Submitted:
16 August 2024
Posted:
20 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Single cell RNA (scRNA) Sequencing has Revealed the Importance of Innate Immunity, Especially Macrophages
1.2. The Clue of Antibody Dependent Enhancement (ADE) of Infection into Macrophages
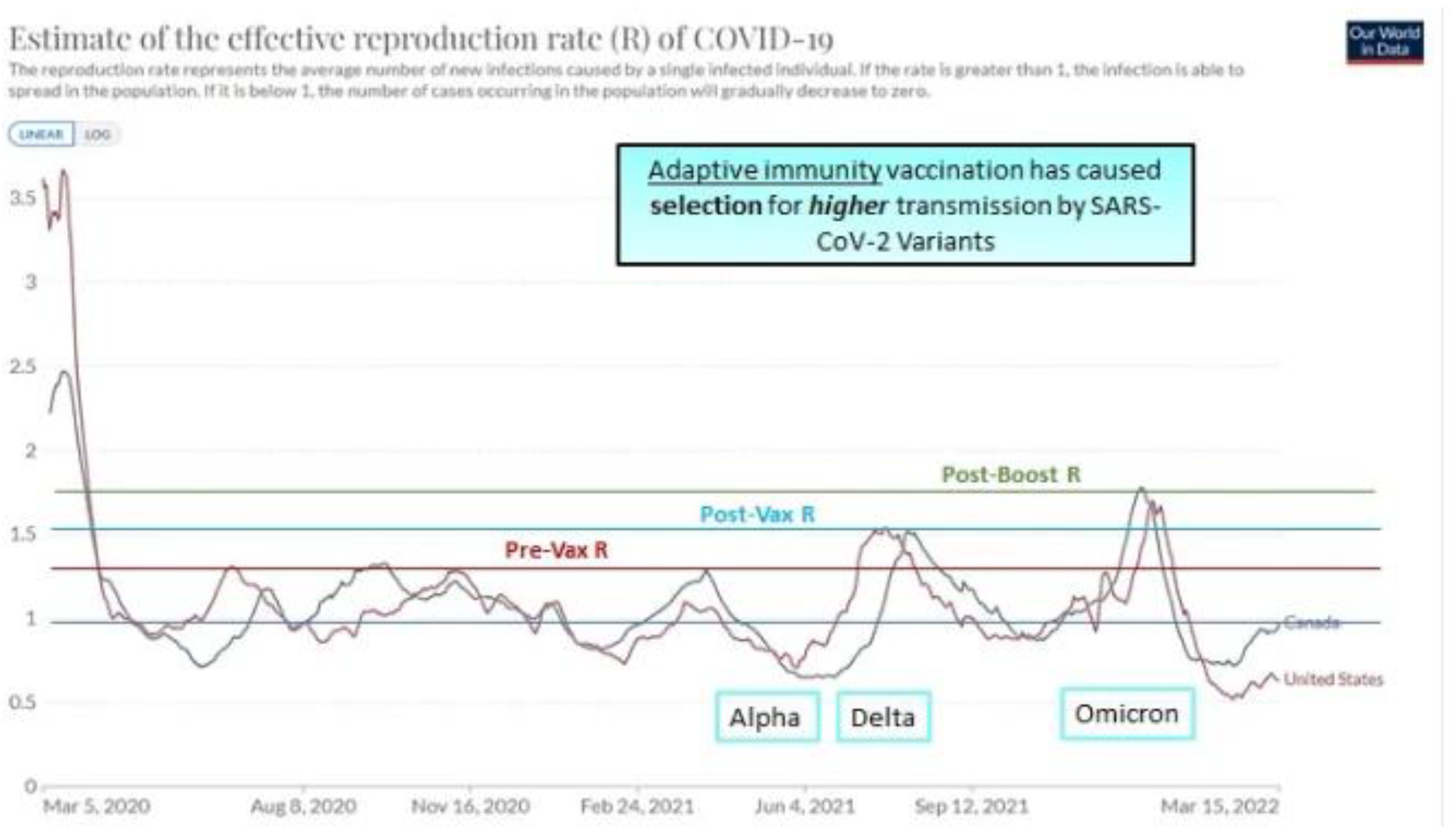
1.3. Evidence from Canada for Selection of SARS-CoV-2 Variants Correlating with Vaccination Milestones
1.4. Vaccination Increased SARS-CoV-2 Symptomatic Infection Rates in the UK Putatively by ADE
1.5. From First Principles Adaptive Immunity is Not Able to Control Emerging Pathogens or Pandemics
1.6. Aims of the Review
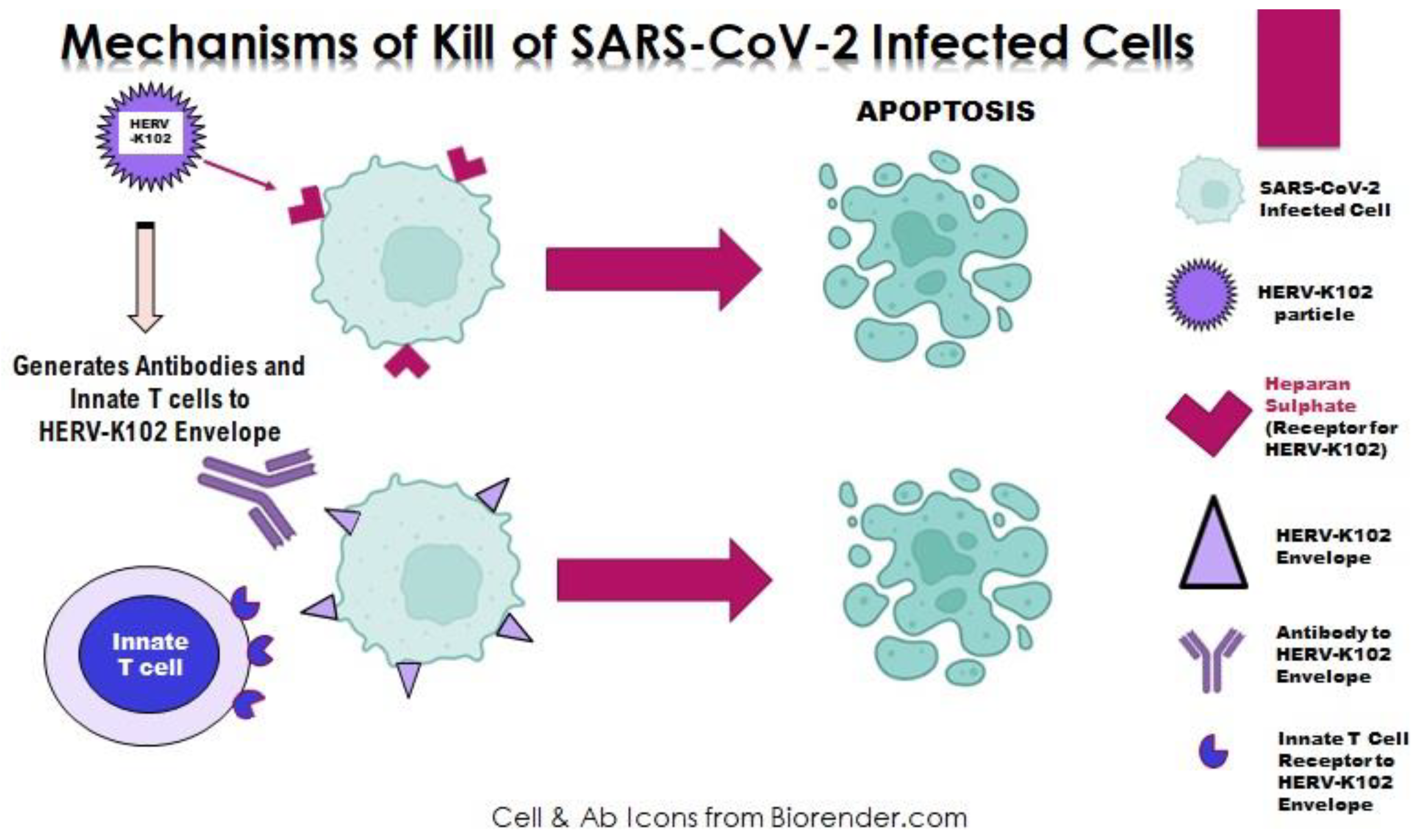
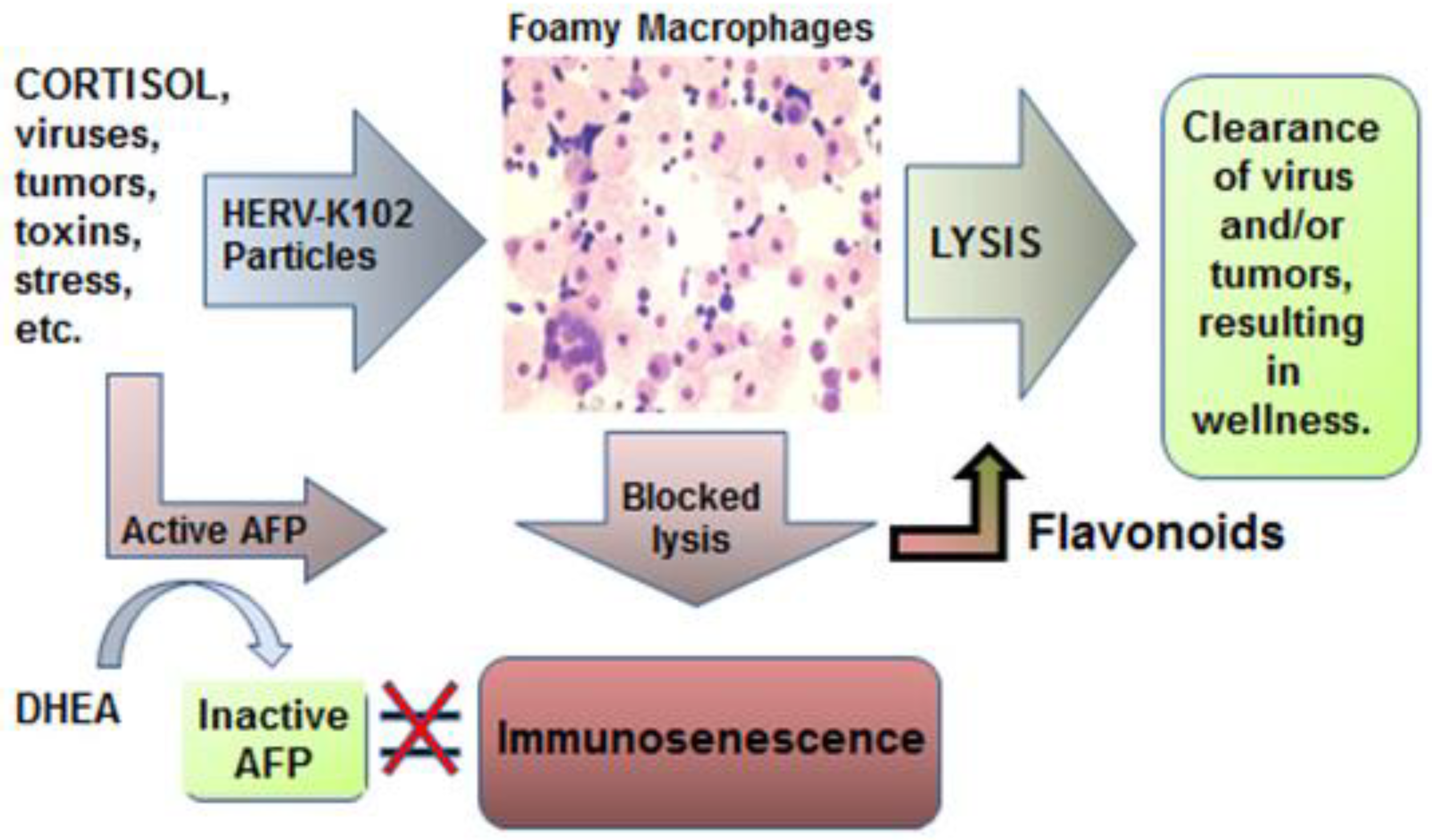
2. The HERV-K102 Protection System of Pro-inflammatory M1-like Foamy Macrophages
2.1. HERV-K102 is a Type 1 HERV-K HML-2 Group Member
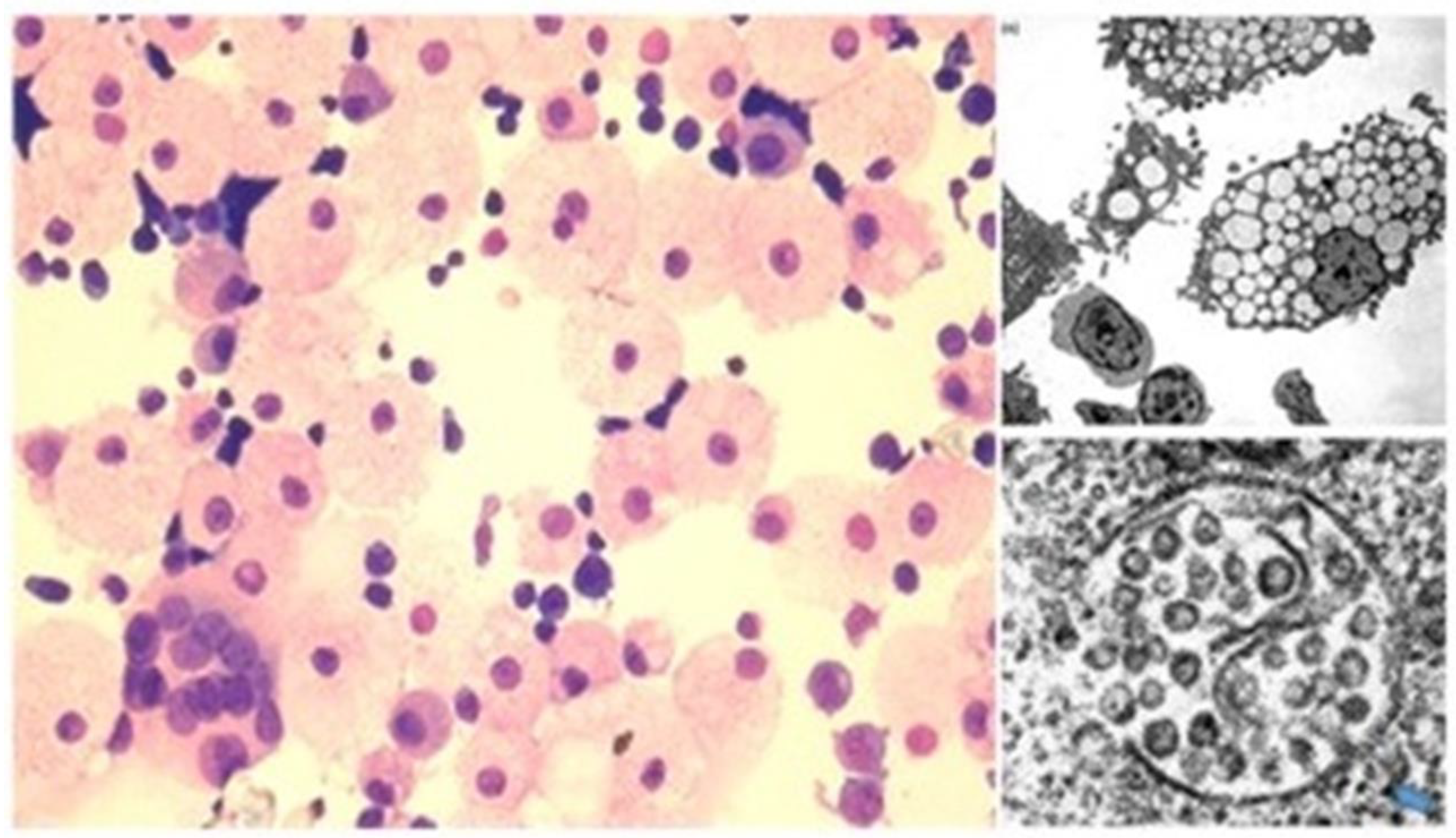
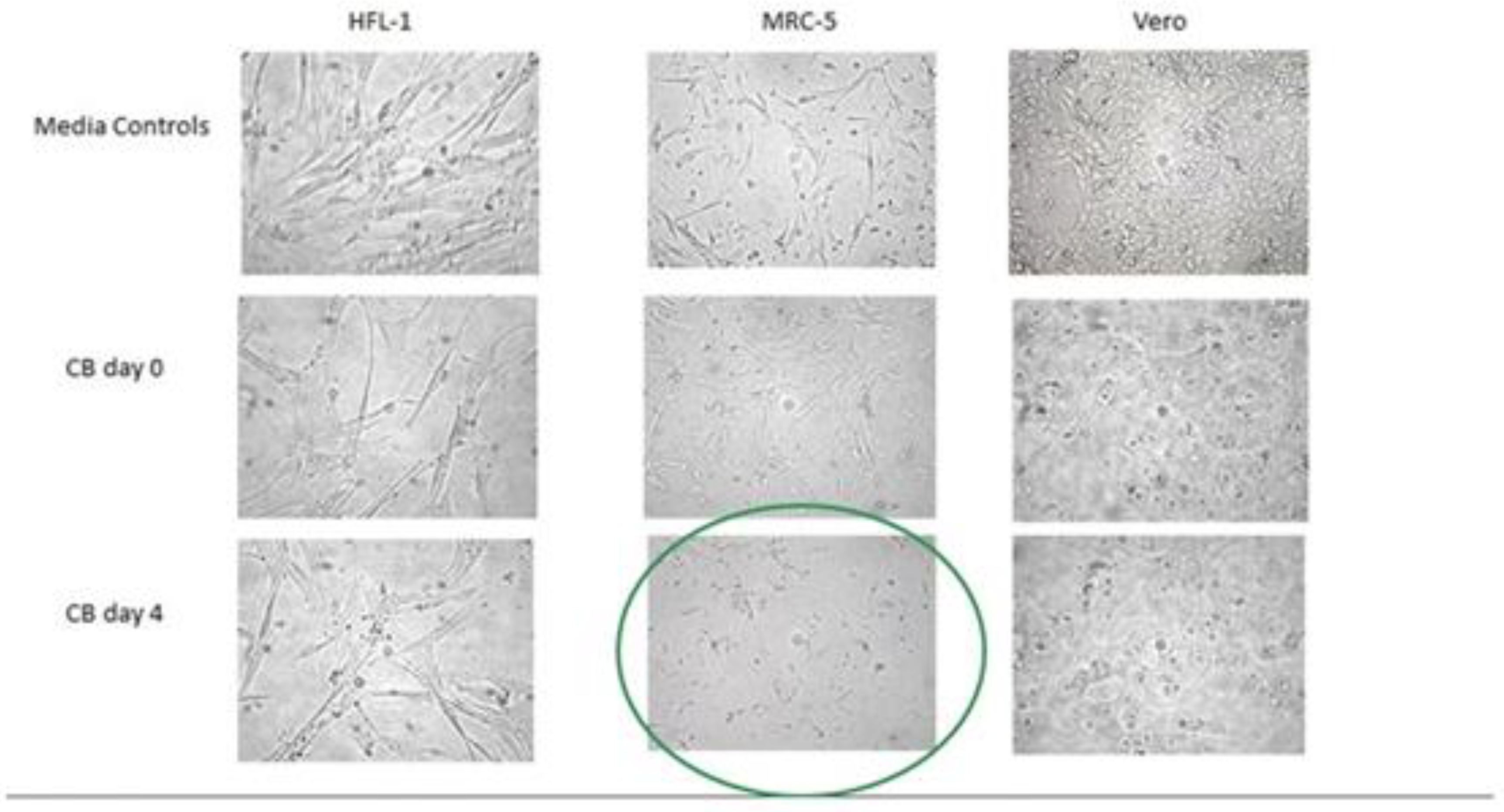
2.2. HERV-K102 as the Elusive Foamy Retrovirus of Humans
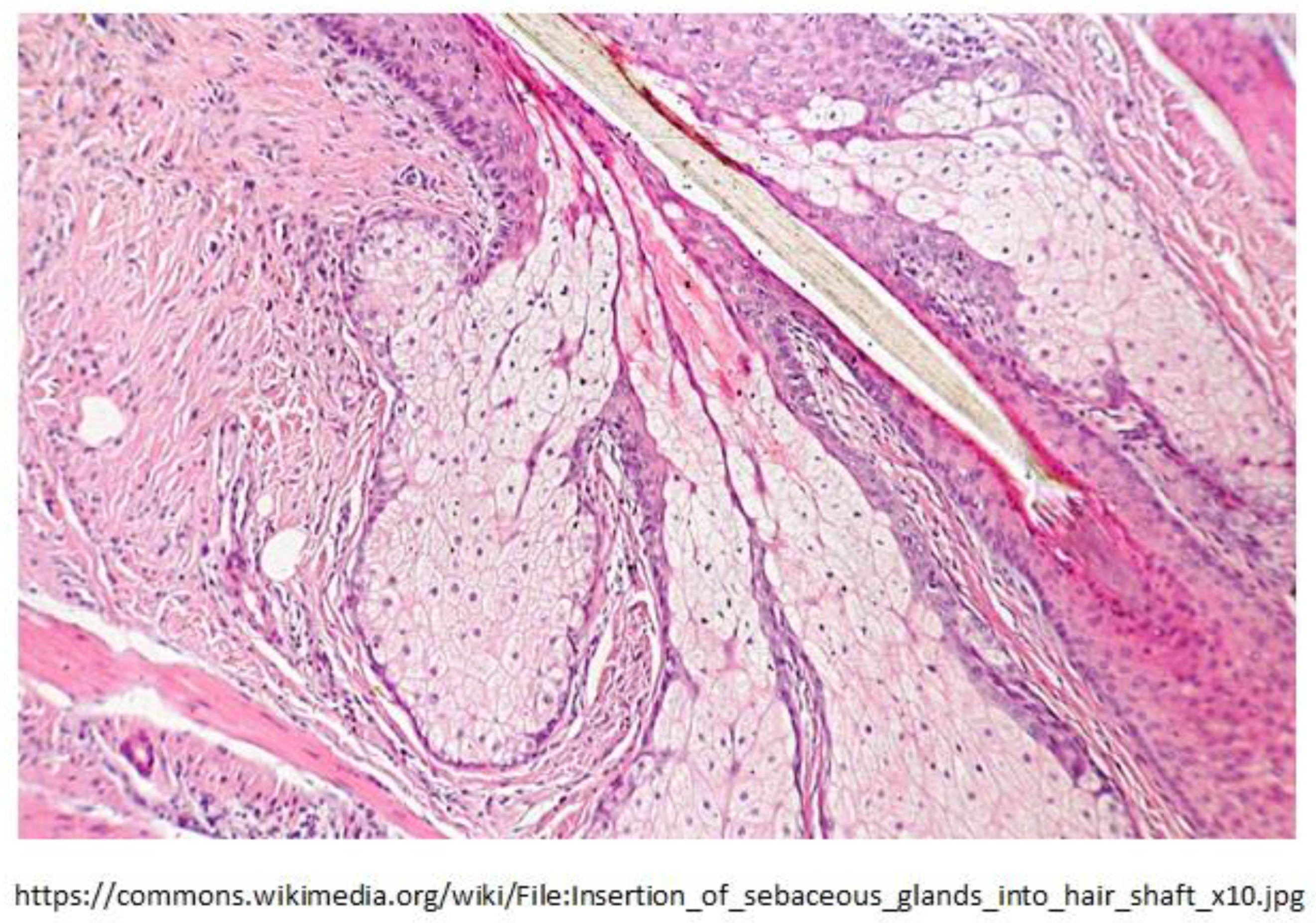
2.3. Sebocytes of Sebaceous Glands Lining the Mucosa Were Discovered to Produce HERV-K102 Particles
2.4. The Origins of HERV-K102 Also Provides a Major Clue to Its Purpose
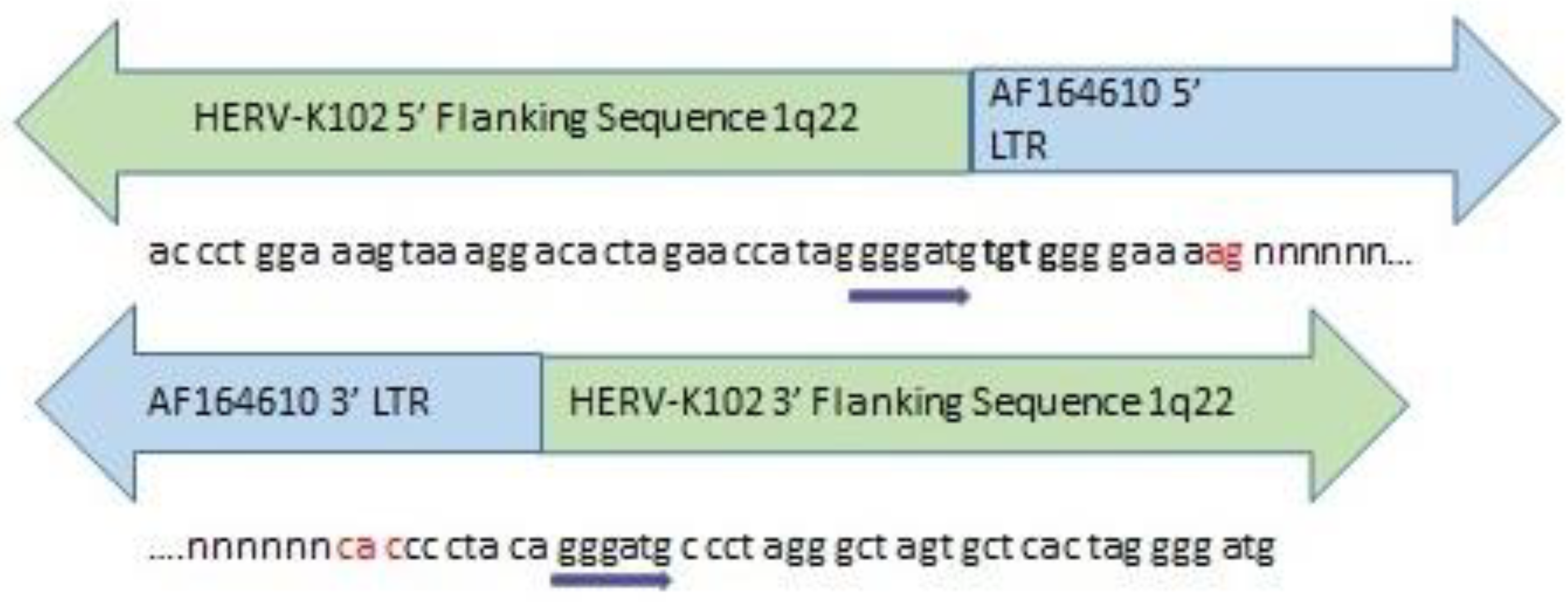
2.4.1. On the Curious Origins of HERV-K102 in Humans
2.5. Evidence for a Role of HERV-K HML-2 Activation in Innate Immunity
2.5.1. The ‘Virus-Antivirus Properties’ Associated with HERV-K HML-2 Activity
2.6. The Concept of Innate T and B Cell Responses Against HERV-K102 Envelope
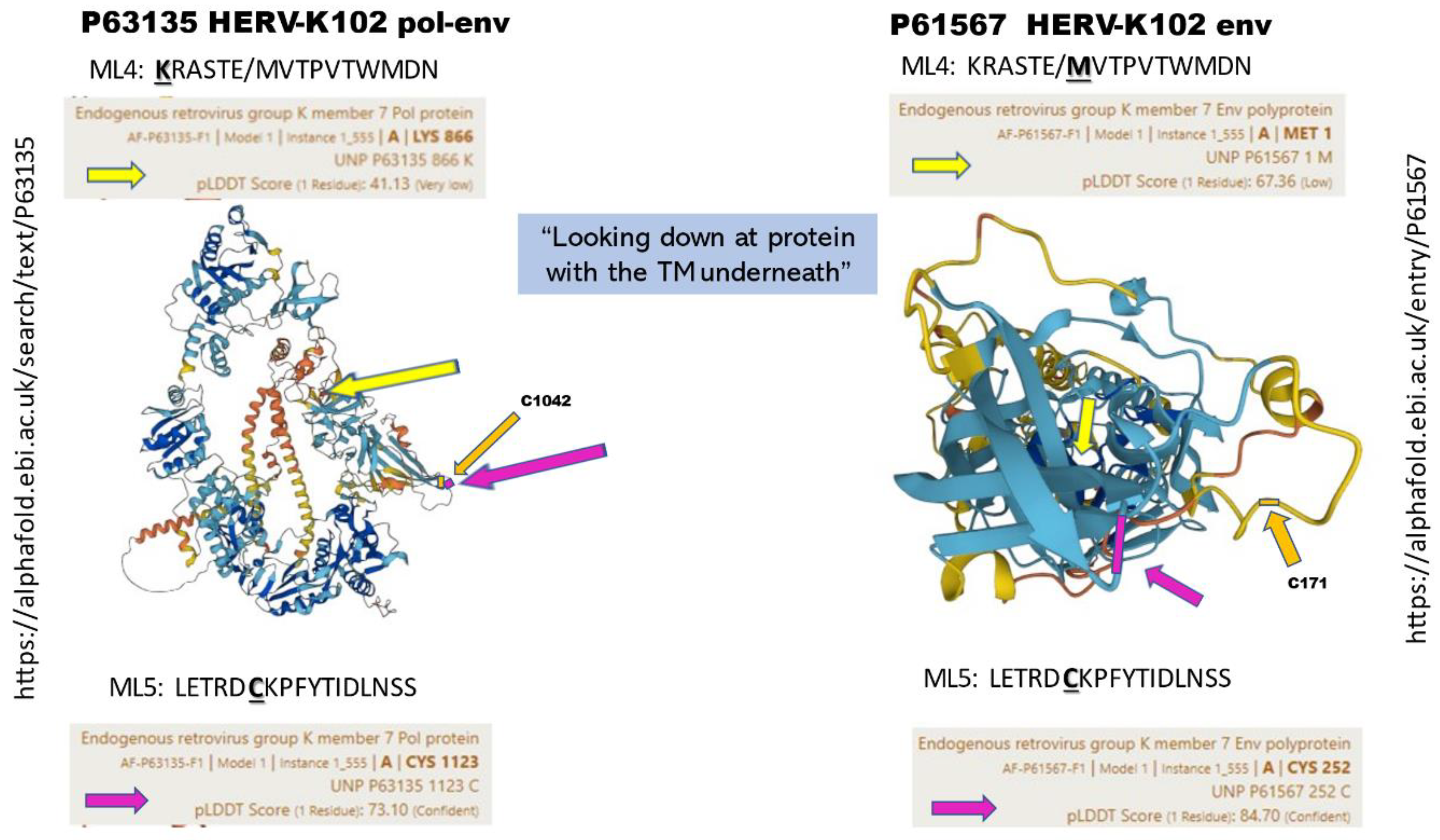
2.6.1. The Concept of SELECT Epitopes Reactive to Innate Antibodies to HERV-K102 Env
2.7. Epigenetic Control of HERV-K102 Expression and Trained Immunity
2.8. When Things Go Wrong with the HERV-K102 Protector System: Immunosenescence
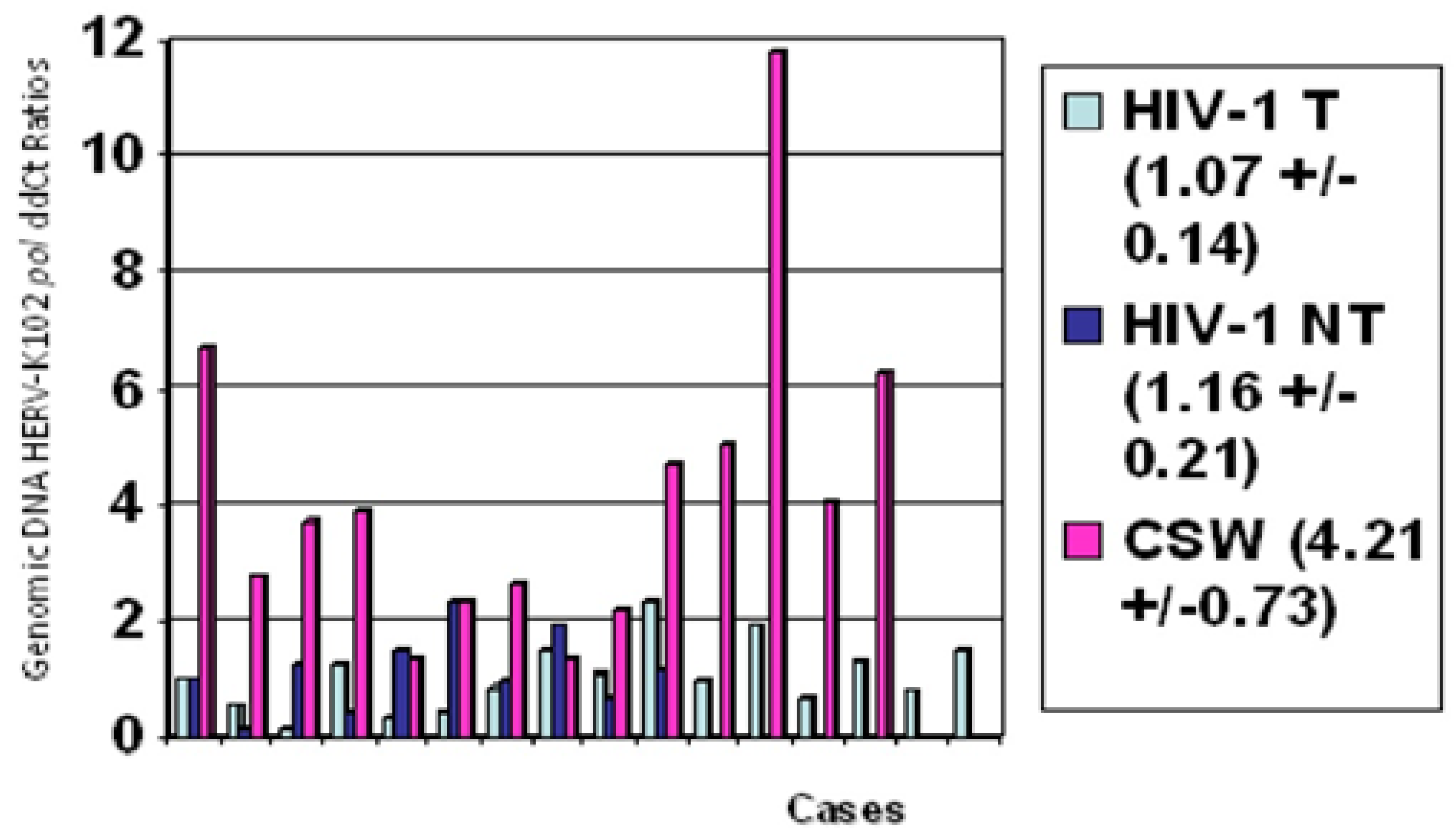
3. Effects of ADE on the Launch of the Critical HERV-K102 Protector System in M1-like Foamy Macrophages
4. Incontrovertible Evidence for ADE As an Impediment to the Safety and Effectiveness of the Adaptive Immunity COVID-19 Vaccines
5. Future Directions
6. Summary and Conclusions
7. Patents
- Cdn Patent Application 2,501,301 March 18, 2005:Patent number CA 2673395 issued October 22, 2013, for screening methods.
- US Patent Application 60/663,263 March 21, 2005:Patent 7,964,341 issued June 21, 2011, for screening methods.
- PCT Application CA2006/000397 March 20, 2006:PCT: WO 2006/096985
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan GC, Cai L, Elowitz M, et al. Challenges and emerging directions in single-cell analysis. Genome Biol. 2017 May 8;18(1):84. [CrossRef]
- Gao S, Yan L, Wang R, et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat Cell Biol. 2018 Jun;20(6):721-734. [CrossRef]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015 Aug;21(8):938-945. [CrossRef]
- https://en.wikipedia.org/wiki/Galactose-alpha-1,3-galactose.
- Mumme H, Thomas BE, Bhasin SS, et al. Single-cell analysis reveals altered tumor microenvironments of relapse- and remission-associated pediatric acute myeloid leukemia. Nat Commun. 2023 Oct 5;14(1):6209. [CrossRef]
- Zhang JT, Zhang J, Wang SR, et al. Spatial downregulation of CD74 signatures may drive invasive component development in part-solid lung adenocarcinoma. iScience. 2023 Aug 21;26(10):107699. [CrossRef]
- Shangguan S, Ehrenberg PK, Geretz A, et al. Monocyte-derived transcriptome signature indicates antibody-dependent cellular phagocytosis as a potential mechanism of vaccine-induced protection against HIV-1. Elife. 2021 Sep 17;10:e69577. [CrossRef]
- Vaccari M, Fourati S, Gordon SN, et al. HIV vaccine candidate activation of hypoxia and the inflammasome in CD14+ monocytes is associated with a decreased risk of SIVmac251 acquisition. Nat Med. 2018 Jun;24(6):847-856. [CrossRef]
- Thomas C, Leleu D, Masson D. Cholesterol and HIF-1α: dangerous liaisons in atherosclerosis. Front Immunol. 2022 Mar 21;13:868958. [CrossRef]
- Laderoute MP, Larocque LJ, Giulivi A, Diaz-Mitoma F. Further evidence that human endogenous retrovirus K102 is a replication competent foamy virus that may antagonize HIV-1 replication. Open AIDS J. 2015 Dec 7;9:112-22. [CrossRef]
- Laderoute MP. Clues to finding correlates of risk/protection for HIV-1 vaccines [version 2; peer review: 2 approved with reservations] F1000 Research 2018, 6:868. [CrossRef]
- Hussein HAM, Thabet AAA, Mohamed TIA, et al. Phenotypical changes of hematopoietic stem and progenitor cells in COVID-19 patients: Correlation with disease status. Cent Eur J Immunol. 2023;48(2):97-110. [CrossRef]
- Sawant J, Patil A, Kurle S. A review: understanding molecular mechanisms of antibody-dependent enhancement in viral infections. Vaccines (Basel). 2023 Jul 14;11(7):1240. [CrossRef]
- Ricke, D.O. Two different antibody-dependent enhancement (ADE) risks for SARS-CoV-2 antibodies. Front Immunol. 2021 Feb 24;12:640093. [CrossRef]
- Gartlan C, Tipton T, Salguero FJ, Sattentau Q, Gorringe A, Carroll MW. Vaccine-associated enhanced disease and pathogenic human coronaviruses. Front Immunol. 2022 Apr 4;13:882972. [CrossRef]
- Zohar T, Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol. 2020 Jul;20(7):392-394. [CrossRef]
- Chen W, Zhang J, Qin X, et al. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomed Pharmacother. 2020 Oct;130:110629. [CrossRef]
- Chen X, Pan Z, Yue S, et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Sig Transduct Target Ther. 2020 5, 180. [CrossRef]
- Hashem AM, Algaissi A, Almahboub SA, et al. Early humoral response correlates with disease severity and outcomes in COVID-19 patients. Viruses. 2020 Dec 4;12(12):1390. [CrossRef]
- Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020 Nov 19;71(16):2027-2034. [CrossRef]
- Shrock E, Fujimura E, Kula T, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020 Nov 27;370(6520):eabd4250. [CrossRef]
- Choteau M, Scohy A, Messe S, et al. Development of SARS-CoV2 humoral response including neutralizing antibodies is not sufficient to protect patients against fatal infection. Sci Rep. 2022 Feb 8;12(1):2077. [CrossRef]
- Legros V, Denolly S, Vogrig M, et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021 Feb;18(2):318-327. [CrossRef]
- Ren L, Fan G, Wu W, et al. Antibody responses and clinical outcomes in adults hospitalized with severe coronavirus disease 2019 (COVID-19): a post hoc analysis of LOTUS China trial. Clin Infect Dis. 2021 May 18;72(10):e545-e551. [CrossRef]
- Wang K, Fan G, Wu W, et al. Longitudinal dynamics of the neutralizing antibody response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Clin Infect Dis. 2021 Aug 2;73(3):e531-e539. [CrossRef]
- Xu X, Nie S, Wang Y, et al. Dynamics of neutralizing antibody responses to SARS-CoV-2 in patients with COVID-19: an observational study. Signal Transduct Target Ther. 2021 May 18;6(1):197. [CrossRef]
- Gao L, Zhou J, Yang S, et al. The dichotomous and incomplete adaptive immunity in COVID-19 patients with different disease severity. Sig Transduct Target Ther. 2021 6, 113. [CrossRef]
- Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020 Sep 17;11(1):4704. [CrossRef]
- Nair S, Chen X. Biology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the humoral immunoresponse: a systematic review of evidence to support global policy-level actions and research. Glob Health J. 2022 Mar;6(1):38-43. [CrossRef]
- Wu F, Liu M, Wang A, et al. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai, China. JAMA Intern Med. 2020 Oct 1;180(10):1356-1362. [CrossRef]
- Dispinseri S, Secchi M, Pirillo MF, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 2021 May 11;12(1):2670. [CrossRef]
- Garcia-Beltran WF, Lam EC, Astudillo MG, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021 Jan 21;184(2):476-488.e11. [CrossRef]
- Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022 Jan 7;375(6576):43-50. [CrossRef]
- Zohar T, Loos C, Fischinger S, et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020 Dec 10;183(6):1508-1519.e12. [CrossRef]
- Tetro, JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020 Mar;22(2):72-73. [CrossRef]
- Moorlag SJCFM, Taks E, Ten Doesschate T, et al. Efficacy of Bacillus Calmette-Guérin vaccination against respiratory tract infections in the elderly during the Covid-19 pandemic. Clin Infect Dis. 2022 Mar 5:ciac182. [CrossRef]
- Bigay J, Le Grand R, Martinon F, Maisonnasse P. Vaccine-associated enhanced disease in humans and animal models: lessons and challenges for vaccine development. Front Microbiol. 2022 Aug 10;13:932408. [CrossRef]
- Munitz A, Edry-Botzer L, Itan M, et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci Rep. 2021 Feb 10;11(1):3461. [CrossRef]
- Ho, M.S., Chen W.J., Chen H.Y., Lin S.F., Wang W.C., Di J. Neutralizing antibody response and SARS severity. Emerg Infect Dis. 2005;11:1730–1737.
- Liu L, Wei Q, Lin Q, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019 Feb 21;4(4):e123158. [CrossRef]
- Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 May;581(7809):465-469. [CrossRef]
- Jensen B, Luebke N, Feldt T, et al. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg Health Eur. 2021 Sep;8:100164. [CrossRef]
- Guerrieri M, Francavilla B, Fiorelli D, et al. Nasal and salivary mucosal humoral immune response elicited by mRNA BNT162b2 COVID-19 vaccine compared to SARS-CoV-2 natural infection. Vaccines (Basel). 2021 Dec 18;9(12):1499. [CrossRef]
- Aksyuk AA, Bansal H, Wilkins D, et al. AZD1222-induced nasal antibody responses are shaped by prior SARS-CoV-2 infection and correlate with virologic outcomes in breakthrough infection. Cell Rep Med. 2022 Dec 15:100882. [CrossRef]
- Kistler KE, Huddleston J, Bedford T. Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2. Cell Host Microbe. 2022 Apr 13;30(4):545-555.e4. [CrossRef]
- Walsh EE, Frenck RW Jr, Falsey AR, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. 2020 Dec 17;383(25):2439-2450. [CrossRef]
- Chu L, McPhee R, Huang W, et al. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine. 2021 May 12;39(20):2791-2799. [CrossRef]
- https://www.cbc.ca/news/health/vaccine-dose-delay-canada-covid-19-research-1.5965996#.
- https://ourworldindata.org.
- Xu S, Huang R, Sy LS, et al. COVID-19 vaccination and non–COVID-19 mortality risk — seven integrated health care organizations, United States, December 14, 2020–July 31, 2021. MMWR Morb Mortal Wkly Rep 2021;70:1520–1524. [CrossRef]
- Laderoute MP. Did the Second Dose of mRNA COVID-19 Vaccines Select for the Alpha and Delta Variants Which Prolonged the Pandemic? June 30, 2022. https://hervk102.substack.com/p/did-the-second-dose-of-mrna-covid.
- https://science.gc.ca/site/science/en/blogs/science-health/surviving-heat-impacts-2021-western-heat-dome-canada.
- Servellita V, Morris MK, Sotomayor-Gonzalez A, et al. Predominance of antibody-resistant SARS-CoV-2 variants in vaccine breakthrough cases from the San Francisco Bay Area, California. Nat Microbiol. 2022 Feb;7(2):277-288. [CrossRef]
- Tartof SY, Slezak JM, Fischer H, Hong V, Ackerson BK, Ranasinghe ON, et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021 Oct 16;398(10309):1407-1416. [CrossRef]
- UK: COVID-19 Weekly Surveillance Reports by Public Health England: COVID-19 Vaccine Surveillance Reports 2021 Weeks 19-38 at https://www.gov.uk/government/publications/covid-19-vaccine-surveillance-report and by UK Health Security Agency: COVID-19 Surveillance Reports for 2021 Week 39 to 2023 Week 23 at https://www.gov.uk/government/publications/covid-19-vaccine-weekly-surveillance-reports.
- https://assets.publishing.service.gov.uk/media/623c6ef0d3bf7f6ab9fe28bf/Vaccine-surveillance-reportweek-12.pdf.
- Shrestha NK, Burke PC, Nowacki AS, Simon JF, Hagen A, Gordon SM. Effectiveness of the coronavirus disease 2019 bivalent vaccine. Open Forum Infect Dis. 2023 Apr 19;10(6):ofad209. [CrossRef]
- Irrgang P, Gerling J, Kocher K, et al. Class switch toward noninflammatory, spike-specific IgG4 antibodies after repeated SARS-CoV-2 mRNA vaccination. Sci Immunol. 2023 Jan 27;8(79):eade2798. [CrossRef]
- https://ourworldindata.org/explorers/coronavirus-data-explorer? The case fatality rate (CFR) is the ratio between confirmed deaths and confirmed cases. Our rolling-average CFR is calculated as the ratio between the 7-day average number of deaths and the 7-day average number of cases 10 days earlier.
- Ziegler CGK, Miao VN, Owings AH, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell. 2021 Sep 2;184(18):4713-4733.e22. [CrossRef]
- Ren X, Wen W, Fan X, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021 Apr 1;184(7):1895-1913.e19. [CrossRef]
- Bost P, Giladi A, Liu Y, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020 Jun 25;181(7):1475-1488.e12. [CrossRef]
- Chua RL, Lukassen S, Trump S, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020 Aug;38(8):970-979. [CrossRef]
- Wendisch D, Dietrich O, Mari T, et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell. 2021 Dec 22;184(26):6243-6261.e27. [CrossRef]
- Grant RA, Morales-Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021 Feb;590(7847):635-641. [CrossRef]
- Delorey TM, Ziegler CGK, Heimberg G, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021 Jul;595(7865):107-113. [CrossRef]
- Laderoute MP, Giulivi A, Larocque L, et al. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS. 2007 Nov 30;21(18):2417-24. [CrossRef]
- Bhardwaj N, Maldarelli F, Mellors J, Coffin JM. HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol. 2014 Oct;88(19):11108-20. [CrossRef]
- Yu SF, Eastman SW, Linial ML. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic. 2006 Aug;7(8):966-77. [CrossRef]
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011 Nov 8;8:90. [CrossRef]
- van der Kuyl, AC. HIV infection and HERV expression: a review. Retrovirology. 2012 Jan 16;9:6. [CrossRef]
- Linial, ML. Foamy viruses are unconventional retroviruses. J Virol. 1999 Mar;73(3):1747-55. [CrossRef]
- Murray SM, Linial ML. Simian foamy virus co-infections. Viruses. 2019 Sep 27;11(10):902. [CrossRef]
- Switzer WM, Salemi M, Shanmugam V, Gao F, Cong M.E, Kuiken C, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. [CrossRef]
- Yu SF, Sullivan MD, Linial ML. Evidence that the human foamy virus genome is DNA. J Virol. 1999 Feb;73(2):1565-72. [CrossRef] [PubMed]
- Meiering CD, Comstock KE, Linial ML. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J Virol. 2000 Feb;74(4):1718-26. [CrossRef]
- Heinkelein M, Hoffmann U, Lücke M, et al. Experimental therapy of allogeneic solid tumors induced in athymic mice with suicide gene-transducing replication-competent foamy virus vectors. Cancer Gene Ther. 2005 Dec;12(12):947-53. [CrossRef]
- Mikovits JA, Hoffman PM, Rethwilm A, Ruscetti FW. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J Virol. 1996 May;70(5):2774-80. [CrossRef]
- Laderoute MP. What you need to know about the HERV-K102 innate immunity protector system of macrophages against RNA pandemic viruses. hervk102.substack.com, January 24, 2022. https://hervk102.substack.com/p/what-you-need-to-know-about-the-herv.
- Plochmann K, Horn A, Gschmack E, et al. Heparan sulfate is an attachment factor for foamy virus entry. J Virol. 2012 Sep;86(18):10028-35. [CrossRef]
- Robinson-McCarthy LR, McCarthy KR, Raaben M, et al. Reconstruction of the cell entry pathway of an extinct virus. PLoS Pathog. 2018 Aug 6;14(8):e1007123. [CrossRef]
- https://commons.wikimedia.org/wiki/File:Insertion_of_sebaceous_glands_into_hair_shaft_x10.jpg.
- https://www.ncbi.nlm.nih.gov/geoprofiles/.
- Nelson AM, Zhao W, Gilliland KL, Zaenglein AL, Liu W, Thiboutot DM. Neutrophil gelatinase-associated lipocalin mediates 13-cis retinoic acid-induced apoptosis of human sebaceous gland cells. J Clin Invest. 2008 Apr;118(4):1468-78. [CrossRef]
- Nelson AM, Zhao W, Gilliland KL, Zaenglein AL, Liu W, Thiboutot DM. Isotretinoin temporally regulates distinct sets of genes in patient skin. J Invest Dermatol. 2009 Apr;129(4):1038-42. [CrossRef]
- Stec M, Weglarczyk K, Baran J, et al. Expansion and differentiation of CD14+CD16(-) and CD14+ +CD16+ human monocyte subsets from cord blood CD34+ hematopoietic progenitors. J Leukoc Biol. 2007 Sep;82(3):594-602. [CrossRef]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006 Nov 15;177(10):7303-11. [CrossRef]
- Maslova A, Ramirez RN, Ma K, et al. Immunological genome project. deep learning of immune cell differentiation. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25655-25666. [CrossRef]
- Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014 Sep 26;345(6204):1251086. [CrossRef]
- Arts RJW, Joosten LAB, Netea MG. The potential role of trained immunity in autoimmune and autoinflammatory disorders. Front Immunol. 2018 Feb 20;9:298. [CrossRef]
- Lv JJ, Wang H, Cui HY, et al. Blockade of macrophage CD147 protects against foam cell formation in atherosclerosis. Front Cell Dev Biol. 2021 Jan 8;8:609090. [CrossRef]
- Zhang MF, Cai XL, Jing KP, et al. Differentiation model establishment and differentiation-related protein screening in primary cultured human sebocytes. Biomed Res Int. 2018 Apr 5;2018:7174561. [CrossRef]
- Bekkering S, Quintin J, Joosten LA, van der Meer JW, Netea MG, Riksen NP. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler Thromb Vasc Biol. 2014 Aug;34(8):1731-8. [CrossRef]
- Bekkering S, Arts RJW, Novakovic B, et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell. 2018 Jan 11;172(1-2):135-146.e9. [CrossRef]
- Wang J, Liu YM, Hu J, Chen C. Trained immunity in monocyte/macrophage: novel mechanism of phytochemicals in the treatment of atherosclerotic cardiovascular disease. Front Pharmacol. 2023 Feb 21;14:1109576. [CrossRef]
- Keating ST, Groh L, Thiem K, et al. Rewiring of glucose metabolism defines trained immunity induced by oxidized low-density lipoprotein. J Mol Med (Berl). 2020 Jun;98(6):819-831. [CrossRef]
- Manghera M, Ferguson-Parry J, Lin R, Douville RN. NF-κB and IRF1 induce endogenous retrovirus K expression via interferon-stimulated response elements in its 5' long terminal repeat. J Virol. 2016 Sep 29;90(20):9338-49. [CrossRef]
- Manghera M, Douville RN. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology. 2013 Feb 9;10:16. [CrossRef]
- Russ E, Mikhalkevich N, Iordanskiy S. Expression of human endogenous retrovirus group K (HERV-K) HML-2 correlates with immune activation of macrophages and type I interferon response. Microbiol Spectr. 2023 Mar 2;11(2):e0443822. [CrossRef]
- Fischer H, Fumicz J, Rossiter H, et al. Holocrine secretion of sebum is a unique DNase2-dependent mode of programmed cell death. J Invest Dermatol. 2017 Mar;137(3):587-594. [CrossRef]
- Törőcsik D, Kovács D, Póliska S, et al.. Genome wide analysis of TLR1/2- and TLR4-activated SZ95 sebocytes reveals a complex immune-competence and identifies serum amyloid A as a marker for activated sebaceous glands. PLoS One. 2018 Jun 21;13(6):e0198323. [CrossRef]
- Magiorkinis G, Blanco-Melo D, Belshaw R. The decline of human endogenous retroviruses: extinction and survival. Retrovirology. 2015 Feb 2;12:8. [CrossRef]
- Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016 Mar 4;351(6277):1083-7. [CrossRef]
- Kim YJ, Han K. Endogenous retrovirus-mediated genomic variations in chimpanzees. Mob Genet Elements. 2015 Feb 3;4(6):1-4. [CrossRef]
- Romano CM, Ramalho RF, Zanotto PM. Tempo and mode of ERV-K evolution in human and chimpanzee genomes. Arch Virol. 2006 Nov;151(11):2215-28. [CrossRef]
- Langergraber KE, Prüfer K, Rowney C,et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15716-21. [CrossRef]
- Katzourakis A, Pereira V, Tristem M. Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol. 2007 Oct;81(19):10712-7. [CrossRef]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004 Sep;2(9):E275. [CrossRef]
- Gifford RJ. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 2012 Feb;28(2):89-100. [CrossRef]
- O'Neil SP, Novembre FJ, Hill AB, et al. Progressive infection in a subset of HIV-1-positive chimpanzees. J Infect Dis. 2000 Oct;182(4):1051-62. [CrossRef]
- Khan N, de Manuel M, Peyregne S, et al. Multiple genomic events altering hominin SIGLEC biology and innate immunity predated the common ancestor of humans and archaic hominins. Genome Biol Evol. 2020 Jul 1;12(7):1040-1050. [CrossRef]
- Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos Trans R Soc Lond B Biol Sci. 2013 Aug 12;368(1626):20120496. [CrossRef]
- Murphy WJ, Frönicke L, O'Brien SJ, Stanyon R. The origin of human chromosome 1 and its homologs in placental mammals. Genome Res. 2003 Aug;13(8):1880-8. [CrossRef]
- Posth C, Wißing C, Kitagawa K, et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat Commun. 2017 Jul 4;8:16046. [CrossRef]
- Petr HC, Wißing C, Kitagawa K, et al. The evolutionary history of Neanderthal and Denisovan Y chromosomes. Science. 2020 Sep 25;369(6511):1653-1656. [CrossRef]
- Hajdinjak M, Mafessoni F, Skov L, et al. Initial upper palaeolithic humans in Europe had recent Neanderthal ancestry. Nature. 2021 Apr;592(7853):253-257. [CrossRef]
- Enard D, Petrov DA. Evidence that RNA viruses drove adaptive introgression between neanderthals and modern humans. Cell. 2018 Oct 4;175(2):360-371.e13. [CrossRef]
- Gouy A, Excoffier L. Polygenic patterns of adaptive introgression in modern humans are mainly shaped by response to pathogens. Mol Biol Evol. 2020 May 1;37(5):1420-1433. [CrossRef]
- Bücking R, Cox MP, Hudjashov G, Saag L, Sudoyo H, Stoneking M. Archaic mitochondrial DNA inserts in modern day nuclear genomes. BMC Genomics. 2019 Dec 26;20(1):1017. [CrossRef]
- Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986 Jun;58(3):937-44. [CrossRef]
- Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A. 1996 May 28;93(11):5177-84. [CrossRef]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004 Oct 5;101 Suppl 2(Suppl 2):14572-9. [CrossRef]
- Simpson GR, Patience C, Löwer R, et al. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology. 1996 Aug 15;222(2):451-6. [CrossRef]
- Grow EJ, Flynn RA, Chavez SL, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015 Jun 11;522(7555):221-5. [CrossRef]
- Morgan D, Brodsky I. Human endogenous retrovirus (HERV-K) particles in megakaryocytes cultured from essential thrombocythemia peripheral blood stem cells. Exp Hematol. 2004 Jun;32(6):520-5. [CrossRef]
- Morozov VA, Dao Thi VL, Denner J. The transmembrane protein of the human endogenous retrovirus--K (HERV-K) modulates cytokine release and gene expression. PLoS One. 2013 Aug 7;8(8):e70399. [CrossRef]
- Padow M, Lai L, Fisher RJ, et al. Analysis of human immunodeficiency virus type 1 containing HERV-K protease. AIDS Res Hum Retroviruses. 2000 Dec 10;16(18):1973-80. [CrossRef]
- Brinzevich D, Young GR, Sebra R, et al. HIV-1 interacts with human endogenous retrovirus K (HML-2) envelopes derived from human primary lymphocytes. J Virol. 2014 Jun;88(11):6213-23. [CrossRef]
- Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A. Human endogenous retrovirus K Gag coassembles with HIV-1 Gag and reduces the release efficiency and infectivity of HIV-1. J Virol. 2012 Oct;86(20):11194-208. [CrossRef]
- Monde K, Terasawa H, Nakano Y, et al. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology. 2017 Apr 26;14(1):27. [CrossRef]
- Minkoff JM, TenOever B. Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. 2023 Jan 11:1–17. [CrossRef]
- Su HC, Jing H, Zhang Y, Casanova JL. Interfering with interferons: a critical mechanism for critical COVID-19 pneumonia. Annu Rev Immunol. 2023 Apr 26;41:561-585. [CrossRef]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008 Nov;1143:1-20. [CrossRef]
- Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020 Sep;20(9):537-551. [CrossRef]
- Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021 Sep;21(9):548-569. [CrossRef]
- Russ E, Iordanskiy S. Endogenous retroviruses as modulators of innate immunity. Pathogens. 2023 Jan 19;12(2):162. [CrossRef]
- Guo Y, Yang C, Liu Y, et al. High expression of HERV-K (HML-2) might stimulate interferon in COVID-19 patients. Viruses 2022, 14, 996. [CrossRef]
- Kenney DJ, O'Connell AK, Turcinovic J, et al. Humanized mice reveal a macrophage-enriched gene signature defining human lung tissue protection during SARS-CoV-2 infection. Cell Rep. 2022 Apr 19;39(3):110714. [CrossRef]
- Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res. 2001 Jun;7(6):1553-60.
- Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003 Mar 13;22(10):1528-35. [CrossRef]
- Wang-Johanning F, Radvanyi L, Rycaj K, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008 Jul 15;68(14):5869-77. [CrossRef]
- Wang-Johanning F, Rycaj K, Plummer JB, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012 Feb 8;104(3):189-210. [CrossRef]
- Jones RB, Garrison KE, Mujib S, et al. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Invest. 2012 Dec;122(12):4473-89. [CrossRef]
- Michaud HA, de Mulder M, SenGupta D, et al. Trans-activation, post-transcriptional maturation, and induction of antibodies to HERV-K (HML-2) envelope transmembrane protein in HIV-1 infection. Retrovirology. 2014 Jan 28;11:10. [CrossRef]
- Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug;596(7873):583-589. [CrossRef]
- Varadi M, Velankar S. The impact of AlphaFold Protein Structure Database on the fields of life sciences. Proteomics. 2022 Nov 16:e2200128. [CrossRef]
- Apostolou E, Rizwan M, Moustardas P, et al. Saliva antibody-fingerprint of reactivated latent viruses after mild/asymptomatic COVID-19 is unique in patients with myalgic-encephalomyelitis/chronic fatigue syndrome. Front Immunol. 2022 Oct 20;13:949787. [CrossRef]
- Risch H. Rise in Aggressive ‘Turbo Cancers” -And especially Among Younger People. September 11, 2023. https://www.theepochtimes.com/epochtv/dr-harvey-risch-rise-in-aggressive-turbo-cancers-and-especially-among-younger-people-atlnow-5489582?utm_source=prtnrhard&utm_campaign=vigilantf&src_src=prtnrhard&src_cmp=vigilantf.
- Arru G, Galleri G, Deiana GA, et al. HERV-K modulates the immune response in ALS patients. Microorganisms. 2021 Aug 23;9(8):1784. [CrossRef]
- Lo Presti E, De Gaetano A, Pioggia G, Gangemi S. Comprehensive analysis of the ILCs and unconventional T cells in virus infection: profiling and dynamics associated with COVID-19 disease for a future monitoring system and therapeutic opportunities. Cells. 2022 Feb 4;11(3):542. [CrossRef]
- Bhat RK, Rudnick W, Antony JM, et al. Human endogenous retrovirus-K(II) envelope induction protects neurons during HIV/AIDS. PLoS One. 2014 Jul 2;9(7):e97984. [CrossRef]
- Garcia-Montojo M, Simula ER, Fathi S, et al. Antibody response to HML-2 may be protective in amyotrophic lateral sclerosis. Ann Neurol. 2022 Nov;92(5):782-792. [CrossRef]
- Simula ER, Arru G, Zarbo IR, Solla P, Sechi LA. TDP-43 and HERV-K envelope-specific immunogenic epitopes are recognized in ALS patients. Viruses. 2021 Nov 18;13(11):2301. [CrossRef]
- Xue B, Sechi LA, Kelvin DJ. Human endogenous retrovirus K (HML-2) in health and disease. Front Microbiol. 2020 Jul 17;11:1690. [CrossRef]
- Tokuyama M, Gunn BM, Venkataraman A, et al. Antibodies against human endogenous retrovirus K102 envelope activate neutrophils in systemic lupus erythematosus. J Exp Med. 2021 Jul 5;218(7):e20191766. [CrossRef]
- Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011 May 19;9(5):355-61. [CrossRef]
- Mitroulis I, Ruppova K, Wang B, et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018 Jan 11;172(1-2):147-161.e12. [CrossRef]
- Cirovic B, de Bree LCJ, Groh L, et al. BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe. 2020 Aug 12;28(2):322-334.e5. [CrossRef]
- Al B, Suen TK, Placek K, Netea MG. Innate (learned) memory, J Allerg Clin Immunol. 2023 Jun 14. [CrossRef]
- Zhang B, Moorlag SJ, Dominguez-Andres J, et al. Single-cell RNA sequencing reveals induction of distinct trained-immunity programs in human monocytes. J Clin Invest. 2022 Apr 1;132(7):e147719. [CrossRef]
- Li W, Moorlag SJCFM, Koeken VACM, et al. A single-cell view on host immune transcriptional response to in vivo BCG-induced trained immunity. Cell Rep. 2023 May 30;42(5):112487. [CrossRef]
- Drummer C 4th, Saaoud F, Shao Y, et al. Trained immunity and reactivity of macrophages and endothelial cells. Arterioscler Thromb Vasc Biol. 2021 Mar;41(3):1032-1046. [CrossRef]
- Cheng SC, Quintin J, Cramer RA, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014 Sep 26;345(6204):1250684. [CrossRef]
- Ieronymaki E, Daskalaki MG, Lyroni K, Tsatsanis C. Insulin signaling and insulin resistance facilitate trained immunity in macrophages through metabolic and epigenetic changes. Front Immunol. 2019 Jun 12;10:1330. [CrossRef]
- Oh J, Weng S, Felton SK, et al. 1,25(OH)2 vitamin D inhibits foam cell formation and suppresses macrophage cholesterol uptake in patients with type 2 diabetes mellitus. Circulation. 2009 Aug 25;120(8):687-98. [CrossRef]
- Coleman PS, Parlo RA. Warburg's ghost-cancer's self-sustaining phenotype: the aberrant carbon flux in cholesterol-enriched tumor mitochondria via deregulated cholesterogenesis. Front Cell Dev Biol. 2021 Mar 12;9:626316. [CrossRef]
- Arts RJW, Moorlag SJCFM, Novakovic B, et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100.e5. [CrossRef]
- Viola A, Munari F, Sánchez-Rodríguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol. 2019 Jul 3;10:1462. [CrossRef]
- Gomez Marti JL, Wells A, Brufsky AM. Dysregulation of the mevalonate pathway during SARS-CoV-2 infection: An in silico study. J Med Virol. 2021 Apr;93(4):2396-2405. [CrossRef]
- Gao L, Chen Q, Zhou X, Fan L. The role of hypoxia-inducible factor 1 in atherosclerosis. J Clin Pathol. 2012 Oct;65(10):872-6. [CrossRef]
- Rius J, Guma M, Schachtrup C, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008 Jun 5;453(7196):807-11. [CrossRef]
- Jiang G, Li T, Qiu Y, Rui Y, Chen W, Lou Y. RNA interference for HIF-1alpha inhibits foam cells formation in vitro. Eur J Pharmacol. 2007 May 21;562(3):183-90. [CrossRef]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014 Feb;13(2):140-56. [CrossRef]
- Appelberg S, Gupta S, Svensson Akusjärvi S, et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect. 2020 Dec;9(1):1748-1760. [CrossRef]
- Orekhov AN, Oishi Y, Nikiforov NG, et al. Modified LDL particles activate inflammatory pathways in monocyte-derived macrophages: transcriptome analysis. Curr Pharm Des. 2018;24(26):3143-3151. [CrossRef]
- Zeboudj L, Giraud A, Guyonnet L, et al. Selective EGFR (epidermal growth factor receptor) deletion in myeloid cells limits atherosclerosis-brief report. Arterioscler Thromb Vasc Biol. 2018 Jan;38(1):114-119. [CrossRef]
- Wang L, Huang Z, Huang W, et al. Inhibition of epidermal growth factor receptor attenuates atherosclerosis via decreasing inflammation and oxidative stress. Sci Rep. 2017 Apr 4;8:45917. [CrossRef]
- Tallam A, Perumal TM, Antony PM, et al. Gene regulatory network inference of immunoresponsive gene 1 (IRG1) identifies interferon regulatory factor 1 (IRF1) as its transcriptional regulator in mammalian macrophages. PLoS One. 2016 Feb 12;11(2):e0149050. [CrossRef]
- Zhang M, Liu K, Zhang Q, et al. Alpha fetoprotein promotes polarization of macrophages towards M2-like phenotype and inhibits macrophages to phagocytize hepatoma cells. Front Immunol. 2023 Feb 23;14:1081572.
- Platanitis E, Decker T. Regulatory networks involving STATs, IRFs, and NFκB in inflammation. Front Immunol. 2018 Nov 13;9:2542. [CrossRef]
- Tan L, Lu J, Liu L, Li L. Fatty acid binding protein 3 deficiency limits atherosclerosis development via macrophage foam cell formation inhibition. Exp Cell Res. 2021 Oct 1;407(1):112768. [CrossRef]
- Wang K, Chen W, Zhang Z, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020 Dec 4;5(1):283. [CrossRef]
- Geng J, Chen L, Yuan Y, et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct Target Ther. 2021 Sep 25;6(1):347. [CrossRef]
- Helal MA, Shouman S, Abdelwaly A, et al. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J Biomol Struct Dyn. 2022 Feb;40(3):1109-1119. [CrossRef]
- Laderoute, M.P. A new paradigm about HERV-K102 particle production and blocked release to explain cortisol mediated immunosenescence and age-associated risk of chronic disease. Discov Med. 2015 Dec;20(112):379-91.
- Laderoute, M. The paradigm of immunosenescence in atherosclerosis-cardiovascular disease (ASCVD). Discov Med. 2020 Jan-Feb;29(156):41-51.
- Laderoute MP. The Characterization of a Novel, Widespread, PNA-Reactive Tumor Associated Antigen; The Alpha-fetoprotein Receptor/ Binding Protein. Ph.D. Thesis, University of Alberta, Edmonton, Alberta, Canada, January 7, 1991. https://era.library.ualberta.ca/items/6f548eb6-49a2-456c-b472-41f68976077f.
- Laderoute MP, Pilarski LM. The inhibition of apoptosis by alpha-fetoprotein (AFP) and the role of AFP receptors in anti-cellular senescence. Anticancer Res. 1994 Nov-Dec;14(6B):2429-38.
- Nakabayashi H, Koyama Y, Sakai M, Li HM, Wong NC, Nishi S. Glucocorticoid stimulates primate but inhibits rodent alpha-fetoprotein gene promoter. Biochem Biophys Res Commun. 2001 Sep 14;287(1):160-72. [CrossRef]
- Phillips AC, Carroll D, Gale CR, Lord JM, Arlt W, Batty GD. Cortisol, DHEA sulphate, their ratio, and all-cause and cause-specific mortality in the Vietnam Experience Study. Eur J Endocrin. 2010 Aug; 163(2):285-292. doi.org/10.1530/EJE-10-0299.
- Laderoute M. Ivermectin may prevent and reverse immunosenescence by antagonizing alpha-fetoprotein and downmodulating PI3K/Akt/mTOR hyperactivity. Open Heart, April 29, 2021. https://openheart.bmj.com/content/8/1/e001655.responses#ivermectin-may-prevent-and-reverse-immunosenescence-by-antagonizing-alpha-fetoprotein-and-downmodulating-pi3k-akt-mtor-hyperactivity.
- Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987 Jun;61(6):2059-62. [CrossRef]
- Kory P, Meduri GU, Iglesias J, Varon J, Cadegiani FA, Marik PE. "MATH+" Multi-Modal Hospital Treatment Protocol for COVID-19 Infection: Clinical and Scientific Rationale. J Clin Med Res. 2022 Feb;14(2):53-79. [CrossRef]
- Draghici S, Nguyen TM, Sonna LA, et al. COVID-19: disease pathways and gene expression changes predict methylprednisolone can improve outcome in severe cases. Bioinformatics. 2021 Sep 9;37(17):2691-2698. [CrossRef]
- Leech M, Metz C, Hall P, et al. Macrophage migration inhibitory factor in rheumatoid arthritis: evidence of proinflammatory function and regulation by glucocorticoids. Arthritis Rheum. 1999 Aug;42(8):1601-8. [CrossRef]
- Dias SSG, Soares VC, Ferreira AC, et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020 Dec 16;16(12):e1009127. [CrossRef]
- Singh N, Bharara Singh A. S2 subunit of SARS-CoV-2 interacts with tumor suppressor protein p53 and BRCA: an in silico study. Transl Oncol. 2020 Oct;13(10):100814. [CrossRef]
- Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999 Feb;19(2):1279-88. [CrossRef]
- Desterke C, Turhan AG, Bennaceur-Griscelli A, Griscelli F. PPARγ cistrome repression during activation of lung monocyte-macrophages in severe COVID-19. iScience. 2020 Oct 23;23(10):101611. [CrossRef]
- Verway M, Bouttier M, Wang TT, et al. Vitamin D induces interleukin-1β expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013;9(6):e1003407. [CrossRef]
- Borsche L, Glauner B, von Mendel J. COVID-19 mortality risk correlates inversely with vitamin D3 status, and a mortality rate close to zero could theoretically be achieved at 50 ng/ml 25(OH)D3: results of a systematic review and meta-analysis. Nutrients. 2021 Oct 14;13(10):3596. [CrossRef]
- Grant WB, Al Anouti F, Boucher BJ, et al. A narrative review of the evidence for variations in serum 25-hydroxyvitamin D concentration thresholds for optimal health. Nutrients. 2022 Feb 2;14(3):639. [CrossRef]
- Office for National Statistics (UK). Deaths involving COVID-19 by vaccination status, England: Deaths occurring between 1 January 2021 and 31 May 2022. Age-standardised mortality rates and raw death numbers for deaths involving COVID-19 by vaccination status, broken down by age and /or sex group. https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/bulletins/deathsinvolvingcovid19byvaccinationstatusengland/deathsoccurringbetween1january2021and31may2022.
- Fenton N, Neil M, Craig C, McLachlan S. What the ONS mortality COVID-19 surveillance data can tell us about vaccine safety and efficacy, November 10, 2022 preprint. https://d7694293-ffb8-4ed0-a014-.3581d49070e4.usrfiles.com/ugd/d76942_bd64e0ebb4754477afd3d9f00bb3dc0f.pdf.
- Fenton N, Campbell J. Is ONS data on mortality by vaccine status fit for purpose? https://www.youtube.com/watch?v=W-N-17_j_44, January 22, 2023.
- Graña C, Ghosn L, Evrenoglou T, et al. Efficacy and safety of COVID-19 vaccines. Cochrane Database Syst Rev. 2022 Dec 7;12(12):CD015477. [CrossRef]
- https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/968513/Weekly_Flu_and_COVID-19_report_w10.pdf. Note: page 73/81; Table 10. Cumulative data up to week 9, 2021 for week ending March 7.
- Brogna C, Cristoni S, Marino G, et al. Detection of recombinant spike protein in the blood of individuals vaccinated against SARS-CoV-2: possible molecular mechanisms. Proteomics Clin Appl. 2023 Nov;17(6):e2300048. [CrossRef]
- Sahanic S, Hilbe R, Dünser C, et al. SARS-CoV-2 activates the TLR4/MyD88 pathway in human macrophages: a possible correlation with strong pro-inflammatory responses in severe COVID-19. Heliyon. 2023 Nov 17;9(11):e21893. [CrossRef]
- McCullough, Peter: at U.S. Sen. Ron Johnson Roundtable Discussion. COVID-19 Vaccines: What they are, how they work, and possible causes of injuries. December 7, 2022. https://twitter.com/SenRonJohnson/status/160050489272825037. https://rumble.com/v1ze4d0-covid-19-vaccines-what-they-are-how-they-work-and-possible-causes-of-injuri.html.
- Seneff S, Nigh G, Kyriakopoulos AM, McCullough PA. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and microRNAs. Food Chem Toxicol. 2022 Apr 15;164:113008. [CrossRef]
- Hulscher N, Hodkinson R, Makis W, McCullough P. Autopsy proven fatal COVID-19 vaccine-induced myocarditis. Preprints 2023, 2023071198. [CrossRef]
- Schwab C, Domke LM, Hartmann L, Stenzinger A, Longerich T, Schirmacher P. Autopsy-based histopathological characterization of myocarditis after anti-SARS-CoV-2-vaccination. Clin Res Cardiol. 2023 Mar;112(3):431-440. [CrossRef]
- Kleen TO, Galdon AA, MacDonald AS, Dalgleish AG. Mitigating coronavirus induced dysfunctional immunity for at-risk populations in COVID-19: trained immunity, BCG and "new old friends". Front Immunol. 2020 Sep 4;11:2059. [CrossRef]
- Desingu PA, Nagarajan K. Unveiling HERV-K113-ENV as SARS-CoV-2 severity admissible biomarker by mining transcriptome data. J Med Virol. 2023 Jan;95(1):e28149. [CrossRef]
- Tovo PA, Garazzino S, Daprà V, et al. COVID-19 in children: expressions of type I/II/III interferons, TRIM28, SETDB1, and endogenous retroviruses in mild and severe cases. Int J Mol Sci. 2021 Jul 13;22(14):7481. [CrossRef]
- Temerozo JR, Fintelman-Rodrigues N, Dos Santos MC, et al. Human endogenous retrovirus K in the respiratory tract is associated with COVID-19 physiopathology. Microbiome. 2022 Apr 22;10(1):65. [CrossRef]
- Utrero-Rico A, González-Cuadrado C, Chivite-Lacaba M, et al. Alterations in circulating monocytes predict COVID-19 severity and include chromatin modifications still detectable six months after recovery. Biomedicines. 2021 Sep 17;9(9):1253. [CrossRef]
- Lieberman NAP, Peddu V, Xie H, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020 Sep 8;18(9):e3000849. [CrossRef]
- Pereira PC, de Lima CJ, Fernandes AB, Zângaro RA, Villaverde AB. Cardiopulmonary and hematological effects of infrared LED photobiomodulation in the treatment of SARS-COV2. J Photochem Photobiol B. 2023 Jan;238:112619. [CrossRef]
- McCullough PA, Alexander PE, Armstrong R, et al. Multifaceted highly targeted sequential multidrug treatment of early ambulatory high-risk SARS-CoV-2 infection (COVID-19). Rev Cardiovasc Med. 2020 Dec 30;21(4):517-530. [CrossRef]
- Wimalawansa SJ. Infections and autoimmunity-the immune system and vitamin D: a systematic review. Nutrients. 2023 Sep 2;15(17):3842. [CrossRef]
- Zhan SH, Deverman BE, Chan YA. SARS-CoV-2 is well adapted for humans. What does this mean for reemergence? BioRxiv May 2 2020 (preprint). https://www.biorxiv.org/content/10.1101/2020.05.01.073262v1.
- Wahl A, De C, Abad Fernandez M, et al. Precision mouse models with expanded tropism for human pathogens. Nat Biotechnol. 2019 Oct;37(10):1163-1173. [CrossRef]
- Maxmen A, Mallaaty S. The COVID lab-leak hypothesis: what scientists do and don’t know. Nature, June 8, 2021. https://www.nature.com/articles/d41586-021-01529-3.
- Kaiser, J. House approves ban on gain-of-function pathogen research. 15 Nov 2023. https://www.science.org/content/article/house-approves-ban-gain-function-pathogen-research.
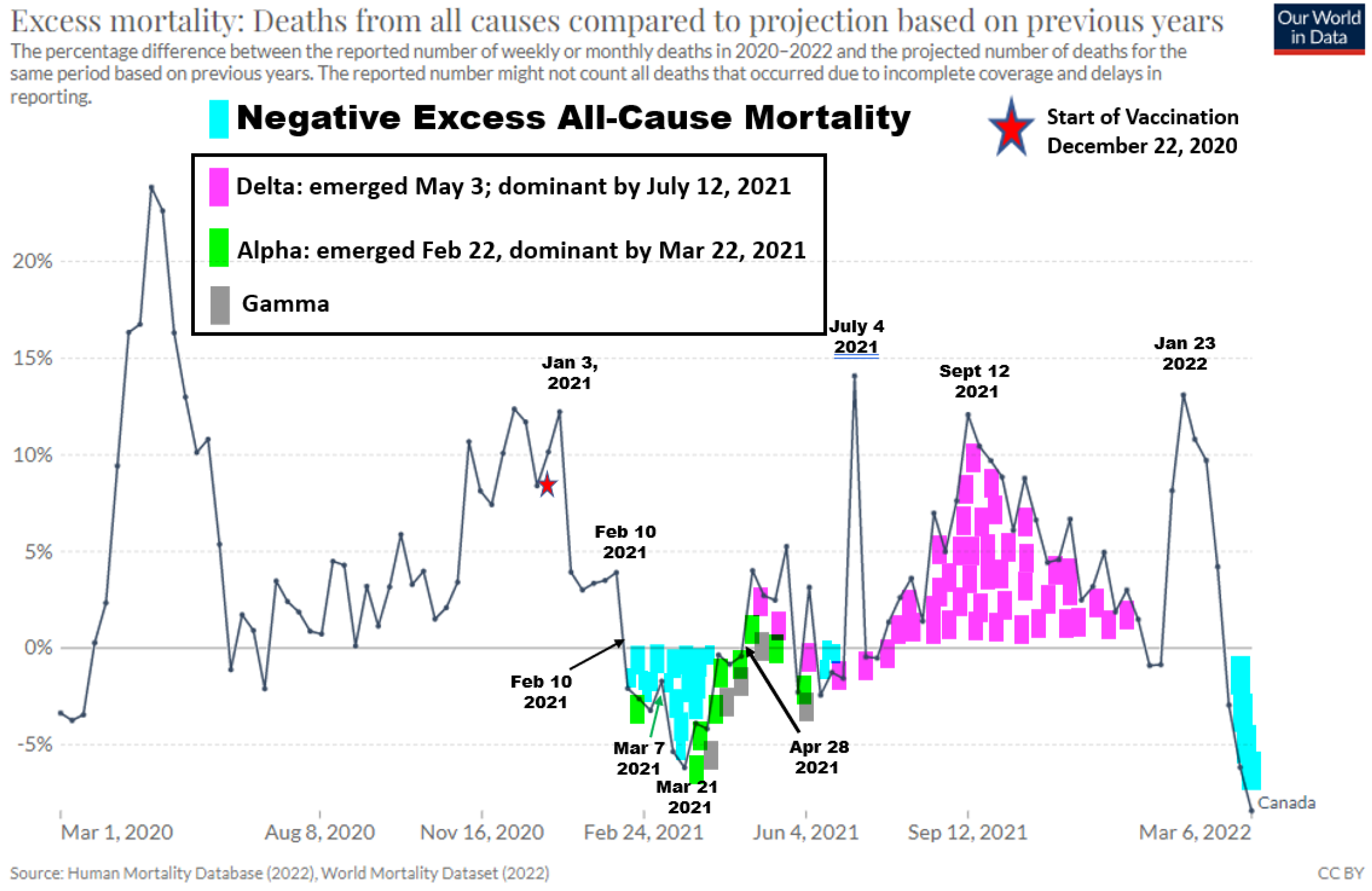

| DATE | % At Least One Dose | % Two Doses | % One Dose | Two / One RATIO | EVENTS | NOTES |
| 22-Dec | 0.071 | None | 0.071 | N/A | ||
| 29-Dec | 0.19 | None | 0.19 | N/A | ||
| 03-Jan | 0.3 | None | 0.30 | N/A | EACM Decreases | |
| 10-Jan | 0.84 | 0.1 | 0.74 | 0.135 | ||
| 17-Jan | 1.5 | 0.6 | 0.9 | 0.667 | EACM temporarily flattened | |
| 24-Jan | 2.03 | 0.15 | 1.88 | 0.080 | ||
| 31-Jan | 2.3 | 0.3 | 2 | 0.150 | ||
| 07-Feb | 2.4 | 0.47 | 1.93 | 0.244 | ||
| 08-Feb | 2.4 | 0.5 | 1.9 | 0.263 | ||
| 10-Feb | 2.5 | 0.6 | 1.9 | 0.316 | Enters Neg EACM | |
| 14-Feb | 2.59 | 0.81 | 1.78 | 0.455 | ||
| 21-Feb | 2.9 | 1.1 | 1.8 | 0.611 | ||
| 22-Feb | 3 | 1.2 | 1.8 | 0.667 | Alpha Emerges | |
| 25-Feb | 3.2 | 1.3 | 1.9 | 0.684 | ||
| 26-Feb | 3.4 | 1.4 | 2 | 0.700 | ||
| 27-Feb | 3.5 | 1.4 | 2.1 | 0.667 | NACI Intervention around Feb 27 | |
| 28-Feb | 3.63 | 1.41 | 2.22 | 0.635 | ||
| 07-Mar | 4.85 | 1.52 | 3.33 | 0.456 | Uptick in Neg EACM | |
| 08-Mar | 5.1 | 1.6 | 3.5 | 0.457 | ||
| 11-Mar | 5.67 | 1.59 | 4.08 | 0.390 | ||
| 14-Mar | 6.5 | 1.6 | 4.9 | 0.327 | ||
| 15-Mar | 6.8 | 1.6 | 5.2 | 0.308 | ||
| 21-Mar | 8.83 | 1.7 | 7.13 | 0.238 | Lowest Neg EACM(Nadir) | |
| 22-Mar | 9.2 | 1.7 | 7.5 | 0.227 | Alpha Dominates | |
| 28-Mar | 11.81 | 1.81 | 10 | 0.181 | Uptick in Neg EACM | |
| 04-Apr | 15.07 | 1.92 | 13.15 | 0.146 | ||
| 11-Apr | 19.04 | 2.19 | 16.85 | 0.130 | Uptick in Neg EACM | |
| 18-Apr | 24 | 2.5 | 21.5 | 0.116 | ||
| 19-Apr | 25 | 2.5 | 22.5 | 0.111 | ||
| 25-Apr | 29.18 | 2.75 | 26.43 | 0.104 | ||
| 26-Apr | 30 | 2.8 | 27.2 | 0.103 | ||
| 28-Apr | 31 | 2.9 | 28.1 | 0.103 | Exit Neg EACM | |
| 02-May | 33.58 | 3.05 | 30.53 | 0.100 | Delta Emerges | |
| 03-May | 34 | 3.1 | 30.9 | 0.100 | ||
| 09-May | 39 | 3.4 | 35.6 | 0.096 | ||
| 16-May | 45 | 3.8 | 41.2 | 0.092 | ||
| 17-May | 46 | 3.9 | 42.1 | 0.093 | Max Alpha at 59% | |
| 23-May | 51 | 4.05 | 46.95 | 0.086 | ||
| 30-May | 56.69 | 5.45 | 51.24 | 0.106 | ||
| 02-Jun | 58.8 | 6.11 | 52.69 | 0.116 | ||
| 11-Jun | 63.87 | 10.82 | 53.05 | 0.204 | ||
| 14-Jun | 64.86 | 13.11 | 51.75 | 0.253 | ||
| 20-Jun | 66.29 | 18.85 | 47.44 | 0.397 | ||
| 26-Jun | 67.38 | 26.47 | 40.91 | 0.647 | ||
| 04-Jul | 68.31 | 35.02 | 33.29 | 1.052 | 50% of the Eligible Receive 2nd Dose | |
| 12-Jul | 69.27 | 44.33 | 24.94 | 1.777 | Delta Dominant | |
| 19-Jul | 69.99 | 50.61 | 19.38 | 2.611 | 50% Fully Vaccinated | |
| 25-Jul | 71 | 55 | 16 | 3.438 | ||
| 01-Aug | 71 | 59 | 12 | 4.917 |
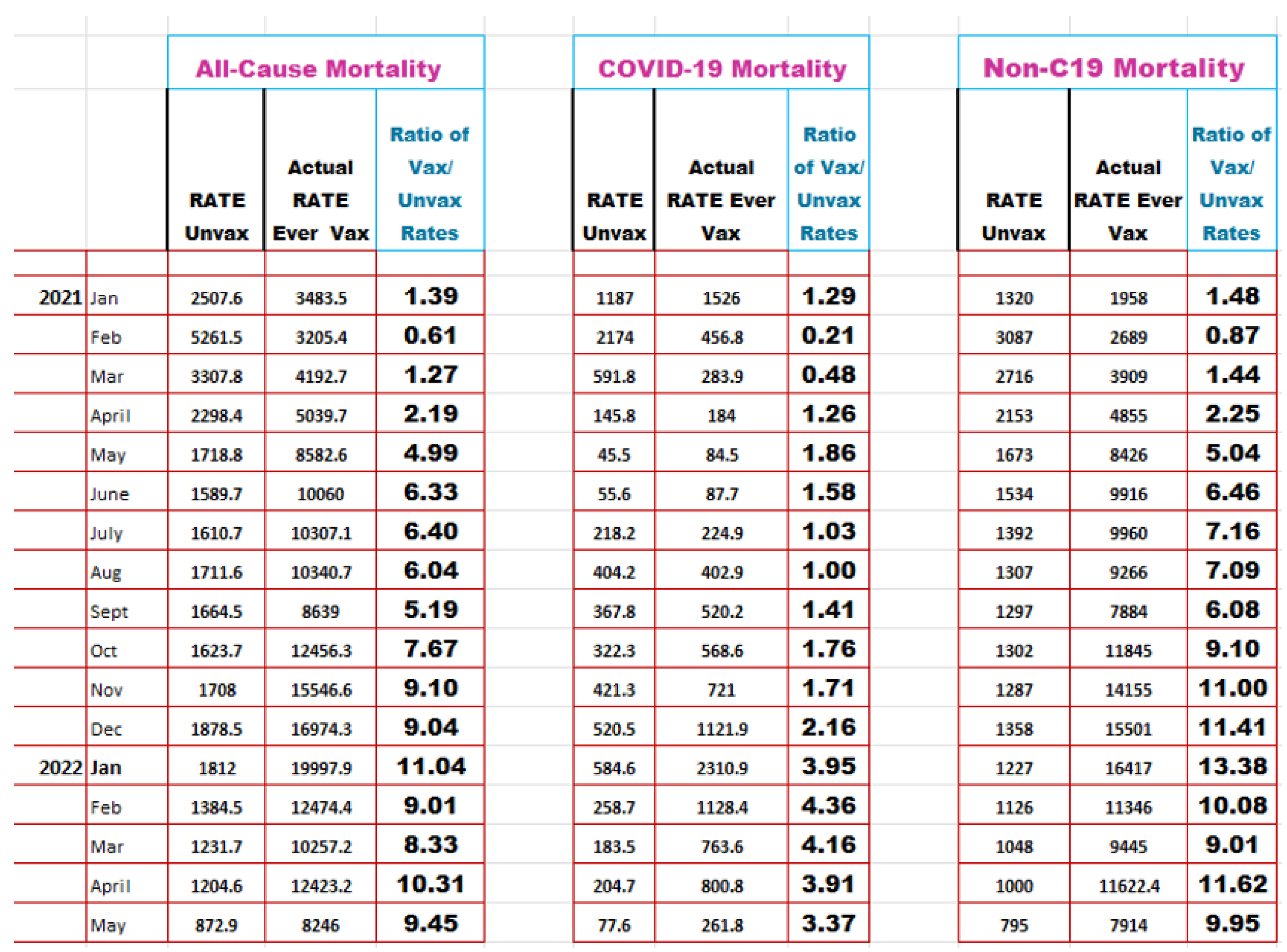 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




