microbes that renders the fungus avirulent once the endosymbiont is knocked out (Itabangi et al., 2022). Now, this seems to be an extension of relationship that is established between R. microsporus and Burkholderia rhizoxinica. This is a strong evidence that bacteria and fungi can formulate subtle relationships. The question now becomes, are the pairings seen here established to such inter dependence interactional levels or these are superficial but vital interactions?
Additionally, Cryptococcus neoformans and Klebsiella aerogenes seem to form significant relationship where this bacterial species induces melanin production by C. neoformans through secretion of dopamine by the bacterium, thereby leading to enhanced protection of C. neoformans from macrophages. Enhanced melanization of Cryptococcus may also confer a protective effect against antifungal treatment (Rella et al., 2012, Frases et al., 2006). But we have not yet interrogated the interactions mechanisms of the identified pairings here and thus can’t attest to the impact of these on microbial virulence or patient prognosis. Finally, we also note that the Acremonium + K. pneumoniae pairings are common among patients during and Post TB treatment. Whether this contributes to poor prognosis in this these patients remains unclear. But at least, it’s important that efforts are made to investigate for such events especially in patients that show poor prognosis. This comes off a background, where a lot of efforts and attention are put on anti-TB drug resistance mostly when such cases appear. However, what we have not investigated yet is how far these associations influence patient prognosis.
On the other hand, poly fungal and Poly non-TB bacterial aetiological pairings were also noted, these mono kingdoms mixed infections can be uncommon. This is also crippled by the attention given to the common suspects. However, what these findings are showing here is that a lot of patients might be misdiagnosed most of the time and this calls for a revised diagnostic approach especially with the number of at-risk individuals dramatically increasing. Unfortunately, for many of the fungal-bacterial poly microbial pairings highlighted above, no data about their interactions are available regarding the interplay with the hosts and how these influences the immune system. Thus, there is a need to examine the important features of the fungal bacterial interactions and the interplay with the immune system. Indeed, perturbations of the microbiome and weakening of the immune system are conditions facilitating the transition of opportunistic microbes from commensal to a pathogenic state, mediating the initiation of infection.
Indeed, the interaction between microbial pathogens and the host induces the activation of several virulence factors and adaptation mechanisms. In fungal infections, the virulence factors include morphological transitions (e.g. yeast-to-hyphae), phenotypic switching (e.g., white to opaque state in C. albicans), biofilm formation, increased adhesion capacity, and environmental pH modulation. Whilst bacteria have also developed mechanisms to hide or escape from the immune system. Some of these mechanisms are similar to those used by fungi. Biofilm formation is also an important feature used by bacteria to evade the immune response. Other factors include the secretion of proteins, quorum sensing regulation, production of antigenic exotoxins, pore-forming toxins, and capsular polysaccharides. This similarity in the evasion approach of the immune system suggests a possibility of major cross talk between the two microbial kingdoms.
4.1. Conclusions
Our study provides a strong evidence and further confirms that bacteria and fungi can co-exist and possibly formulate subtle relationships, with significant mono and di-kingdom aetiological pairings observed in out patient population. However, these findings further demonstrate some rare aetiological pairings from what have been shown before. Additionally, our data may indicate possible underlying synergistic interactions and relationships that can be addictive, mutualistic, commensalitic and endosymbiotic among others between bacteria and fungi. Yet, how deep these interactions go may require further investigations in order to identify specific interaction signatures and their impact on pathogenicity and pathogenesis by either microbial consortium.
4.2. Limitations
The main limitation of the study has been lack of access to advanced techniques to investigate the different subtle relationships identified here
Author Contributions
HI: EBR, MJC, RM, and JK developed the study concept. HI, EBR, NE, MJC, and AL, developed the study design. HI, EBR, NIK, MA, BM, RCM, MM, JKM, PN, EN, JM, JSI, BM, KK, FS, provided input into biological sample collection. JB, JM, JSI, JKM, JK, MC, RM, ERB and LA provided technical insight for the project direction. HI drafted the paper, and all authors read and commented on the paper and agreed with the final version.
Funding
This project was funded by the European Developing Countries Clinical Trails Partnership (EDCTP) with support from the European Union (Grant Ref: TMA 2019 CDF- 2789) under the Project Name Metabolic and molecular ecological evolution of opportunistic pulmonary fungal co-infections (MeMoF).
Data Availability Statement
All data sets generated during this study are available from the corresponding author and can be shared according to the data sharing act of Mbarara University of science and Technology.
Acknowledgments
We recognize all technical investments into the project by our collaborators specially Robin May and Mike Cox from University of Birmingham.
Conflicts of Interest
The authors declare no conflict of interest.
References
- ALLISON, D. L., WILLEMS, H. M. E., JAYATILAKE, J., BRUNO, V. M., PETERS, B. M. & SHIRTLIFF, M. E. Candida-Bacteria Interactions: Their Impact on Human Disease. Microbiol Spectr 2016, 4. [CrossRef]
- AMIRI, M. R. J., SIAMI, R. & KHALEDI, A. Tuberculosis Status and Coinfection of Pulmonary Fungal Infections in Patients Referred to Reference Laboratory of Health Centers Ghaemshahr City during 2007-2017. Ethiop J Health Sci 2018, 28, 683–690. [CrossRef] [PubMed]
- BERGERON, A. C., SEMAN, B. G., HAMMOND, J. H., ARCHAMBAULT, L. S., HOGAN, D. A. & WHEELER, R. T. Candida albicans and Pseudomonas aeruginosa Interact To Enhance Virulence of Mucosal Infection in Transparent Zebrafish. Infect Immun 2017, 85. [CrossRef] [PubMed]
- BURKI, T. WHO publish fungal priority pathogens list. Lancet Microbe 2023, 4, e74. [Google Scholar] [CrossRef] [PubMed]
- CASADEVALL, A. & PIROFSKI, L. A. The damage-response framework of microbial pathogenesis. Nat Rev Microbiol 2003, 1, 17–24. [CrossRef] [PubMed]
- CHONG, W. H., SAHA, B. K., ANANTHAKRISHNAN, R. & CHOPRA, A. State-of-the-art review of secondary pulmonary infections in patients with COVID-19 pneumonia. Infection 2021, 49, 591–605. [CrossRef] [PubMed]
- DIXON, E. F. & HALL, R. A. Noisy neighbourhoods: quorum sensing in fungal-polymicrobial infections. Cell Microbiol 2015, 17, 1431–1441. [CrossRef]
- DU, Q., REN, B., ZHOU, X., ZHANG, L. & XU, X. Cross-kingdom interaction between Candida albicans and oral bacteria. Front Microbiol 2022, 13, 911623. [CrossRef] [PubMed]
- ICHELBERGER, K. R. & CASSAT, J. E. Metabolic Adaptations During Staphylococcus aureus and Candida albicans Co-Infection. Front Immunol 2021, 12, 797550. [CrossRef]
- FEKKAR, A., LAMPROS, A., MAYAUX, J., POIGNON, C., DEMERET, S., CONSTANTIN, J. M., MARCELIN, A. G., MONSEL, A., LUYT, C. E. & BLAIZE, M. Occurrence of Invasive Pulmonary Fungal Infections in Patients with Severe COVID-19 Admitted to the ICU. Am J Respir Crit Care Med 2021, 203, 307–317. [CrossRef]
- FRASES, S., CHASKES, S., DADACHOVA, E. & CASADEVALL, A. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl Environ Microbiol 2006, 72, 1542–1550. [CrossRef] [PubMed]
- HANSON, A., ENOCH, A. S. & WOKEM, G. N. Tuberculosis-Candida Co-Infection in Patients having Pulmonary Tuberculosis Attending DOTs Clinic in Rumuigbo Model Primary Health Centre in Port Harcourt, Nigeria. International Journal of TROPICAL DISEASE & Health 2019, 35, 1–7. [CrossRef]
- HUGHES, S., TROISE, O., DONALDSON, H., MUGHAL, N. & MOORE, L. S. P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect 2020, 26, 1395–1399. [CrossRef] [PubMed]
- ITABANGI, H., AMPAIRE, L., NJOVU, I. K., NALUMAGA, P. P., MUSINGUZI, B., KASSAZA, K., KIGULI, J. M., BAZIRA, J., MWESIGYE, J. & IRAMIOT, J. S. Characterization of fungal-bacterial co-infections among presumptive pulmonary tuberculosis cases in Western Uganda, a project protocol. [CrossRef]
- ITABANGI, H., SEPHTON-CLARK, P. C. S., TAMAYO, D. P., ZHOU, X., STARLING, G. P., MAHAMOUD, Z., INSUA, I., PROBERT, M., CORREIA, J., MOYNIHAN, P. J., GEBREMARIAM, T., GU, Y., IBRAHIM, A. S., BROWN, G. D., KING, J. S., BALLOU, E. R. & VOELZ, K. A bacterial endosymbiont of the fungus Rhizopus microsporus drives phagocyte evasion and opportunistic virulence. Curr Biol 2022, 32, 1115–1130.e6. [CrossRef] [PubMed]
- JABBARI AMIRI, M. R., AGHILI, S. R., SHOKOHI, T., HEDAYATI, M. T., ABASTABAR, M., ALIYALI, M., JABBARI AMIRI, M. & HASANPOUR, H. Invasive forms of Candida and Aspergillus in sputum samples of pulmonary tuberculosis patients attending the tuberculosis reference laboratory in Ghaemshahr, Northern Iran: An analysis of samples collected during the past 10years. Int J Mycobacteriol 2016, 5 (Suppl 1), S179–s180. [CrossRef] [PubMed]
- KAPITAN, M., NIEMIEC, M. J., STEIMLE, A., FRICK, J. S. & JACOBSEN, I. D. Fungi as Part of the Microbiota and Interactions with Intestinal Bacteria. Curr Top Microbiol Immunol 2019, 422, 265–301. [CrossRef] [PubMed]
- MACALPINE, J., ROBBINS, N. & COWEN, L. E. Bacterial-fungal interactions and their impact on microbial pathogenesis. Mol Ecol 2023, 32, 2565–2581. [CrossRef]
- MATSUO, K., HAKU, A., BI, B., TAKAHASHI, H., KAMADA, N., YAGUCHI, T., SAIJO, S., YONEYAMA, M. & GOTO, Y. Fecal microbiota transplantation prevents Candida albicans from colonizing the gastrointestinal tract. Microbiol Immunol 2019, 63, 155–163. [CrossRef]
- NADEEM, S. G., HAKIM, S. T. & KAZMI, S. U. Use of CHROMagar Candida for the presumptive identification of Candida species directly from clinical specimens in resource-limited settings. Libyan J Med 2010, 5. [CrossRef]
- NASEEF, H. A., MOHAMMAD, U., AL-SHAMI, N., SAHOURY, Y., ABUKHALIL, A. D., DREIDI, M., ALSAHOURI, I. & FARRAJ, M. Bacterial and fungal co-infections among ICU COVID-19 hospitalized patients in a Palestinian hospital: a retrospective cross-sectional study. F1000Res 2022, 11, 30. [CrossRef]
- NEELY, A. N., LAW, E. J. & HOLDER, I. A. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun 1986, 52, 200–204. [CrossRef]
- NJOVU, I. K., MUSINGUZI, B., MWESIGYE, J., KASSAZA, K., TURIGURWA, J., NUWAGIRA, E., BAZIRA, J., KABANDA, T., MPEIRWE, M., AMPAIRE, L., MUTEKANGA, A., KIGULI, J., ACHAN, B. & ITABANGI, H. Status of pulmonary fungal pathogens among individuals with clinical features of pulmonary tuberculosis at Mbarara University Teaching Hospital in Southwestern Uganda. Ther Adv Infect Dis 2021, 8, 20499361211042477. [CrossRef] [PubMed]
- NJOVU, I. K., NALUMAGA, P. P., AMPAIRE, L., NUWAGIRA, E., MWESIGYE, J., MUSINGUZI, B., KASSAZA, K., TASEERA, K., KIGULI MUKASA, J., BAZIRA, J., IRAMIOT, J. S., BAGUMA, A., BONGOMIN, F., KWIZERA, R., ACHAN, B., COX, M. J., KING, J. S., MAY, R., BALLOU, E. R. & ITABANGI, H. Investigating Metabolic and Molecular Ecological Evolution of Opportunistic Pulmonary Fungal Coinfections: Protocol for a Laboratory-Based Cross-Sectional Study. JMIR Res Protoc 2023, 12, e48014. [CrossRef] [PubMed]
- NOGUEIRA, F., SHARGHI, S., KUCHLER, K. & LION, T. Pathogenetic Impact of Bacterial-Fungal Interactions. Microorganisms 2019, 7. [CrossRef] [PubMed]
- PARUMS, D.V. Editorial: The World Health Organization (WHO) Fungal Priority Pathogens List in Response to Emerging Fungal Pathogens During the COVID-19 Pandemic. Med Sci Monit 2022, 28, e939088. [Google Scholar] [CrossRef] [PubMed]
- PELEG, A. Y., HOGAN, D. A. & MYLONAKIS, E. Medically important bacterial-fungal interactions. Nat Rev Microbiol 2010, 8, 340–349. [CrossRef]
- RAWSON, T. M., MOORE, L. S. P., ZHU, N., RANGANATHAN, N., SKOLIMOWSKA, K., GILCHRIST, M., SATTA, G., COOKE, G. & HOLMES, A. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin Infect Dis 2020, 71, 2459–2468. [CrossRef]
- REECE, E., SEGURADO, R., JACKSON, A., MCCLEAN, S., RENWICK, J. & GREALLY, P. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: an Irish registry analysis. BMC Pulm Med 2017, 17, 70. [CrossRef] [PubMed]
- RELLA, A., YANG, M. W., GRUBER, J., MONTAGNA, M. T., LUBERTO, C., ZHANG, Y. M. & DEL POETA, M. Pseudomonas aeruginosa inhibits the growth of Cryptococcus species. Mycopathologia 2012, 173, 451–461. [CrossRef]
- ROTHE, K., FEIHL, S., SCHNEIDER, J., WALLNÖFER, F., WURST, M., LUKAS, M., TREIBER, M., LAHMER, T., HEIM, M., DOMMASCH, M., WASCHULZIK, B., ZINK, A., QUERBACH, C., BUSCH, D. H., SCHMID, R. M., SCHNEIDER, G. & SPINNER, C. D. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis 2021, 40, 859–869. [CrossRef] [PubMed]
- ROUX, D., GAUDRY, S., DREYFUSS, D., EL-BENNA, J., DE PROST, N., DENAMUR, E., SAUMON, G. & RICARD, J. D. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit Care Med 2009, 37, 1062–1067. [CrossRef] [PubMed]
- SCHÖNHERR, F. A., SPARBER, F., KIRCHNER, F. R., GUIDUCCI, E., TRAUTWEIN-WEIDNER, K., GLADIATOR, A., SERTOUR, N., HETZEL, U., LE, G. T. T., PAVELKA, N., D'ENFERT, C., BOUGNOUX, M. E., CORTI, C. F. & LEIBUNDGUT-LANDMANN, S. The intraspecies diversity of C. albicans triggers qualitatively and temporally distinct host responses that determine the balance between commensalism and pathogenicity. Mucosal Immunol 2017, 10, 1335–1350. [CrossRef] [PubMed]
- SHIRTLIFF, M. E., PETERS, B. M. & JABRA-RIZK, M. A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiology Letters 2009, 299, 1–8. [CrossRef]
- SIDDHARTHAN, T., GRIGSBY, M., MORGAN, B., KALYESUBULA, R., WISE, R. A., KIRENGA, B. & CHECKLEY, W. Prevalence of chronic respiratory disease in urban and rural Uganda. Bull World Health Organ 2019, 97, 318–327. [CrossRef]
- SILVEIRA, F. & PATERSON, D. L. Pulmonary fungal infections. Curr Opin Pulm Med 2005, 11, 242–246. [CrossRef]
- SMITH, J. A. & KAUFFMAN, C. A. Pulmonary fungal infections. Respirology 2012, 17, 913–926. [CrossRef]
- SREENATH, K., BATRA, P., VINAYARAJ, E. V., BHATIA, R., SAIKIRAN, K., SINGH, V., SINGH, S., VERMA, N., SINGH, U. B., MOHAN, A., BHATNAGAR, S., TRIKHA, A., GULERIA, R. & CHAUDHRY, R. Coinfections with Other Respiratory Pathogens among Patients with COVID-19. Microbiol Spectr 2021, 9, e0016321. [CrossRef]
- TACCONELLI, E., CARRARA, E., SAVOLDI, A., HARBARTH, S., MENDELSON, M., MONNET, D. L., PULCINI, C., KAHLMETER, G., KLUYTMANS, J., CARMELI, Y., OUELLETTE, M., OUTTERSON, K., PATEL, J., CAVALERI, M., COX, E. M., HOUCHENS, C. R., GRAYSON, M. L., HANSEN, P., SINGH, N., THEURETZBACHER, U. & MAGRINI, N. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018, 18, 318–327. [CrossRef]
- TOBÓN, A. M. & GÓMEZ, B. L. Pulmonary Histoplasmosis. Mycopathologia 2021, 186, 697–705. [CrossRef]
- TODD, O. A., NOVERR, M. C. & PETERS, B. M. Candida albicans Impacts Staphylococcus aureus Alpha-Toxin Production via Extracellular Alkalinization. mSphere 2019, 4. [CrossRef] [PubMed]
- VAN GEMERT, F., KIRENGA, B., CHAVANNES, N., KAMYA, M., LUZIGE, S., MUSINGUZI, P., TURYAGARUKA, J., JONES, R., TSILIGIANNI, I., WILLIAMS, S., DE JONG, C. & VAN DER MOLEN, T. Prevalence of chronic obstructive pulmonary disease and associated risk factors in Uganda (FRESH AIR Uganda): a prospective cross-sectional observational study. Lancet Glob Health 2015, 3, e44–e51. [CrossRef] [PubMed]
- YAMAMURA, D. & XU, J. Update on Pulmonary Cryptococcosis. Mycopathologia 2021, 186, 717–728. [CrossRef] [PubMed]
- ZHAO, Z., SONG, J., YANG, C., YANG, L., CHEN, J., LI, X., WANG, Y. & FENG, J. Prevalence of Fungal and Bacterial Co-Infection in Pulmonary Fungal Infections: A Metagenomic Next Generation Sequencing-Based Study. Front Cell Infect Microbiol 2021, 11, 749905. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).