Submitted:
04 December 2023
Posted:
05 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Method
2.1. Definition of the Atom and Special Groups
2.2. Calculation of the Atom- and Special-Group Contributions
2.3. Calculation of Descriptors logPow and logKoa
2.4. Cross-Validation Calculations
3. Sources
3.1. Sources of logPow Values
3.2. Sources of logKoa Values
3.3. Sources of logKaw Values
4. Results
4.1. Partition Coefficient logPow
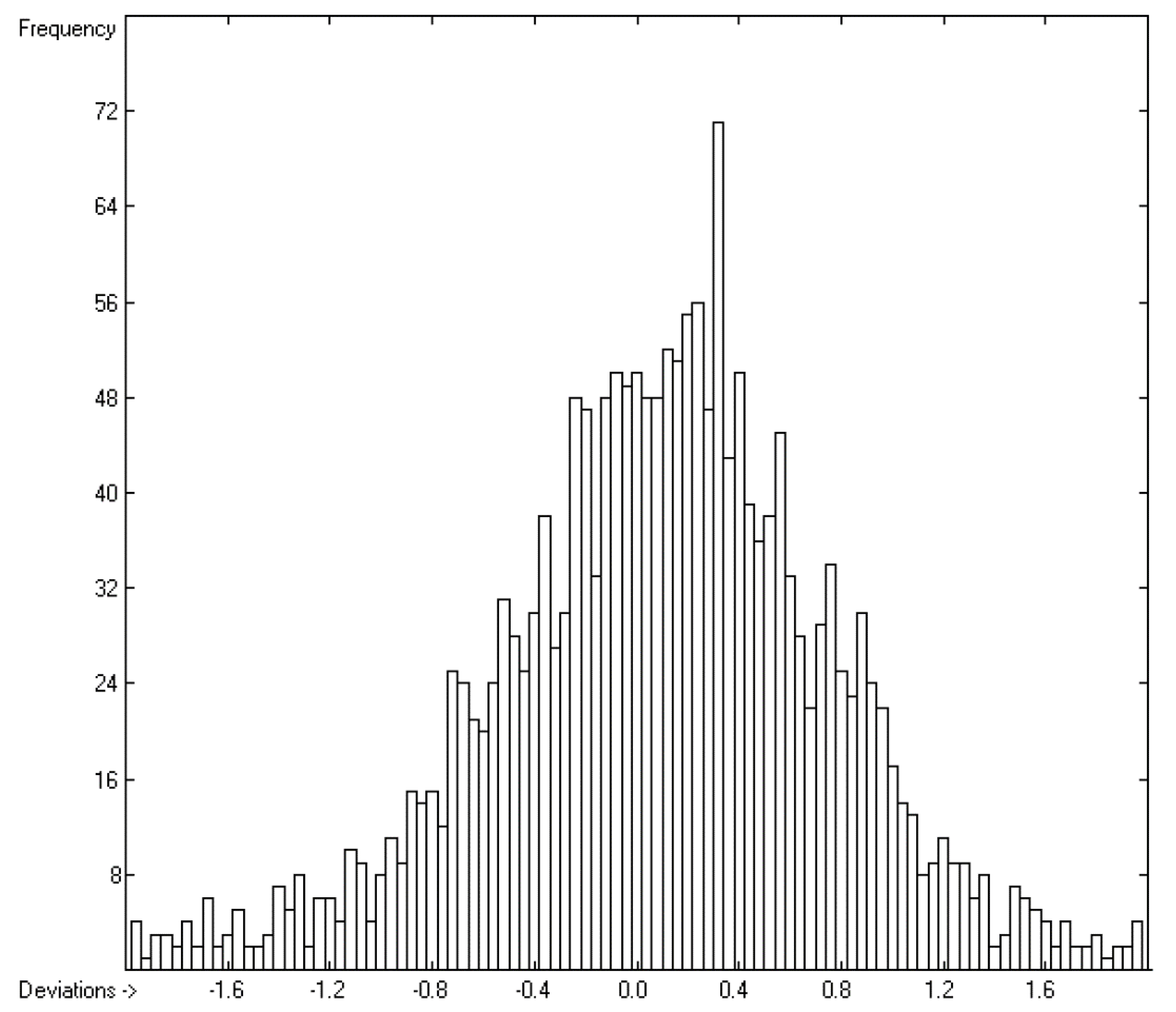
4.2. Partition Coefficient logKoa
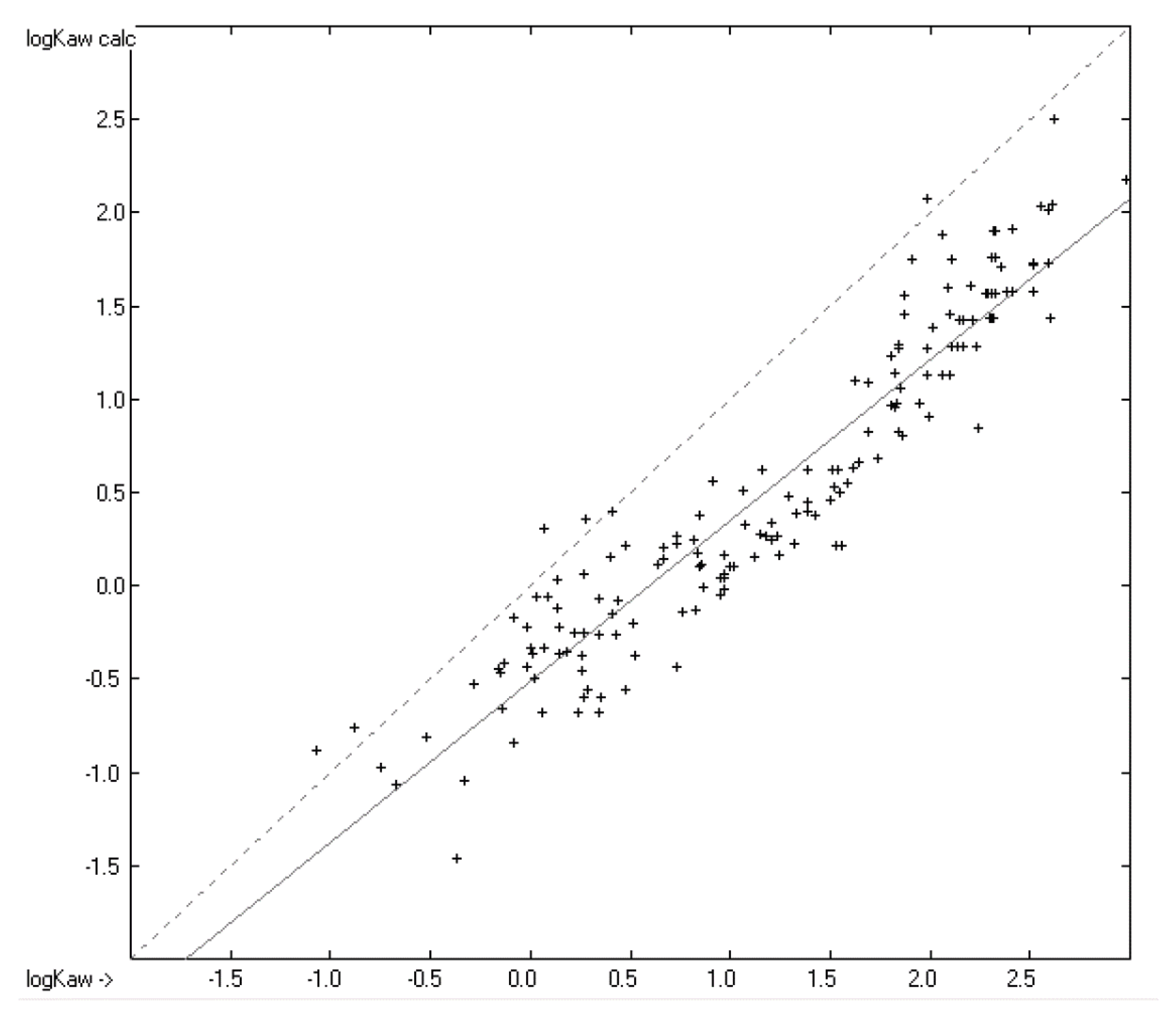
4.3. Partition Coefficient logKaw
4.4. Interpretation of the Special-Groups Contributions for logPow and logKoa, and ultimately for logKaw
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Simonich, S. L.; Hites, R. A. Organic Pollutant Accumulation in Vegetation. Environ. Sci. Technol. 1995, 29, 2905−2914. [CrossRef]
- McLachlan, M. S. Bioaccumulation of Hydrophobic Chemicals in Agricultural Food Chains. Environ. Sci. Technol. 1996, 30, 252−259. [CrossRef]
- Doucette, W. J.; Shunthirsasingham, C.; Dettenmaier, E. M.; Zaleski, R. T.; Fantke, P.; Arnot, J. A. A Review of Measured Bioaccumulation Data on Terrestrial Plants for Organic Chemicals:Metrics, Variability, and the Need for Standardized Measurement Protocols. Environ. Toxicol. Chem. 2018, 37, 21−33. [CrossRef]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional structure-directed quantitative structure-activity relationships I. Partition coefficients as a measure of hydrophobicity. J. Computer. Chem. 1986, 7, 565–577. [CrossRef]
- Ghose, A.K.; Pritchett, A.; Crippen, G.M. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships III: Modeling hydrophobic interactions. J. Comput. Chem. 1988, 9, 80–90. [CrossRef]
- Klopman, G.; Li, J.-Y.; Wang, S.; Dimayuga, M. Computer automated log P calculations based on an extended group contribution approach. J. Chem. Inf. Comput. Sci. 1994, 34, 752–781. [CrossRef]
- Visvanadhan, V. N.; Ghose, A.K.; Revankar, G.R.; Robins, R.K. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inf. Comput. Sci. 1989, 29, 163–172. [CrossRef]
- Leo, A.J. Calculating log Poct from structures. Chem. Rev. 1993, 93, 1281–1306. [CrossRef]
- Wang, R.; Fu, Y.; Lai, L. A new atom-additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 1997, 37, 615–621. [CrossRef]
- Hou, T.J.; Xu, X.J. ADME evaluation in drug discovery. 2. Prediction of partition coefficient by atom-additive approach based on atom-weighted solvent accessible surface areas. J. Chem. Inf. Comput. Sci. 2003, 43, 1058–1067. [CrossRef]
- Naef, R. A Generally Applicable Computer Algorithm Based on the Group Additivity Method for the Calculation of Seven Molecular Descriptors: Heat of Combustion, LogPO/W, LogS, Refractivity, Polarizability, Toxicity and LogBB of Organic Compounds; Scope and Limits of Applicability. Molecules 2015, 20, 18279-18351. [CrossRef]
- Chen, J.; Harner, T.; Schramm, K.W.; Quan, X.; Xue, X.; Wu, W.; Kettrup, A. Quantitative relationships between molecular structures, environmental temperatures and octanol/air partition coefficients of PCDD/Fs. Sci. Total Environ. 2002, 300, 155-166. [CrossRef]
- Chen, J.; Harner, T.; Yang, P.; Quan, X.; Chen, S.; Schramm, K.W.; Kettrup, A. Quantitative predictive models for octanol/air partition coefficients of polybrominated diphenyl ethers at different temperatures. Chemosphere 2003, 51, 577-584. [CrossRef]
- Chen, J.; Harner, T.; Schramm, K.W.; Quan, X.; Xue, X.; Kettrup, A. Quantitative relationships between molecular structures, environmental temperatures and octanol/air partition coefficients of polychlorinated biphenyls. Comput. Biol. Chem. 2003, 27, 405-421. [CrossRef]
- Hongxia, Z.; Jingwen, C.; Xie, Q.; Baocheng, Q.; Xinmiao, L. Octanol/air partition coefficients of polybrominated biphenyls. Chemosphere 2009, 74, 1490-1494. [CrossRef]
- Staikova, M.; Wania, F.; Donaldson, D. Molecular polarizability as a single parameter predictor of vapour pressures and octanoleair partitioning coefficients of non-polar compounds: a priori approach and results. Atmos. Environ. 2004, 38, 213-225. [CrossRef]
- Zhao, H.; Zhang, Q.; Chen, J.; Xue, X.; Liang, X. Prediction of octanol/air partition coefficients of semivolatile organic compounds based on molecular connectivity index. Chemosphere 2005, 59, 1421-1426. [CrossRef]
- Zeng, X.L.; Zhang, X.L.; Wang, Y. Qspr modeling of n-octanol/air partition coefficients and liquid vapor pressures of polychlorinated dibenzo-p-dioxins. Chemosphere 2013, 91, 229-232. [CrossRef]
- Liu, H.; Shi, J.; Liu, H.; Wang, Z. Improved 3D-QSPR analysis of the predictive octanol/air partition coefficients of hydroxylated and methoxylated polybrominated diphenyl ethers. Atmos. Environ. 2013, 77, 840-845. [CrossRef]
- Jiao, L.; Gao, M.; Wang, X.; Li, H. QSPR study on the octanol/air partition coefficient of polybrominated diphenyl ethers by using molecular distance-edge vector index. Chem. Cent. J. 2014, 8. [CrossRef]
- Chen, Y.; Cai, X.; Jiang, L.; Li, Y. Prediction of octanol-air partition coefficients for polychlorinated biphenyls (PCBs) using 3D-SQAR models. Ecotoxicol. Environ. Saf. 2016, 124, 202-212. [CrossRef]
- Fu, Z.; Chen, J.; Li, X.; Wang, Y.; Yu, H. Comparison of prediction methods for octanol-air partition coefficients of diverse organic compounds. Chemosphere 2016, 148, 118-125. [CrossRef]
- Jin, X.; Fu, Z.; Li, X.; Chen, J. Development of polyparameter linear free energy relationship models for octanol/air partition coefficients of diverse chemicals. Environ. Sci.: Process. Impact. 2017, 19, 300-306. [CrossRef]
- Li, X.; Chen, J.; Zhang, L.; Qiao, X.; Huang, L. The fragment constant method for predicting octanol/air partition coefficients of persistent organic pollutants at different temperatures. J. Phys. Chem. Ref. Data 2006, 35, 1365-1384. [CrossRef]
- Ebert, R.-U.; Kühne, R.; Schüürmann, G. Henry’s Law Constant ─ A General-Purpose Fragment Model to Predict Log Kaw from Molecular Structure. Environ. Sci. Technol. 2023, 57, 1, 160–167. [CrossRef]
- Hardtwig, E. Fehler- Und Ausgleichsrechnung; Bibliographisches Institut AG: Mannheim, Germany, 1968.
- Sangster, J. Octanol-water partition coefficients of simple organic compounds. J. Phys. Chem. Ref. Data 1989, 18, 1111–1229. [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [CrossRef]
- Tewari, Y. B.; Miller, M. M.; Wasik, St. P.; Martire, D. E. Aqueous Solubility and Octanol/Water Partition Coefficient of Organic Compounds at 25.0 °C. J. Chem. Eng. Data 1982, 27, 451-454. [CrossRef]
- Abraham, M. H.; Chadha, H. S.; Whiting, G. S.; Mitchell, R. C. Hydrogen Bonding. 32. An Analysis of Water-Octanol and Water-Alkane Partitioning and the delta log P Parameter of Seiler. J. Pharm. Sci. 1994, 83(8) 1085 -1100. [CrossRef]
- Czerwinski, St. E.; Skvorak, J. P.; Maxwell, D. M.; Lenz, D. E.; Baskin, St. I. Organophosphorus Compounds on Biodistribution and Percutaneous Toxicity. J. Biochem. Mol. Tox. 2006, 20(5) 241-246. [CrossRef]
- Boddu, V. M.; Abburi, K.; Maloney, St. W.; Damavarapu, R. Thermophysical Properties of an Insensitive Munitions Compound, 2,4-Dinitroanisole. J. Chem. Eng. Data 2008, 53, 1120–1125. [CrossRef]
- Li, X.-J.; Shan, G.; Liu, H.; Wang, Z.-Y. Determination of lgKow and QSPR Study on Some Fluorobenzene Derivatives. Chin. J. Struct. Chem. 2009, 28(10) 1236-1241.
- Saranjampour, P.; Vebrosky, E. N.; Armbrust, K. L. Salinity Impacts on Water Solubility and n-Octanol/Water Partition Coefficients of Selected Pesticides and Oil Constituents. Environ Toxicol Chem 2017, 36, 2274–2280. [CrossRef]
- Ebert, R.-U.; Kühne, R.; Schüürmann, G. Octanol/Air Partition Coefficient. A General-Purpose Fragment Model to Predict Log Koa from Molecular Structure. Environ. Sci. Technol. 2023, 57, 976−984. [CrossRef]
- Puzyn, T.; Falandysz, J.; Rostkowski, P.; Piliszek, S.; Wilczynska, A. Computational estimation of logarithm of octanol/air partition coefficients and subcooled vapour pressures for each of 75 chloronaphtalene congeners. Phys.-Chem. Prop., Distr. Model. Organohal. Compds. 2004, 66, 2354- 2360. [CrossRef]
- Odabasi, M.; Cetin, E.; Sofuoglu, A. Determination of octanol–air partition coefficients and supercooled liquid vapor pressures of PAHs as a function of temperature: Application to gas–particle partitioning in an urban atmosphere. Atm. Environ. 2006, 40, 6615-6625. [CrossRef]
- Xu, Sh.; Kropscott, B.; Method for Simultaneous Determination of Partition Coefficients for Cyclic Volatile Methylsiloxanes and Dimethylsilanediol. Anal. Chem. 2012, 84, 1948−1955. [CrossRef]
- Easterbrook, K. D.; Vona, M. A,; Osthoff, H. D. Measurement of Henry’s law constants of ethyl nitrate in deionized water, synthetic sea salt solutions, and n-octanol. Chemosphere, 2024, 346, 140482. [CrossRef]
- Sander, R. Compilation of Henry’s law constants (version 5.0.0) for water as solvent. Atmos. Chem. Phys., 2023, 23, 10901–12440. [CrossRef]
- Allen, G.; Dwek, R.A. An n.m.r. study of keto-enol tautomerism in β-diketones. J. Chem. Soc. B 1966, 161–163. [CrossRef]
- COSMOlogic GmbH Co. KG. A Dassault Systèmes company, version 19.0.4, COSMOthermX, 2019. www.cosmologic.de.
- US EPA. Estimation Programs Interface Suite for Microsoft Windows, v. 4.11, module KOAWIN v. 1.11, United States Environmental Protection Agency: Washington, DC, USA, 2015.
- Naef, R.; Acree, W.E., Jr. Calculation of Five Thermodynamic Molecular Descriptors by Means of a General Computer Algorithm Based on the Group-Additivity Method: Standard Enthalpies of Vaporization, Sublimation and Solvation, and Entropy of Fusion of Ordinary Organic Molecules and Total Phase-Change Entropy of Liquid Crystals. Molecules 2017, 22, 1059. [CrossRef]
- Naef, R.; Acree, W.E. Application of a General Computer Algorithm Based on the Group-Additivity Method for the Calculation of Two Molecular Descriptors at Both Ends of Dilution: Liquid Viscosity and Activity Coefficient inWater at Infinite Dilution. Molecules 2018, 23, 5. [CrossRef]
- Naef, R.; Acree, W.E., Jr. Calculation of the Surface Tension of Ordinary Organic and Ionic Liquids by Means of a Generally Applicable Computer Algorithm Based on the Group-Additivity Method. Molecules 2018, 23, 1224. [CrossRef]
- Naef, R. Calculation of the Isobaric Heat Capacities of the Liquid and Solid Phase of Organic Compounds at 298.15K by Means of the Group-Additivity Method. Molecules 2020, 25, 1147. [CrossRef]
- Naef, R.; Acree, W.E., Jr. Calculation of the Vapour Pressure of Organic Molecules by Means of a Group-Additivity Method and Their Resultant Gibbs Free Energy and Entropy of Vaporization at 298.15 K. Molecules 2021, 26, 1045. [CrossRef]
- Naef, R.; Acree,W.E., Jr. Revision and Extension of a Generally Applicable Group-Additivity Method for the Calculation of the Standard Heat of Combustion and Formation of Organic Molecules. Molecules 2021, 26, 6101. [CrossRef]
- Naef, R.; Acree, W.E., Jr. Revision and Extension of a Generally Applicable Group Additivity Method for the Calculation of the Refractivity and Polarizability of Organic Molecules at 298.15 K. Liquids, 2022, 2, 327–377. [CrossRef]
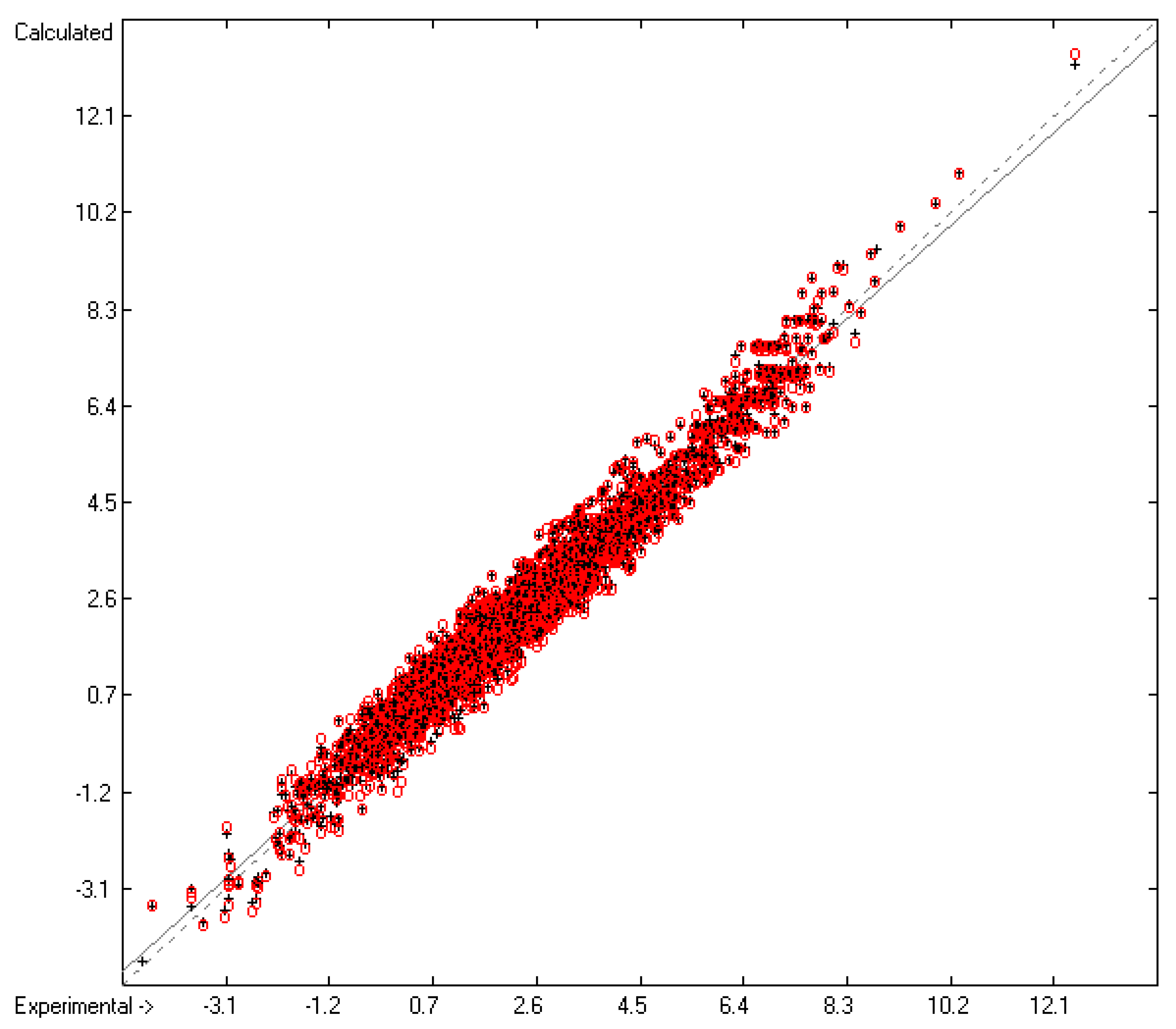
 =0.39; S =0.42; experimental values range from -4.6 to +12.53).
=0.39; S =0.42; experimental values range from -4.6 to +12.53).
 =0.39; S =0.42; experimental values range from -4.6 to +12.53).
=0.39; S =0.42; experimental values range from -4.6 to +12.53).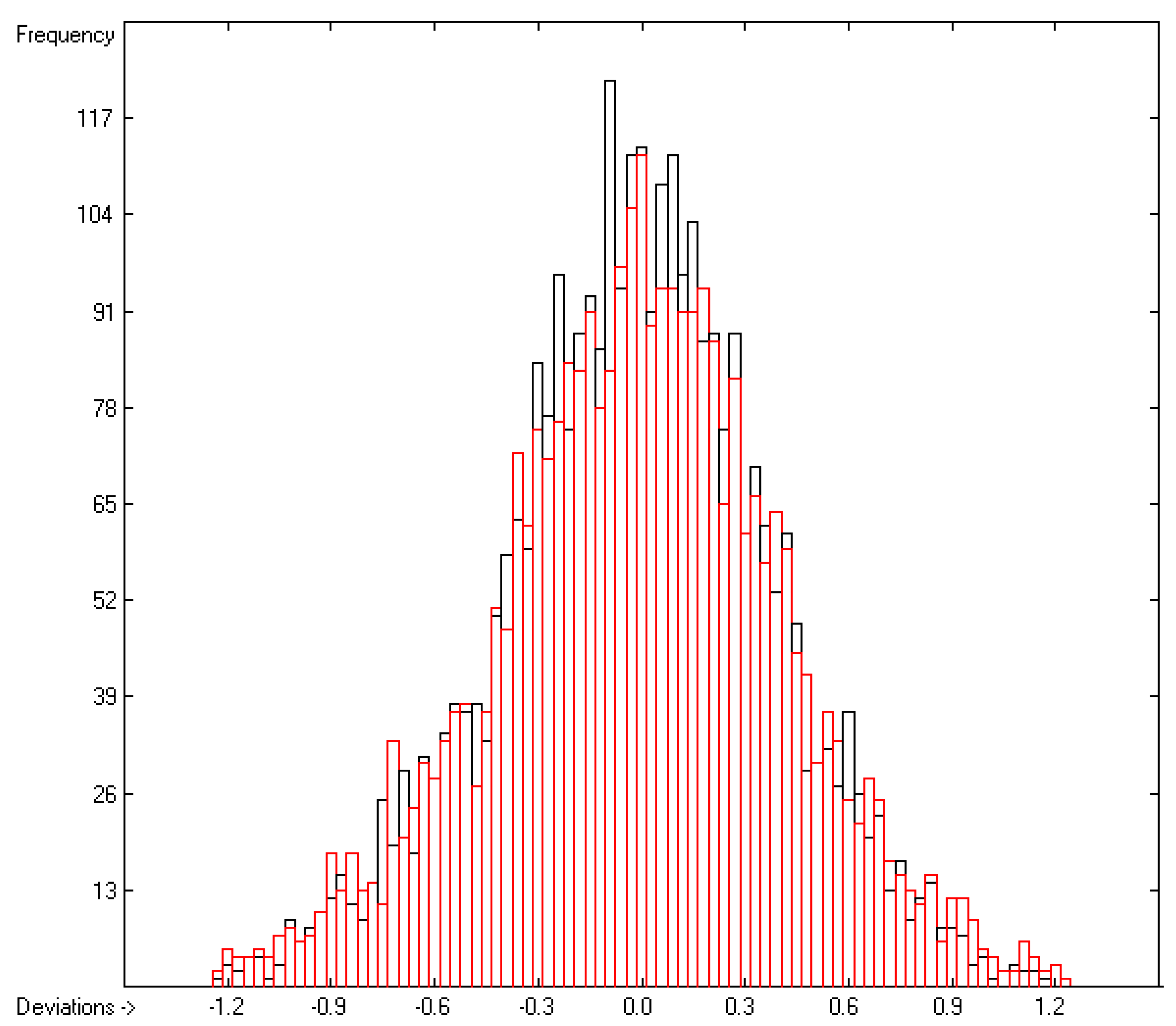
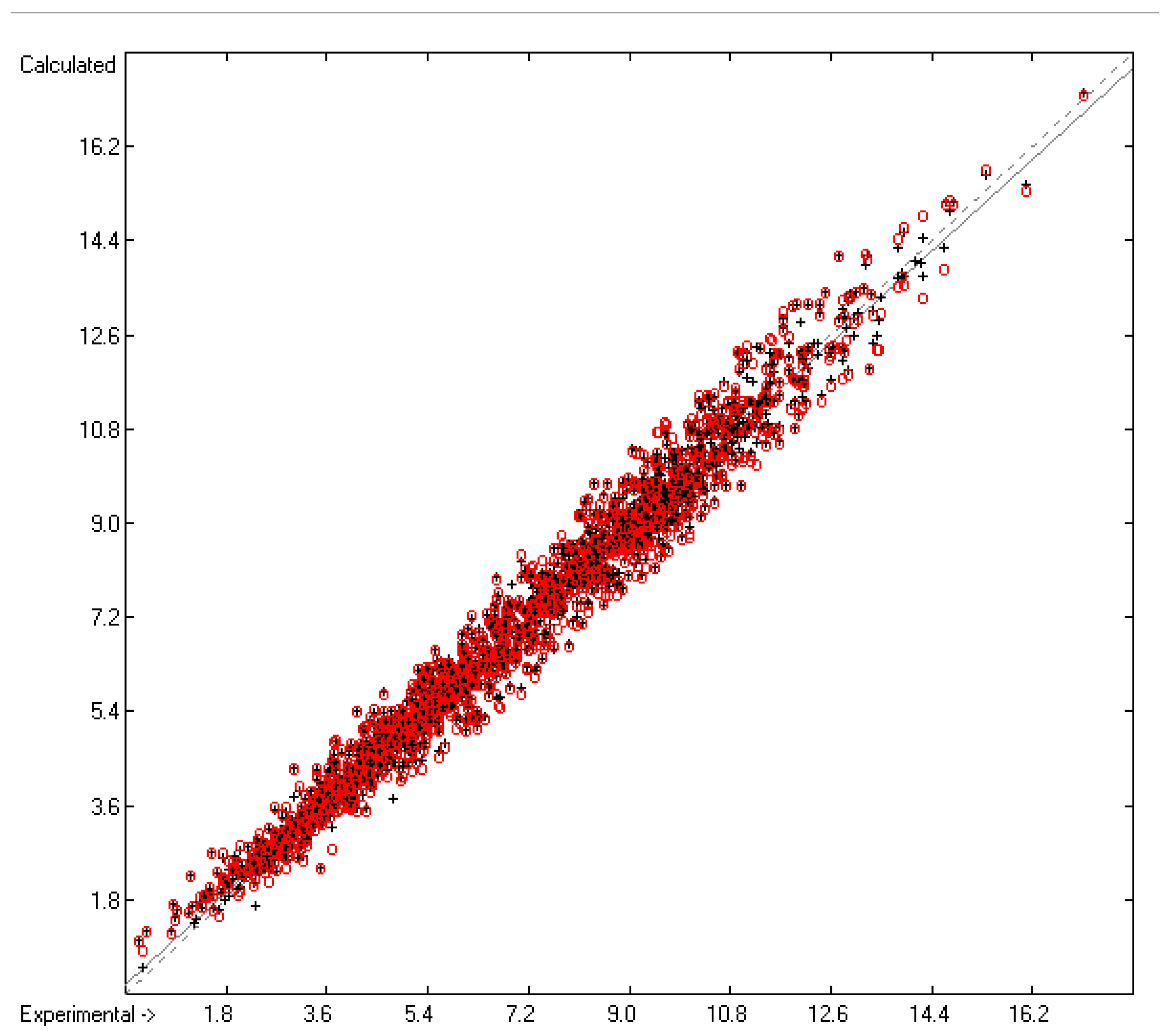
 =0.44; S =0.48; experimental values range from 0.28 to 17.15).
=0.44; S =0.48; experimental values range from 0.28 to 17.15).
 =0.44; S =0.48; experimental values range from 0.28 to 17.15).
=0.44; S =0.48; experimental values range from 0.28 to 17.15).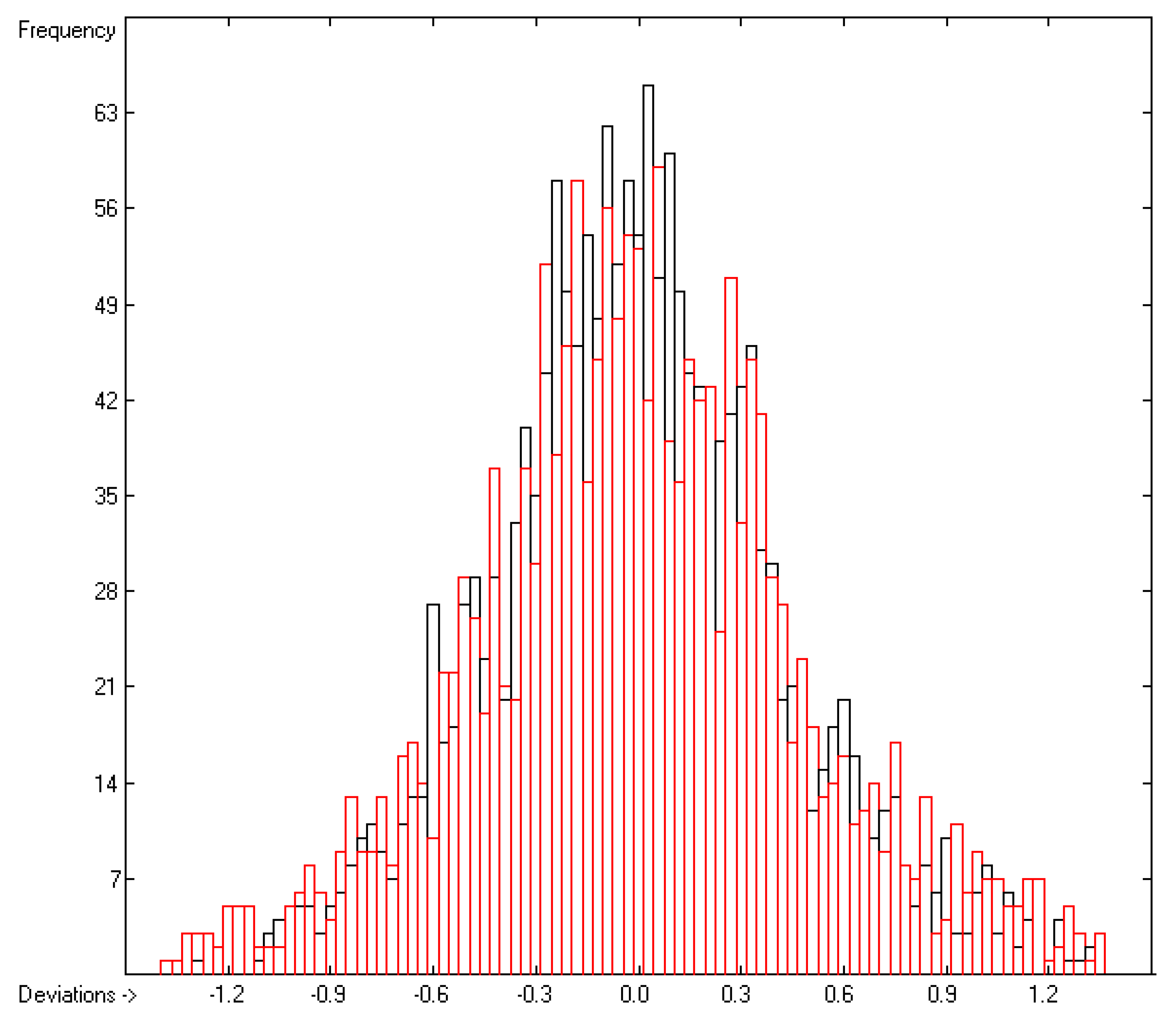

| Atom type | Neighbours | Meaning |
| H | H Acceptor | Correction value for intramolecular H bridge between acidic H (on O, N or S) and basic acceptor (O, N or F) |
| (COH)n | n>1 | Correction value for each additional hydroxy group |
| (COOH)n | n>1 | Correction value for each additional carboxylic acid group |
| Alkane | No of C atoms | Correction value for each C atom in a pure alkane |
| Unsaturated HC | No of C atoms | Correction value for each C atom in an aromatic hydrocarbon |
| Endocyclic bonds | No of single bonds | Correction value for each single endocyclic bond |
| Entry | Atom Type | Neighbours | Contribution | Occurrences | Molecules |
|---|---|---|---|---|---|
| 1 | Const | 0.73 | 3332 | 3332 | |
| 2 | B(-) | F4 | 2.71 | 10 | 10 |
| 3 | C sp3 | H3C | 0.27 | 2614 | 1498 |
| 4 | C sp3 | H3N | 0.14 | 457 | 320 |
| 5 | C sp3 | H3N(+) | -1.35 | 2 | 2 |
| 6 | C sp3 | H3O | -0.26 | 375 | 285 |
| 7 | C sp3 | H3P | -0.3 | 4 | 4 |
| 8 | C sp3 | H3S | -0.34 | 61 | 53 |
| 9 | C sp3 | H3Si | 0.76 | 44 | 5 |
| 10 | C sp3 | H2C2 | 0.44 | 3262 | 1046 |
| 11 | C sp3 | H2CN | 0.42 | 741 | 429 |
| 12 | C sp3 | H2CN(+) | -0.86 | 32 | 25 |
| 13 | C sp3 | H2CO | -0.1 | 799 | 604 |
| 14 | C sp3 | H2CS | -0.33 | 97 | 69 |
| 15 | C sp3 | H2CF | -0.29 | 5 | 5 |
| 16 | C sp3 | H2CCl | 0.33 | 84 | 67 |
| 17 | C sp3 | H2CBr | 0.41 | 54 | 48 |
| 18 | C sp3 | H2CJ | 1.08 | 6 | 6 |
| 19 | C sp3 | H2CP | 2.77 | 1 | 1 |
| 20 | C sp3 | H2N2 | 2.05 | 3 | 3 |
| 21 | C sp3 | H2NO | 0.46 | 4 | 4 |
| 22 | C sp3 | H2NS | 0.72 | 3 | 3 |
| 23 | C sp3 | H2O2 | -0.17 | 6 | 6 |
| 24 | C sp3 | H2S2 | -0.86 | 6 | 6 |
| 25 | C sp3 | HC3 | 0.45 | 417 | 269 |
| 26 | C sp3 | HC2N | 0.58 | 200 | 157 |
| 27 | C sp3 | HC2N(+) | -0.73 | 25 | 24 |
| 28 | C sp3 | HC2O | 0.1 | 383 | 241 |
| 29 | C sp3 | HC2S | -0.21 | 8 | 8 |
| 30 | C sp3 | HC2F | -0.36 | 2 | 2 |
| 31 | C sp3 | HC2Cl | 0.69 | 64 | 22 |
| 32 | C sp3 | HC2Br | 0.81 | 26 | 22 |
| 33 | C sp3 | HCN2 | 1.2 | 6 | 5 |
| 34 | C sp3 | HCNO | 1.15 | 17 | 17 |
| 35 | C sp3 | HCNS | 0.9 | 25 | 25 |
| 36 | C sp3 | HCO2 | -0.02 | 31 | 22 |
| 37 | C sp3 | HCOS | 0.6 | 3 | 3 |
| 38 | C sp3 | HCOCl | 0.19 | 3 | 1 |
| 39 | C sp3 | HCOBr | 1.03 | 1 | 1 |
| 40 | C sp3 | HCOP | 0.31 | 1 | 1 |
| 41 | C sp3 | HCF2 | -0.02 | 2 | 2 |
| 42 | C sp3 | HCCl2 | 0.93 | 13 | 12 |
| 43 | C sp3 | HOF2 | -0.04 | 1 | 1 |
| 44 | C sp3 | C4 | 0.54 | 144 | 111 |
| 45 | C sp3 | C3N | 0.71 | 37 | 36 |
| 46 | C sp3 | C3N(+) | -0.43 | 6 | 6 |
| 47 | C sp3 | C3O | 0.04 | 54 | 52 |
| 48 | C sp3 | C3S | -0.1 | 17 | 17 |
| 49 | C sp3 | C3F | 0.94 | 4 | 4 |
| 50 | C sp3 | C3Cl | 0.8 | 21 | 8 |
| 51 | C sp3 | C3Br | 0.59 | 5 | 4 |
| 52 | C sp3 | C2N2 | -1.17 | 1 | 1 |
| 53 | C sp3 | C2NO | 0.52 | 5 | 5 |
| 54 | C sp3 | C2O2 | 1.65 | 5 | 5 |
| 55 | C sp3 | C2F2 | 0.67 | 2 | 2 |
| 56 | C sp3 | C2Cl2 | 0.84 | 9 | 9 |
| 57 | C sp3 | CNO2 | 1.46 | 1 | 1 |
| 58 | C sp3 | CF3 | 0.86 | 80 | 76 |
| 59 | C sp3 | CF2Cl | 1.1 | 3 | 2 |
| 60 | C sp3 | CFCl2 | 1.1 | 3 | 2 |
| 61 | C sp3 | CCl3 | 1.6 | 23 | 21 |
| 62 | C sp3 | CCl2Br | 0 | 1 | 1 |
| 63 | C sp3 | CBr3 | 2.44 | 1 | 1 |
| 64 | C sp3 | OF3 | 0.8 | 2 | 2 |
| 65 | C sp3 | SF3 | 1.04 | 8 | 8 |
| 66 | C sp3 | SFCl2 | 1.9 | 1 | 1 |
| 67 | C sp3 | SCl3 | 0.76 | 3 | 3 |
| 68 | C sp2 | H2=C | 0.25 | 97 | 87 |
| 69 | C sp2 | H2=N | -0.62 | 1 | 1 |
| 70 | C sp2 | HC=C | 0.24 | 449 | 285 |
| 71 | C sp2 | HC=N | -1.98 | 18 | 18 |
| 72 | C sp2 | HC=N(+) | 0.94 | 10 | 10 |
| 73 | C sp2 | H=CN | -0.08 | 146 | 109 |
| 74 | C sp2 | H=CN(+) | -0.6 | 18 | 18 |
| 75 | C sp2 | HC=O | -0.73 | 45 | 45 |
| 76 | C sp2 | H=CO | 0.32 | 14 | 13 |
| 77 | C sp2 | H=CS | 0.02 | 17 | 16 |
| 78 | C sp2 | H=CCl | 0.51 | 8 | 6 |
| 79 | C sp2 | H=CBr | 0.59 | 1 | 1 |
| 80 | C sp2 | HN=N | -0.06 | 65 | 52 |
| 81 | C sp2 | HN=O | -0.63 | 16 | 15 |
| 82 | C sp2 | HO=O | -0.4 | 10 | 10 |
| 83 | C sp2 | H=NS | -0.51 | 4 | 4 |
| 84 | C sp2 | C2=C | 0.38 | 160 | 133 |
| 85 | C sp2 | C2=N | -0.25 | 105 | 102 |
| 86 | C sp2 | C2=N(+) | 2.45 | 1 | 1 |
| 87 | C sp2 | C2=O | -0.86 | 242 | 194 |
| 88 | C sp2 | C=CN | 0.76 | 76 | 64 |
| 89 | C sp2 | C=CN(+) | -0.56 | 3 | 3 |
| 90 | C sp2 | C=CO | 0.64 | 41 | 36 |
| 91 | C sp2 | C=CS | -0.16 | 17 | 15 |
| 92 | C sp2 | C=CF | -0.01 | 3 | 3 |
| 93 | C sp2 | C=CCl | 0.81 | 31 | 21 |
| 94 | C sp2 | C=CBr | 0.94 | 4 | 4 |
| 95 | C sp2 | C=CJ | 0.89 | 1 | 1 |
| 96 | C sp2 | C=CP | 0 | 1 | 1 |
| 97 | C sp2 | =CN2 | 1.36 | 19 | 19 |
| 98 | C sp2 | =CN2(+) | 0.74 | 11 | 11 |
| 99 | C sp2 | CN=N | 0.24 | 67 | 63 |
| 100 | C sp2 | CN=N(+) | -0.67 | 1 | 1 |
| 101 | C sp2 | CN=O | -0.69 | 449 | 364 |
| 102 | C sp2 | C=NO | -0.76 | 1 | 1 |
| 103 | C sp2 | =CNO | -0.01 | 4 | 4 |
| 104 | C sp2 | =CNO(+) | -0.37 | 2 | 2 |
| 105 | C sp2 | CN=S | -0.36 | 8 | 8 |
| 106 | C sp2 | C=NS | 0.07 | 5 | 4 |
| 107 | C sp2 | =CNS | 0.37 | 4 | 4 |
| 108 | C sp2 | =CNCl | 1.94 | 1 | 1 |
| 109 | C sp2 | =CNBr | 0.7 | 5 | 3 |
| 110 | C sp2 | C=NCl | 1.75 | 1 | 1 |
| 111 | C sp2 | CO=O | -0.13 | 700 | 613 |
| 112 | C sp2 | CO=O(-) | -2.16 | 35 | 35 |
| 113 | C sp2 | C=OS | -0.99 | 4 | 4 |
| 114 | C sp2 | C=OCl | 0.28 | 4 | 4 |
| 115 | C sp2 | =COCl | 1.27 | 1 | 1 |
| 116 | C sp2 | =CS2 | 0 | 3 | 3 |
| 117 | C sp2 | =CSBr | -2.41 | 1 | 1 |
| 118 | C sp2 | =CF2 | 0.26 | 1 | 1 |
| 119 | C sp2 | =CCl2 | 1.21 | 12 | 10 |
| 120 | C sp2 | =CBr2 | 1.36 | 1 | 1 |
| 121 | C sp2 | N2=N | 0.79 | 26 | 25 |
| 122 | C sp2 | N2=N(+) | 0.74 | 1 | 1 |
| 123 | C sp2 | N2=O | 0.07 | 135 | 134 |
| 124 | C sp2 | N=NO | 0.11 | 1 | 1 |
| 125 | C sp2 | N2=S | 0.11 | 9 | 8 |
| 126 | C sp2 | N=NS | 0.24 | 25 | 24 |
| 127 | C sp2 | N=NCl | 1.13 | 3 | 3 |
| 128 | C sp2 | N=NBr | 0.24 | 3 | 2 |
| 129 | C sp2 | NO=O | 0.2 | 117 | 114 |
| 130 | C sp2 | =NOS | -0.19 | 1 | 1 |
| 131 | C sp2 | N=OS | 0.05 | 7 | 7 |
| 132 | C sp2 | NO=S | 0.97 | 1 | 1 |
| 133 | C sp2 | =NS2 | -1.65 | 2 | 2 |
| 134 | C sp2 | NS=S | -1.02 | 5 | 3 |
| 135 | C sp2 | =NSCl | 1.17 | 1 | 1 |
| 136 | C sp2 | O2=O | 0 | 3 | 3 |
| 137 | C sp2 | O=OCl | -0.13 | 3 | 3 |
| 138 | C aromatic | H:C2 | 0.25 | 9963 | 2133 |
| 139 | C aromatic | H:C:N | -0.49 | 283 | 193 |
| 140 | C aromatic | H:C:N(+) | 0.22 | 33 | 27 |
| 141 | C aromatic | H:N2 | -0.91 | 9 | 9 |
| 142 | C aromatic | :C3 | 0.25 | 389 | 170 |
| 143 | C aromatic | C:C2 | 0.32 | 2023 | 1351 |
| 144 | C aromatic | C:C:N | -0.38 | 74 | 62 |
| 145 | C aromatic | C:C:N(+) | -3.29 | 4 | 3 |
| 146 | C aromatic | :C2N | 0.39 | 653 | 534 |
| 147 | C aromatic | :C2N(+) | -0.15 | 194 | 161 |
| 148 | C aromatic | :C2:N | -0.09 | 93 | 72 |
| 149 | C aromatic | :C2:N(+) | -3.54 | 19 | 19 |
| 150 | C aromatic | :C2O | 0.57 | 1076 | 742 |
| 151 | C aromatic | :C2S | 0.08 | 208 | 170 |
| 152 | C aromatic | :C2F | 0.27 | 126 | 86 |
| 153 | C aromatic | :C2Cl | 0.78 | 1718 | 565 |
| 154 | C aromatic | :C2Br | 0.9 | 248 | 111 |
| 155 | C aromatic | :C2J | 1.26 | 50 | 34 |
| 156 | C aromatic | :C2P | 1.08 | 1 | 1 |
| 157 | C aromatic | C:N2 | -1.81 | 9 | 9 |
| 158 | C aromatic | :C:N2 | -0.13 | 1 | 1 |
| 159 | C aromatic | :CN:N | 0.49 | 38 | 34 |
| 160 | C aromatic | :CN:N(+) | -0.83 | 1 | 1 |
| 161 | C aromatic | :C:NO | 0.97 | 21 | 15 |
| 162 | C aromatic | :C:NS | -0.16 | 5 | 5 |
| 163 | C aromatic | :C:NF | -0.23 | 4 | 3 |
| 164 | C aromatic | :C:NCl | 0.16 | 18 | 16 |
| 165 | C aromatic | :C:NBr | 0.06 | 1 | 1 |
| 166 | C aromatic | N:N2 | -0.05 | 51 | 41 |
| 167 | C aromatic | N:N2(+) | 0 | 1 | 1 |
| 168 | C aromatic | :N2O | 1.53 | 8 | 8 |
| 169 | C aromatic | :N2S | 0.8 | 3 | 3 |
| 170 | C aromatic | :N2Cl | 0.89 | 6 | 6 |
| 171 | C(+) aromatic | H:N2 | 0.21 | 25 | 25 |
| 172 | C sp | H#C | -0.27 | 28 | 28 |
| 173 | C sp | C#C | 0.2 | 86 | 57 |
| 174 | C sp | C#N | -0.7 | 136 | 130 |
| 175 | C sp | N#N | 0.04 | 3 | 3 |
| 176 | C sp | #NS | -0.59 | 5 | 5 |
| 177 | C sp | =N=O | 0.64 | 4 | 4 |
| 178 | C sp | =N=S | 1.53 | 27 | 26 |
| 179 | N sp3 | H2C | -1.57 | 86 | 84 |
| 180 | N sp3 | H2C(pi) | -1.05 | 326 | 292 |
| 181 | N sp3 | H2N | -0.85 | 20 | 20 |
| 182 | N sp3 | H2S | -1.55 | 34 | 34 |
| 183 | N sp3 | HC2 | -1.3 | 74 | 73 |
| 184 | N sp3 | HC2(pi) | -0.93 | 225 | 203 |
| 185 | N sp3 | HC2(2pi) | -0.47 | 311 | 272 |
| 186 | N sp3 | HCN | -1.1 | 4 | 3 |
| 187 | N sp3 | HCN(pi) | -0.49 | 14 | 13 |
| 188 | N sp3 | HCN(2pi) | 1.65 | 42 | 42 |
| 189 | N sp3 | HCO(pi) | -1.32 | 9 | 9 |
| 190 | N sp3 | HCS | -1.69 | 4 | 4 |
| 191 | N sp3 | HCS(pi) | -0.98 | 47 | 47 |
| 192 | N sp3 | HCP | -1.78 | 3 | 3 |
| 193 | N sp3 | HCP(pi) | -0.41 | 1 | 1 |
| 194 | N sp3 | C3 | -1.03 | 122 | 108 |
| 195 | N sp3 | C3(pi) | -0.73 | 153 | 138 |
| 196 | N sp3 | C3(2pi) | -0.72 | 149 | 136 |
| 197 | N sp3 | C3(3pi) | -0.75 | 23 | 23 |
| 198 | N sp3 | C2N | -1.57 | 1 | 1 |
| 199 | N sp3 | C2N(pi) | -1.41 | 31 | 28 |
| 200 | N sp3 | C2N(2pi) | -0.67 | 51 | 47 |
| 201 | N sp3 | C2N(3pi) | -0.44 | 10 | 10 |
| 202 | N sp3 | C2O(pi) | -0.31 | 5 | 5 |
| 203 | N sp3 | C2S | -1.42 | 5 | 5 |
| 204 | N sp3 | C2S(pi) | 0.03 | 7 | 6 |
| 205 | N sp3 | C2S(2pi) | 0.76 | 2 | 2 |
| 206 | N sp3 | C2P | -0.33 | 5 | 3 |
| 207 | N sp3 | CN2(2pi) | 1.36 | 1 | 1 |
| 208 | N sp3 | CS2 | 0.27 | 1 | 1 |
| 209 | N sp3 | CS2(pi) | -0.29 | 1 | 1 |
| 210 | N sp2 | H=C | -0.67 | 12 | 11 |
| 211 | N sp2 | C=C | -0.72 | 200 | 180 |
| 212 | N sp2 | C=N | 0.01 | 13 | 12 |
| 213 | N sp2 | =CN | 0.49 | 96 | 78 |
| 214 | N sp2 | C=N(+) | -6.61 | 1 | 1 |
| 215 | N sp2 | =CN(+) | -1.02 | 2 | 2 |
| 216 | N sp2 | =CO | -0.64 | 47 | 41 |
| 217 | N sp2 | C=O | -1.05 | 2 | 2 |
| 218 | N sp2 | =CS | -1.44 | 5 | 4 |
| 219 | N sp2 | N=N | -0.78 | 25 | 18 |
| 220 | N sp2 | N=O | 0.16 | 40 | 37 |
| 221 | N aromatic | C2:C(+) | 0 | 50 | 25 |
| 222 | N aromatic | :C2 | 0.38 | 354 | 258 |
| 223 | N aromatic | :C:N | -0.35 | 4 | 2 |
| 224 | N(+) sp3 | H3C | -1.03 | 26 | 26 |
| 225 | N(+) sp3 | H2C2 | 1.2 | 5 | 5 |
| 226 | N(+) sp3 | HC3 | 2.68 | 1 | 1 |
| 227 | N(+) sp3 | C4 | 3.03 | 1 | 1 |
| 228 | N(+) sp2 | C=CO(-) | -2.3 | 10 | 10 |
| 229 | N(+) sp2 | CO=O(-) | 0.27 | 235 | 198 |
| 230 | N(+) sp2 | NO=O(-) | -0.19 | 2 | 2 |
| 231 | N(+) sp2 | O2=O(-) | 0.44 | 55 | 29 |
| 232 | N(+) aromatic | H:C2 | 2.5 | 3 | 3 |
| 233 | N(+) aromatic | C:C2 | -0.48 | 7 | 6 |
| 234 | N(+) aromatic | :C2O(-) | 1.73 | 19 | 19 |
| 235 | N(+) sp | =C=N(-) | 1.8 | 1 | 1 |
| 236 | N(+) sp | =N2(-) | 0 | 1 | 1 |
| 237 | O | HC | -0.96 | 481 | 344 |
| 238 | O | HC(pi) | -0.72 | 627 | 557 |
| 239 | O | HN | -0.15 | 11 | 11 |
| 240 | O | HN(pi) | -0.24 | 6 | 6 |
| 241 | O | C2 | 0.06 | 156 | 115 |
| 242 | O | C2(pi) | -0.13 | 726 | 588 |
| 243 | O | C2(2pi) | -0.51 | 301 | 280 |
| 244 | O | CN | 0.4 | 3 | 3 |
| 245 | O | CN(pi) | 0.82 | 4 | 4 |
| 246 | O | CN(+)(pi) | 0.01 | 55 | 29 |
| 247 | O | CN(2pi) | 0.53 | 13 | 12 |
| 248 | O | CS | -0.13 | 13 | 8 |
| 249 | O | CS(pi) | -0.1 | 3 | 3 |
| 250 | O | CP | 0.23 | 132 | 68 |
| 251 | O | CP(pi) | -0.49 | 36 | 26 |
| 252 | O | CSi | -0.15 | 8 | 2 |
| 253 | O | N2(2pi) | 1.91 | 5 | 5 |
| 254 | O | NP(pi) | -1.95 | 14 | 14 |
| 255 | O | Si2 | 0.09 | 18 | 4 |
| 256 | S2 | HC | 0.65 | 14 | 12 |
| 257 | S2 | HC(pi) | 0.14 | 31 | 31 |
| 258 | S2 | C2 | 1.39 | 48 | 45 |
| 259 | S2 | C2(pi) | 0.98 | 68 | 63 |
| 260 | S2 | C2(2pi) | 0.98 | 55 | 54 |
| 261 | S2 | CN | 0 | 3 | 3 |
| 262 | S2 | CN(2pi) | 2.3 | 1 | 1 |
| 263 | S2 | CS | 0.87 | 2 | 1 |
| 264 | S2 | CS(pi) | 1.97 | 4 | 2 |
| 265 | S2 | CP | 1.12 | 17 | 15 |
| 266 | S2 | CP(pi) | 0.48 | 3 | 2 |
| 267 | S2 | N2 | -2.2 | 2 | 2 |
| 268 | S2 | N2(2pi) | 5.96 | 1 | 1 |
| 269 | S4 | C2=O | -1.13 | 11 | 11 |
| 270 | S4 | C2=O2 | -0.5 | 16 | 16 |
| 271 | S4 | CO=O2 | -0.48 | 2 | 1 |
| 272 | S4 | CN=O2 | -0.05 | 85 | 80 |
| 273 | S4 | C=O2F | 0.24 | 2 | 2 |
| 274 | S4 | NO=O2 | 0 | 3 | 3 |
| 275 | S4 | N2=O2 | 0.77 | 5 | 5 |
| 276 | S4 | O2=O | 0.83 | 2 | 2 |
| 277 | S4 | O2=O2 | 0.5 | 2 | 2 |
| 278 | S4 | O2=O2(-) | -1.14 | 3 | 3 |
| 279 | P4 | CO2=O | -1.11 | 2 | 2 |
| 280 | P4 | CO2=S | 0.26 | 1 | 1 |
| 281 | P4 | CO=OS | -2.58 | 1 | 1 |
| 282 | P4 | CO=OF | -0.88 | 3 | 3 |
| 283 | P4 | COS=S | -2.04 | 1 | 1 |
| 284 | P4 | O3=O | -0.56 | 29 | 29 |
| 285 | P4 | O3=S | 1.12 | 18 | 18 |
| 286 | P4 | O2S=S | 0.7 | 12 | 11 |
| 287 | P4 | O=OS2 | -0.54 | 2 | 2 |
| 288 | P4 | N3=O | -0.31 | 1 | 1 |
| 289 | P4 | N2O=O | 0.24 | 2 | 2 |
| 290 | P4 | NO=OS | -1.5 | 2 | 2 |
| 291 | Si | C4 | -0.51 | 1 | 1 |
| 292 | Si | C3O | -1.7 | 2 | 1 |
| 293 | Si | C2O2 | 0.13 | 17 | 4 |
| 294 | Si | O4 | 0 | 2 | 2 |
| 295 | Halide | 1.1 | 20 | 19 | |
| 296 | H | H Acceptor | 0.51 | 164 | 154 |
| 297 | (COH)n | n>1 | 0.26 | 137 | 74 |
| 298 | (COOH)n | n>1 | -0.15 | 26 | 25 |
| 299 | Alkane | No of C atoms | 0.09 | 290 | 32 |
| 300 | Unsaturated HC | No of C atoms | 0.02 | 1584 | 135 |
| 301 | Endocyclic bonds | No of single bds | -0.14 | 2338 | 384 |
| A | Based on | Valid groups | 214 | 3332 | |
| B | Goodness of fit | R2 | 0.9648 | 3246 | |
| C | Deviation | Average | 0.31 | 3246 | |
| D | Deviation | Standard | 0.39 | 3246 | |
| E | K-fold cv | K | 10 | 3164 | |
| F | Goodness of fit | Q2 | 0.9599 | 3164 | |
| G | Deviation | Average (cv) | 0.33 | 3164 | |
| H | Deviation | Standard (cv) | 0.42 | 3164 |
| Entry | Atom Type | Neighbours | Contribution | Occurrences | Molecules |
|---|---|---|---|---|---|
| 1 | Const | 1.46 | 1900 | 1900 | |
| 2 | C sp3 | H3C | -0.07 | 1800 | 875 |
| 3 | C sp3 | H3N | 3.42 | 131 | 87 |
| 4 | C sp3 | H3N(+) | 1.42 | 1 | 1 |
| 5 | C sp3 | H3O | 2.24 | 292 | 219 |
| 6 | C sp3 | H3S | 1.51 | 30 | 26 |
| 7 | C sp3 | H3P | -0.42 | 3 | 3 |
| 8 | C sp3 | H3Si | 0.42 | 68 | 11 |
| 9 | C sp3 | H2C2 | 0.43 | 1732 | 538 |
| 10 | C sp3 | H2CN | 3.91 | 191 | 129 |
| 11 | C sp3 | H2CN(+) | 1.64 | 6 | 5 |
| 12 | C sp3 | H2CO | 2.61 | 535 | 342 |
| 13 | C sp3 | H2CS | 1.76 | 57 | 44 |
| 14 | C sp3 | H2CP | 2.58 | 3 | 3 |
| 15 | C sp3 | H2CF | -0.77 | 3 | 3 |
| 16 | C sp3 | H2CCl | 0.71 | 75 | 56 |
| 17 | C sp3 | H2CBr | 1.05 | 23 | 18 |
| 18 | C sp3 | H2CJ | 1.13 | 5 | 5 |
| 19 | C sp3 | H2CSi | 2.91 | 4 | 4 |
| 20 | C sp3 | H2N2 | 4.89 | 8 | 3 |
| 21 | C sp3 | H2NO | 5.65 | 9 | 8 |
| 22 | C sp3 | H2NS | 4.67 | 5 | 5 |
| 23 | C sp3 | H2O2 | 4.78 | 6 | 4 |
| 24 | C sp3 | H2S2 | 3.74 | 4 | 4 |
| 25 | C sp3 | HC3 | 0.64 | 268 | 180 |
| 26 | C sp3 | HC2N | 4.08 | 64 | 53 |
| 27 | C sp3 | HC2N(+) | 2.05 | 1 | 1 |
| 28 | C sp3 | HC2O | 2.86 | 169 | 135 |
| 29 | C sp3 | HC2S | 1.76 | 9 | 7 |
| 30 | C sp3 | HC2F | -1.66 | 1 | 1 |
| 31 | C sp3 | HC2Cl | 1.21 | 43 | 17 |
| 32 | C sp3 | HC2Br | 1.31 | 14 | 9 |
| 33 | C sp3 | HC2J | 1.95 | 1 | 1 |
| 34 | C sp3 | HCNO | 8.18 | 3 | 3 |
| 35 | C sp3 | HCNS | 2.08 | 1 | 1 |
| 36 | C sp3 | HCO2 | 5.73 | 6 | 6 |
| 37 | C sp3 | HCF2 | -0.18 | 7 | 7 |
| 38 | C sp3 | HCFCl | 0.02 | 2 | 2 |
| 39 | C sp3 | HCCl2 | 1.18 | 15 | 14 |
| 40 | C sp3 | HCClBr | 0.77 | 1 | 1 |
| 41 | C sp3 | HOF2 | 1.79 | 3 | 3 |
| 42 | C sp3 | C4 | 0.73 | 98 | 84 |
| 43 | C sp3 | C3N | 4.1 | 13 | 13 |
| 44 | C sp3 | C3O | 3.11 | 40 | 37 |
| 45 | C sp3 | C3S | 2.6 | 3 | 3 |
| 46 | C sp3 | C3Cl | 0.87 | 37 | 15 |
| 47 | C sp3 | C2NO | 5.94 | 1 | 1 |
| 48 | C sp3 | C2O2 | 5.94 | 6 | 6 |
| 49 | C sp3 | C2F2 | 0.23 | 58 | 10 |
| 50 | C sp3 | C2Cl2 | 1.24 | 18 | 17 |
| 51 | C sp3 | CNO2 | 9.56 | 1 | 1 |
| 52 | C sp3 | COF2 | 3.06 | 3 | 3 |
| 53 | C sp3 | CF3 | -0.06 | 55 | 51 |
| 54 | C sp3 | CF2Cl | -0.02 | 4 | 3 |
| 55 | C sp3 | CFCl2 | 0.37 | 3 | 2 |
| 56 | C sp3 | CCl3 | 1.62 | 17 | 16 |
| 57 | C sp3 | CBr3 | 0.57 | 1 | 1 |
| 58 | C sp3 | O2F2 | 6.85 | 1 | 1 |
| 59 | C sp3 | OF3 | 1.86 | 3 | 3 |
| 60 | C sp2 | H2=C | -0.19 | 88 | 76 |
| 61 | C sp2 | HC=C | 0.34 | 233 | 141 |
| 62 | C sp2 | HC=N | 0.85 | 8 | 8 |
| 63 | C sp2 | HC=O | 1 | 27 | 27 |
| 64 | C sp2 | H=CN | 1.1 | 19 | 13 |
| 65 | C sp2 | H=CO | 0.48 | 15 | 14 |
| 66 | C sp2 | H=CS | -1.08 | 9 | 7 |
| 67 | C sp2 | H=CCl | 0.44 | 12 | 10 |
| 68 | C sp2 | H=CBr | 0.6 | 3 | 2 |
| 69 | C sp2 | H=CSi | 2.17 | 1 | 1 |
| 70 | C sp2 | HN=N | 1.73 | 53 | 30 |
| 71 | C sp2 | HN=O | 2.27 | 3 | 3 |
| 72 | C sp2 | HO=O | 0.92 | 4 | 4 |
| 73 | C sp2 | H=NS | 2.88 | 1 | 1 |
| 74 | C sp2 | C2=C | 0.8 | 103 | 79 |
| 75 | C sp2 | C2=N | 1.62 | 34 | 30 |
| 76 | C sp2 | C=CN | 1.6 | 19 | 16 |
| 77 | C sp2 | C2=O | 1.08 | 87 | 75 |
| 78 | C sp2 | C=CO | 1.22 | 27 | 26 |
| 79 | C sp2 | C=CP | -0.09 | 1 | 1 |
| 80 | C sp2 | C=CS | -0.41 | 14 | 10 |
| 81 | C sp2 | C=CCl | 0.62 | 39 | 24 |
| 82 | C sp2 | C=CBr | 1.01 | 12 | 5 |
| 83 | C sp2 | =CN2 | 2.98 | 2 | 2 |
| 84 | C sp2 | CN=N | 2.75 | 7 | 7 |
| 85 | C sp2 | CN=O | 2.64 | 93 | 88 |
| 86 | C sp2 | C=NO | 1.26 | 5 | 5 |
| 87 | C sp2 | =CNO | -1.24 | 3 | 3 |
| 88 | C sp2 | C=NS | 0.46 | 6 | 6 |
| 89 | C sp2 | =CNCl | 3.35 | 6 | 3 |
| 90 | C sp2 | CO=O | 1.73 | 244 | 210 |
| 91 | C sp2 | C=OS | -0.61 | 3 | 2 |
| 92 | C sp2 | =CS2 | -0.66 | 1 | 1 |
| 93 | C sp2 | =CF2 | -1.14 | 1 | 1 |
| 94 | C sp2 | =CCl2 | 1.14 | 16 | 14 |
| 95 | C sp2 | N2=N | 3.36 | 9 | 9 |
| 96 | C sp2 | N2=O | 3.65 | 43 | 40 |
| 97 | C sp2 | N=NO | 2.64 | 4 | 4 |
| 98 | C sp2 | N=NS | 0.71 | 7 | 7 |
| 99 | C sp2 | NO=O | 2.81 | 38 | 36 |
| 100 | C sp2 | N=OS | 0.93 | 17 | 17 |
| 101 | C sp2 | NO=S | 4.26 | 1 | 1 |
| 102 | C sp2 | =NOS | 0.68 | 3 | 3 |
| 103 | C sp2 | NS=S | 6.03 | 3 | 2 |
| 104 | C sp2 | =NSCl | -5.44 | 2 | 2 |
| 105 | C sp2 | O2=O | 2.56 | 3 | 3 |
| 106 | C aromatic | H:C2 | 0.31 | 5436 | 1136 |
| 107 | C aromatic | H:C:N | 0.53 | 81 | 49 |
| 108 | C aromatic | H:N2 | 0.17 | 6 | 6 |
| 109 | C aromatic | :C3 | 0.89 | 441 | 148 |
| 110 | C aromatic | C:C2 | 0.79 | 1163 | 657 |
| 111 | C aromatic | C:C:N | 0.68 | 42 | 30 |
| 112 | C aromatic | :C2N | 1.35 | 164 | 146 |
| 113 | C aromatic | :C2N(+) | 2.09 | 96 | 69 |
| 114 | C aromatic | :C2:N | 1.01 | 13 | 10 |
| 115 | C aromatic | :C2O | 1.27 | 769 | 453 |
| 116 | C aromatic | :C2P | 3.53 | 5 | 3 |
| 117 | C aromatic | :C2S | -0.19 | 38 | 33 |
| 118 | C aromatic | :C2Si | -0.25 | 1 | 1 |
| 119 | C aromatic | :C2F | 0.13 | 99 | 41 |
| 120 | C aromatic | :C2Cl | 0.91 | 1844 | 550 |
| 121 | C aromatic | :C2Br | 1.24 | 391 | 143 |
| 122 | C aromatic | :C2J | 2.14 | 10 | 9 |
| 123 | C aromatic | C:N2 | 0.77 | 11 | 10 |
| 124 | C aromatic | :CN:N | 0.8 | 4 | 4 |
| 125 | C aromatic | :C:NO | 1.2 | 28 | 24 |
| 126 | C aromatic | :C:NCl | 0.9 | 14 | 12 |
| 127 | C aromatic | N:N2 | 1.18 | 60 | 36 |
| 128 | C aromatic | :N2O | 1.15 | 11 | 11 |
| 129 | C aromatic | :N2S | -0.6 | 8 | 8 |
| 130 | C aromatic | :N2Cl | 0.43 | 9 | 8 |
| 131 | C sp | H#C | -0.45 | 18 | 17 |
| 132 | C sp | C#C | 0.67 | 18 | 17 |
| 133 | C sp | C#N | 0.73 | 46 | 43 |
| 134 | C sp | N#N | 5.32 | 1 | 1 |
| 135 | C sp | #NP | -5.58 | 1 | 1 |
| 136 | C sp | =N=S | -0.13 | 2 | 2 |
| 137 | N sp3 | H2C | -2.18 | 17 | 16 |
| 138 | N sp3 | H2C(pi) | 1.02 | 57 | 53 |
| 139 | N sp3 | H2N | 3.57 | 5 | 5 |
| 140 | N sp3 | H2S | 1.81 | 1 | 1 |
| 141 | N sp3 | HC2 | -5.94 | 12 | 11 |
| 142 | N sp3 | HC2(pi) | -2.38 | 93 | 70 |
| 143 | N sp3 | HC2(2pi) | 0.08 | 65 | 56 |
| 144 | N sp3 | HCN(pi) | 0.02 | 5 | 4 |
| 145 | N sp3 | HCN(2pi) | 1.2 | 4 | 4 |
| 146 | N sp3 | HCO(pi) | 1.13 | 1 | 1 |
| 147 | N sp3 | HCP | -4.1 | 3 | 3 |
| 148 | N sp3 | HCP(pi) | 1.51 | 1 | 1 |
| 149 | N sp3 | HCS(pi) | -1.54 | 8 | 8 |
| 150 | N sp3 | C3 | -9.44 | 17 | 17 |
| 151 | N sp3 | C3(pi) | -6.39 | 58 | 55 |
| 152 | N sp3 | C3(2pi) | -4.82 | 49 | 45 |
| 153 | N sp3 | C3(3pi) | -3.61 | 9 | 9 |
| 154 | N sp3 | C2N | -5.12 | 1 | 1 |
| 155 | N sp3 | C2N(pi) | -2.54 | 15 | 14 |
| 156 | N sp3 | C2N(+)(pi) | -1.93 | 7 | 2 |
| 157 | N sp3 | C2N(2pi) | -3.84 | 36 | 36 |
| 158 | N sp3 | C2N(3pi) | -0.65 | 13 | 12 |
| 159 | N sp3 | C2P | 0 | 1 | 1 |
| 160 | N sp3 | C2P(pi) | -2.97 | 1 | 1 |
| 161 | N sp3 | C2P(2pi) | -4.07 | 1 | 1 |
| 162 | N sp2 | H=C | 0.51 | 1 | 1 |
| 163 | N sp2 | C=C | -0.97 | 54 | 48 |
| 164 | N sp2 | C=N | 0.61 | 6 | 4 |
| 165 | N sp2 | =CN | 0.03 | 54 | 49 |
| 166 | N sp2 | =CN(+) | 9.74 | 2 | 2 |
| 167 | N sp2 | =CO | -3.65 | 30 | 26 |
| 168 | N sp2 | N=N | -1.3 | 4 | 3 |
| 169 | N sp2 | N=O | -2.02 | 13 | 13 |
| 170 | N aromatic | :C2 | 0.54 | 194 | 109 |
| 171 | N aromatic | :C:N | 0.47 | 4 | 1 |
| 172 | N(+) sp2 | CO=O(-) | -0.36 | 104 | 76 |
| 173 | N(+) sp2 | NO=O(-) | 0 | 9 | 4 |
| 174 | N(+) sp2 | O2=O(-) | -1.09 | 63 | 35 |
| 175 | O | HC | -0.66 | 143 | 121 |
| 176 | O | HC(pi) | 1.39 | 175 | 159 |
| 177 | O | HN(pi) | 4.18 | 2 | 2 |
| 178 | O | HP | 2.11 | 4 | 2 |
| 179 | O | HSi | 1.91 | 3 | 2 |
| 180 | O | C2 | -4.17 | 139 | 105 |
| 181 | O | C2(pi) | -2.68 | 392 | 317 |
| 182 | O | C2(2pi) | -0.92 | 255 | 228 |
| 183 | O | CN(pi) | 0.51 | 20 | 16 |
| 184 | O | CN(+)(pi) | 0.1 | 63 | 35 |
| 185 | O | CN(2pi) | 3.07 | 8 | 8 |
| 186 | O | CO(pi) | -1.03 | 2 | 1 |
| 187 | O | CS | -0.88 | 11 | 6 |
| 188 | O | CP | -1.2 | 183 | 93 |
| 189 | O | CP(pi) | -0.01 | 70 | 54 |
| 190 | O | CSi | -2.38 | 9 | 3 |
| 191 | O | NP(pi) | 4.65 | 1 | 1 |
| 192 | O | P2 | 1.7 | 1 | 1 |
| 193 | O | Si2 | 0 | 21 | 6 |
| 194 | P4 | C3=O | -5.7 | 1 | 1 |
| 195 | P4 | CNO=O | 1.2 | 1 | 1 |
| 196 | P4 | CO2=O | 1.47 | 3 | 3 |
| 197 | P4 | CO2=S | -1.5 | 3 | 3 |
| 198 | P4 | CO=OS | 1.99 | 1 | 1 |
| 199 | P4 | CO=OF | 1.94 | 1 | 1 |
| 200 | P4 | COS=S | -0.86 | 1 | 1 |
| 201 | P4 | NO2=O | 3.42 | 1 | 1 |
| 202 | P4 | NO2=S | 1.88 | 3 | 3 |
| 203 | P4 | NO=OS | 1.2 | 2 | 2 |
| 204 | P4 | O3=O | 0.09 | 29 | 29 |
| 205 | P4 | O3=S | -0.4 | 32 | 30 |
| 206 | P4 | O2=OS | 0.43 | 5 | 5 |
| 207 | P4 | O2=OF | -0.17 | 1 | 1 |
| 208 | P4 | O=OS2 | 1.58 | 3 | 3 |
| 209 | P4 | O2S=S | -0.27 | 18 | 17 |
| 210 | P4 | =OS3 | 1.46 | 1 | 1 |
| 211 | S2 | HC | -1.08 | 2 | 2 |
| 212 | S2 | HC(pi) | 1.54 | 1 | 1 |
| 213 | S2 | C2 | -1.5 | 14 | 14 |
| 214 | S2 | C2(pi) | 0.4 | 41 | 39 |
| 215 | S2 | C2(2pi) | 2.82 | 24 | 23 |
| 216 | S2 | CS | -0.58 | 4 | 2 |
| 217 | S2 | CS(pi) | -2.98 | 2 | 1 |
| 218 | S2 | CP | -0.12 | 33 | 28 |
| 219 | S2 | CP(pi) | 1.78 | 3 | 2 |
| 220 | S4 | C2=O | 0.6 | 2 | 2 |
| 221 | S4 | C2=O2 | 2.13 | 3 | 3 |
| 222 | S4 | CN=O2 | 3.26 | 9 | 9 |
| 223 | S4 | CO=O2 | -0.05 | 1 | 1 |
| 224 | S4 | O2=O | -0.35 | 2 | 2 |
| 225 | S4 | O2=O2 | 0.24 | 3 | 3 |
| 226 | S6 | CF5 | 1.92 | 3 | 3 |
| 227 | Si | C4 | -1.33 | 3 | 3 |
| 228 | Si | C3O | -0.65 | 7 | 4 |
| 229 | Si | C2O2 | 0.1 | 19 | 6 |
| 230 | Si | CO3 | 0 | 3 | 3 |
| 231 | H | H Acceptor | -1.51 | 47 | 45 |
| 232 | (COH)n | n>1 | 0.06 | 22 | 15 |
| 233 | (COOH)n | n>1 | 1.2 | 6 | 6 |
| 234 | Alkane | No of C atoms | -0.05 | 268 | 34 |
| 235 | Unsaturated HC | No of C atoms | -0.03 | 1512 | 140 |
| 236 | Endocyclic bonds | No of single bds | -0.11 | 1109 | 210 |
| A | Based on | Valid groups | 167 | 1900 | |
| B | Goodness of fit | R2 | 0.9765 | 1829 | |
| C | Deviation | Average | 0.34 | 1829 | |
| D | Deviation | Standard | 0.44 | 1829 | |
| E | K-fold cv | K | 10 | 1765 | |
| F | Goodness of fit | Q2 | 0.9717 | 1765 | |
| G | Deviation | Average (cv) | 0.37 | 1765 | |
| H | Deviation | Standard (cv) | 0.48 | 1765 |
| Atom type | C sp3 | C sp3 | C sp3 | C sp3 | C sp2 | O | S4 | Endocycl. Bonds | Const | Sum |
|---|---|---|---|---|---|---|---|---|---|---|
| Neighbors | H2CO | HC3 | C3Cl | C2Cl2 | C=CCl | CS | O2=O2 | n C-C | ||
| Contribution | 2.61 | 0.64 | 0.87 | 1.24 | 0.62 | -0.88 | 0.24 | -0.11 | 1.46 | |
| n Groups | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 9 | ||
| n x Contribution | 5.22 | 1.28 | 1.74 | 1.24 | 1.24 | -1.76 | 0.24 | -0.99 | 1.46 | 9.67 |
| Descriptor | 2-Nitroaniline | 3-Nitroaniline |
|---|---|---|
| logPow | 1.85 (1.70) | 1.37 (1.19) |
| logKoa | 6.46 (5.29) | 7.62 (6.80) |
| Descriptor | Hexanoic Acid | 1,6-Hexanedioic Acid |
|---|---|---|
| logPow | 1.92 (1.91) | 0.08 (0.64) |
| logKoa | 6.31 (6.23) | 10.74 (10.62) |
| Compound | logKaw exp | logKaw calc |
|---|---|---|
| 2-Nitroaniline | -4.77 | -3.59 |
| 3-Nitroaniline | -6.49 | -5.61 |
| Hexanoic Acid | -4.531 | -4.32 |
| 1,6-Hexanedioic Acid | -11.15 | -9.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





