1. Introduction
Recently, a new class of imipridone-derived antitumor molecules including maternal TRAIL-inducing compound ONC201/TIC10 [
1] and newly synthetized derivatives known as TR-compounds (Madera Therapeutics, Chapel Hill, NC USA) [
2] attracted the attention of investigators. The first data set on the potent efficacy of the first-in-class small molecule ONC201 were published in 2013 [
3] and over 10 years, a lot of preclinical work has appeared on tumor cells related to hematological and solid malignancies including prostate, breast, ovarian lung, pancreatic cancer cell lines as well as leukemia, lymphoma, glioma and hepatocellular cancer cells [
4]. The broad specificity of the action of imipridones, high efficiency, and selectivity for tumor cells made it possible to use them in clinical trials [
4,
5,
6]. While the early studies showed that the effect of imipridones on tumor cells occurs through inducing the expression of TRAIL and its receptor DR5 [
3,
7], as well as increasing the sensitivity of cells to exogenous TRAIL [
8], recent observations demonstrated mitochondrial caseinolytic serine protease ClpXP, localized within the mitochondrial matrix as the only intracellular target for imipridones [
2,
9,
10]. ClpP in the mitochondrial matrix forms a complex with the ATP-dependent protein unfoldase ClpX, which belongs to the AAA+ class (ATPase Associated with various cellular Activities) proteases [
11]. The ClpXP complex is involved in mitochondrial quality control; its substrates are many mitochondrial proteins including proteins of electron transport chain (ETC), tricarboxylic acid (TCA) cycle, mitochondrial gene transcription and translation and ribosomal proteins [
12]. Discovery of imipridones-induced activation of ClpP refocused investigators to study massive remodeling and unregulated proteolysis of proteins in mitochondria, accompanied with dramatic morphological changes and fragmentation of mitochondria, degradation of mitochondrial DNA and a rapid decrease in the level of mitochondrial transcription factor TFAM, responsible for maintaining the number of mtDNA copies [
2,
9,
13,
14,
15]. In addition, RNA-sequencing of MB231 cells treated with ONC201 showed changes in the expression of genes responsible for maintenance of the mitochondrial genome and other mitochondrial processes, including oxidative phosphorylation [
13]. In current work, we focused on the mechanism of action of TR-57 (chemically modified derivative of ONC201) with higher (50-100 times) efficacy compared to ONC201 [
2,
16,
17], which is associated with a higher affinity of their binding to ClpP [
2]. Multiple in vitro tests have shown that the antiproliferative effect of TR-57 is rather cytostatic and inhibits cell proliferation without induction of apoptotic cell death [
13,
18,
19,
20] and its cytostatic effect manifested not only by inhibition of tumor cells proliferation in vitro [
2,
21,
22]. Characteristically, most of the studied breast cancer cell lines are characterized by the absence of induction of apoptosis under the influence of ONC201 or TR-57, only blocking of cell growth is observed [
6], and with prolonged incubation with imipridones, the cells transform into a senescence-like phenotype [
21,
23]. However, it is still not completely clear what factors determine the sensitivity of cells to the action of imipridones, and the question of how imipridones-treated tumor cells containing mitochondria with impaired respiratory function maintain their viability remains unexplored.
Here we demonstrated that the mitochondrial component of the antiproliferative effect of TR-57 in cultured SUM159 human breast cancer cells consists of degradation the key structural and functional proteins of mtDNA translation and transcription, associated with decline in the expression of the key proteins of the respiratory Complexes I-IV, leading to suppression of the mitochondrial respiratory chain activity. Surprisingly, loss of the activity of mitochondrial respiratory chain was not associated with depolarization of remodeled mitochondria, and regardless of fragmentation and inhibition of electron transport, drug-treated cells retained mitochondrial membrane potential. Our data indicate that mitochondrial polarization could be supported by reversal of the activity of mitochondrial FoF1-ATPase, which, due to the consumption of glycolytic ATP, maintains the mitochondrial membrane potential necessary to perform mitochondrial functions and sustain the viability of tumor cells.
3. Discussion
Mitochondria are key multifunctional intracellular organelles determining the life and death of all cell types through regulation of intracellular supply of ATP, intracellular Ca2+ and redox signaling, metabolic remodeling, apoptotic cell death and intercellular communication [
31]. Recent discoveries highlight the role and importance of mitochondria for initiation and development of variety of cancers (for Review See [
32]). Oncotic diseases such as leukemia, lymphoma, lung adenocarcinoma, pancreatic ductal adenocarcinoma, as well as cancer stem cells with high metastatic and tumorigenic potential, require large amounts of ATP, fulfilled by upregulation of oxidative phosphorylation [
33]. In addition to the production of ATP, mitochondria are involved in the catabolic processes of de novo synthesis of nucleotides, lipids and amino acids necessary for proliferating cells, the formation of ROS signaling molecules, calcium signaling pathway, cell death regulation [
34]. In addition, at present time, targeting specific mitochondrial metabolism gaining attention as an opportunity for development of the efficient cancer cell therapy [
34].
Imipridones, a new class of antitumor agents, selectively targeting mitochondria through activating the unique mitochondrial caseinolytic serine protease ClpP [
2,
9,
10]. Mitochondrial effects are associated both directly with the proteolysis of mitochondrial proteins and indirectly through ClpP-dependent activation of the integrated stress response [
2,
9,
13]. It has been demonstrated that majority of mitochondrial processes are disrupted by the action of imipridones, such as imipridone mediated degradation of mtDNA, number of structural and functional proteins of electron-transport chain, tricarboxylic acid cycle, purine and amino acid metabolism, folate-mediated one-carbon metabolism and proline biosynthesis [
9,
17,
30,
35].
In our work, we studied the time-dependent effect of TR-57 on the morphological and functional characteristics of mitochondria in triple negative breast cancer cells SUM159. Using SUM159, we demonstrated that TR-57 induces mitochondrial fragmentation, inhibition of the activity of respiratory chain, decline in the number of mtDNA within first 24 hours of exposure of SUM159 cells to 150 nM of TR57. Longer incubation of these cells with TR57 causes neither a further decrease in mitochondrial size nor a change in mitochondrial mass. At the same time, we observed decline in functional state of mitochondria, revealed by inhibition of mitochondrial respiration in a model of digitonin permeabilized cells, oxidizing substrates of ETC Complexes I and II. Manifestation of decrease in the rate of oxygen consumption following TR57 exposure occurs within 24 hours of treatment. Long-term 72-hour incubation with TR-57 leads to complete inhibition of the respiratory chain of mitochondria in SUM159 cells, as we already observed when studying the effect of ONC201 on BT474 cells [
21].
A decrease in the activity of respiratory chain complexes can be due to both inhibition of activity and a decrease in the content of complexes. As was previously shown in glioblastoma cells, lymphoblastic leukemia cell line and breast cancer cells, substances of the imipridone group and TR-compounds induce a decrease in the levels of enzymes of Complex I and Complex II of the respiratory chain [
9,
13,
14,
15,
16]. We have shown that during the first 24 hours of treatment of SUM159 cells, the protein content of all four complexes of the mitochondrial respiratory chain drops by more than by 50%, and after 72 hours with the agent, no more than 10% of tested proteins remain. At the same time, the content of F
OF
1-ATPase subunits decreased by 40% on the first day and did not change during further incubation, which correlated with changes in mitochondrial mass. This suggests that TR-57 does not affect the distribution of F
OF
1-ATPase in mitochondria. A finding of great importance is that ATPIF1, the natural mitochondrial ATPase inhibitor protein eliminated early in TR-57 treatment. Previously, proteomic analysis revealed downregulation of IF1 in NALM-6 and SUM 159 cells treated with ONC201 [
10,
35], which is opposite an increased expression of IF1, observed in various types of carcinomas, and its role in the metabolic shift from oxidative phosphorylation toward glycolysis has been discussed [
36]. It has been shown that IF1 promotes proliferation, migration and invasion of tumor cells [
36], and selective knockdown of IF1 gene in bladder cancer induced cell proliferation and colony formation [
37]. Interestingly, degradation of IF is carried out by an unidentified serine protease [
38], and ClpP is precisely this type of protease, allowing one to explain the elimination of IF1 TR-57-induced activation of ClpP and that IF1 could play an important role in the antitumor effect of TR-57.
In our experiments, exposure of SUM159 cells to imipridone TR-57 resulted in complete inhibition of mitochondrial respiration and downregulation of key ETC proteins. However, TR-57 treated cells maintained the mitochondrial membrane potential at a relatively high level. In SUM159 cells treated with TR-57 for 24 hours and less, with partial decline in expression of mitochondrial ETC complex proteins and incomplete inhibition of the mitochondrial respiratory chain activity was sufficient to maintain mitochondrial membrane potential. With long-term treatment with TR-57 and associated inhibition of respiration and substantial decline in the expression of the key component of mitochondrial ETC, mitochondrial membrane potential maintained by the reversed activity of F
OF
1-ATPase, as suggested by dissipation of mitochondrial potential in the presence of oligomycin, a pharmacological inhibitor of ATPase. Post-TR-57 level of the mitochondrial membrane potential was significantly lower than that maintained by respiration in intact cells, which is in line with the previous observation that the membrane potential, generated by ATP hydrolysis is lower than that supported by respiratory chain alone [
39].
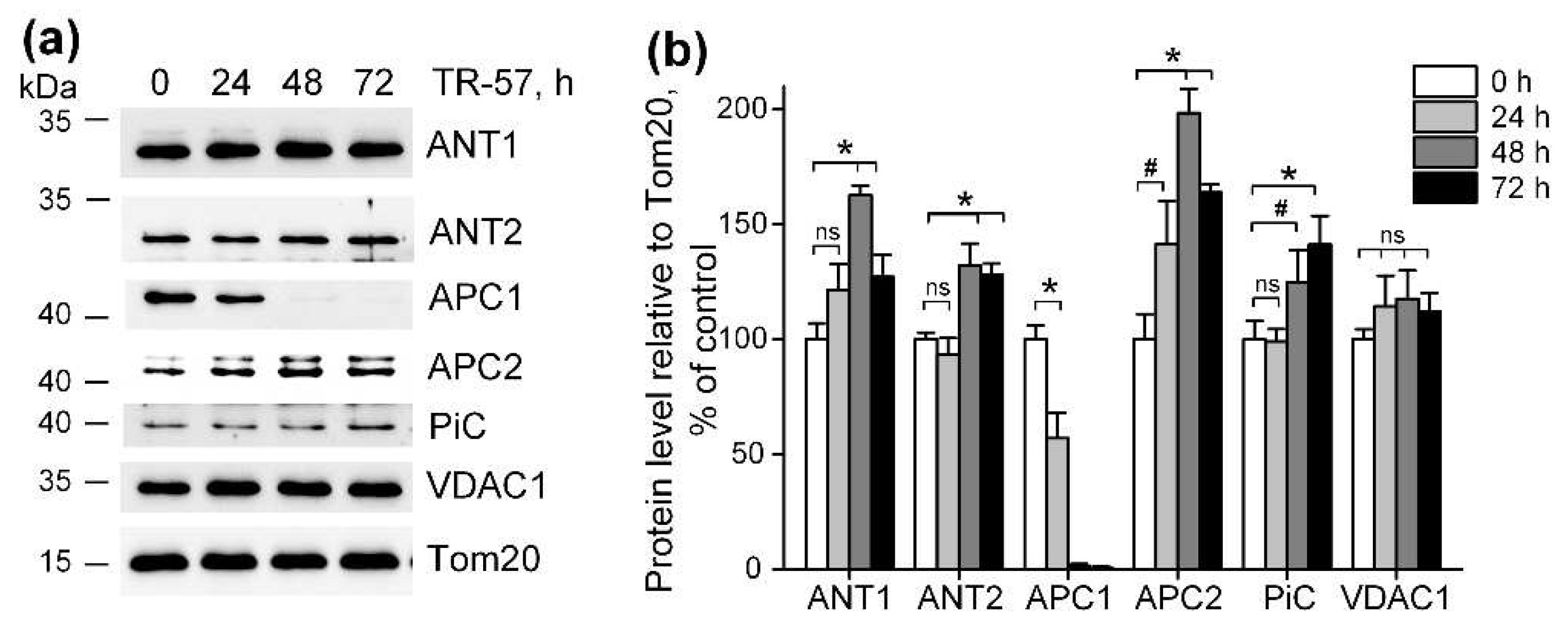
We found that exposure of SUM159 cells to TR-57 for up to 72 hours do not change the expression of VDAC1 and ANT1, ANT2, the major proteins, involved in the transport of ATP into the mitochondria of SUM159 cells. ANTs carry out the electrogenic exchange of ATP for ADP across the membrane; the direction of exchange depends on the concentration of a particular nucleotide on different sides of the membrane [
40]. It have been demonstrated that ANT2 in tumor cells transports glycolytically synthesized ATP in exchange for ADP into the mitochondrial matrix, and even considered as a marker of proliferation in tumor cells [
41]. However, Maldonado et al showed that neither chemical translocase inhibitors nor genetic knockdown of ANT2/3 in tumor cells affected the level of mitochondrial potential maintained by ATP hydrolysis, and concluded that ANT in tumor cells does not participate in transport ATP in mitochondria [
42]. Under conditions of low mitochondrial membrane potential, as well as when it is necessary to quickly replenish the ATP content in mitochondria, ATP transport can occur through calcium-regulated mitochondrial ATP-Mg/Pi carriers (SCaMC, APC1/APC2), presenting in cells as 4 paralogues [
40]. There are publications that the APC1 isoform is mainly present in tumor cells [
43], which is also upregulated in different types of breast cancer cells [
44]. However, in our experiments, we observed a decrease in the level of APC1 during short-term treatment of SUM159 cells with TR-57, and an almost complete disappearance of this protein after 48 hours. At the same time, the APC2 isoform increased almost 2-fold at this time point. The content of the inorganic phosphate transporter, which delivers the phosphate necessary for APC function inside the mitochondria, did not change in the presence of TR-57. This allowed us to conclude that the transport of glycolytic ATP into the mitochondrial matrix of TR-57-treated SUM159 cells occurs through APC2. Apparently, despite the fact that the efficiency of ATP transport through APC2 is an order of magnitude lower than through APC1 [
45], under the conditions of our work APC2 isoform alone could be sufficient to maintain the membrane potential in mitochondria with an impaired respiratory chain.
The mitochondrial membrane potential provides a number of functions important for cell life: oxidative phosphorylation, calcium transport into mitochondria, transport of metabolites across the inner mitochondrial membrane, import of mitochondrial proteins, regulation of mitophagy and cell death [
46]. In tumor cells, mitochondrial potential determines proliferative and metastatic activity and susceptibility to apoptosis [
47]. The question of whether mitochondrial potential is involved in maintaining the viability of TR-57-treated SUM159 cells remains the subject of our further research.
In Summary, the effect of the new antitumor agent TR-57 on mitochondrial population in SUM159 cells resulted in fragmentation of mitochondria and essential decrease in the number of mitochondrial nucleoids/mtDNA. Long-term incubation with TR-57 lead to decreased level of Complexes I-IV proteins and to complete suppression of mitochondrial respiration. However, even long-term exposure of SUM159 cells to TR-57 does not lead to complete dissipation of the mitochondrial membrane potential. Our preliminary data demonstrate enhanced sensitivity of mitochondrial membrane potential (evaluated using TMRM) toward oligomycin. We speculate that increased level of ANT-1/2 and APC2 could result in increased transport of cytoplasmic (glycolytic) ATP into the mitochondrial matrix and used by FOF1-ATPase in reverse mode to support mitochondrial membrane potential in respiratory chain deficient mitochondria. Understanding the mechanism of action of TR-57 on tumor cells opens up new opportunities for its use in pre-clinical and clinical studies in combination with the inhibitors of mitochondrial respiration and membrane potential and/or glycolysis.
4. Materials and Methods
4.1. Chemicals
TR-57 provided by Madera Therapeutics, LLC (Chapel Hill, NC USA). Hoechst 33342, DiOC6(3) and SYBR Green I were obtained from Invitrogen (Waltham, MA, USA), MitoTracker Deep Red 633 and Tetramethylrhodamine methyl ester (TMRM) were from Molecular Probes (Eugenius, OR, USA), DiOC6(3) was from Sigma-Aldrich (St. Louis, MO, USA). DMEM, HEPES, L-glutamine were obtained from PanEco (Russia). Fetal bovine serum was from Gibco (Carlsbad, CA, USA).
Antibodies to ATPIF1, APC2 (SLC25A23), TFAM, GAPDH, Tom20 were purchased from Santa Cruz (CA, USA), antibodies to subunit β, subunit b, subunit c of mitochondrial ATPase, ANT1, ANT2, APC2 (SLC25A24) and Total OXPHOS antibody cocktail were from Abcam (Cambridge, UK), antibody to TUFM was from Invitrogen (Waltham, MA, USA) and antibody to phosphate carrier (SLC25A23) was from FineTest (Wuhan, China). Secondary goat anti rabbit antibody were purchased from PIERCE (USA) and secondary horse anti mouse antibody was from Cell Signaling (Cell Signaling, Danvers, MA, USA).
Other chemicals used in this work were from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. The concentration of the vehicle (DMSO) used as a solvent for hydrophobic agents was kept under 0.5%.
4.2. Cell culture
Human triple negative breast cancer cells SUM159 were obtained from the ATCC (Manassas, VA, USA). SUM159 cells were cultured in DMEM/F12 medium supplemented with 5% fetal bovine serum, 2.4 g/L NaHCO3, 2 mM L-glutamine, 5 µg/mL insulin, 1 μg/ mL hydrocortisone and a 1% mixture of antibiotic-antimycotic in a cell culture incubator (Binder, USA) set at 37°C in a humidified atmosphere of 5%/CO2. SUM159 cells were seeded and cultured overnight in culture dishes (Corning, NY, USA) at density of 15,000-25,000 cells/sm2. Following overnight cell adhesion, incubation media in experimental dishes was replaced with the fresh media supplemented with 150 nM TR-57 and cells were treated with drug for 24, 48 and 72 h.
4.3. Analysis of mitochondrial nucleoids, mitochondrial mass and size
For confocal microscopy experiments, SUM159 cells were plated on 35-mm Petri dishes at density 15,000 cells/cm2, and treated with 150 nM TR-57 for 24, 48 and 72 hours. After treatment, cells were rinsed three times with 2 mL of HBSS and incubated in 2 mL of HBSS, supplemented with 2 µg/mL Hoechst 33342, SYBR Green I at dilution of 1:200000, and 150 nM MitoTracker Deep Red 633 at 37°С for 30 min in CO2-free thermostat. Following staining, the cells were washed 3 times with dye-free HBSS and 3-channel fluorescent images of cells were obtained using laser scanning confocal microscope Leica TCS SP-5 DM6000 CS (Leica Microsystems, Germany), at sequential scanning mode using HCX PL APO lambda blue 63x lens, NA = 1.4, Leica Microsystems, Germany). Excitation and emission were set for Hoechst 33342 405 nm/460 nm, SYBR Green I 488 nm/540 nm, and MitoTracker Deep Red 633 – 633 nm/710 nm. For every sample 5 images from random fields were acquired.
Image analysis was performed using ImageJ software [
24]. We analyzed number of mt-nucleoids per cell, average size of mitochondria as described in [
25]. Briefly, images were separated into three channels: blue (Hoechst 33342), green (SYBG Green I) and red (MitoTracker Deep Red). A mask was created for each channel: mask of nuclei (blue channel), mask of mitochondrial nucleoids (in green channel) and mask of mitochondria (in red channel). Mask of blue channel was used to quantify number of nuclei on the image, green channel – to quantify number of mitochondrial nucleoids and red channel – to quantify average mitochondrial size. Additionally, we analyzed intensity of MitoTracker Deep Red fluorescence as a parameter of mitochondrial mass. For this purpose, we measured fluorescence intensity of MitoTracker Deep Red within mask of mitochondria and divided on the number of nuclei.
4.4. Mitochondrial respiration in permeabilized SUM159 cells
Harvested control and TR-57-treated cells in the amount of 4×106 cell per probe were washed ones with ice-cold PBS, pelleted at 300g for 2 min and resuspended in 1 ml of respiration medium containing 110 mM KCl, 5 mM NaCl, 5 mM KH
2PO
4, 10 mM HEPES (pH 7.4). Incubation medium was supplemented with 5 mM glutamate + 5 mM malate (Complex I substrate) or 10 mM succinate + 1 μg/mL rotenone (Complex II substrate). Cells transferred into 1 mL of closed measuring chamber of multichannel recorder FluoFlux-Æ1 (Econix-Expert Inc., Moscow, Russian Federation, webpage:
http://ionomer.ru) and mitochondrial respiration measured using dissolved oxygen sensor based on the phosphorescence quenching method [
48]. After permeabilization of the cellular plasma membrane with 0.003% digitonin and subsequent additions of 2 mM ADP, 5 μM carboxyatractyloside (CATR), 30 μM 2,4-dinitrophenol (DNP) and 1 mM NaN3 as described in [
21].
4.5. Measurement of mitochondrial membrane potential and mitochondrial mass using flow cytometry
The intact and TR-57-treated SUM159 cells were washed three times with PBS and detached from the surface of the culture plastic using 0.05% trypsin-EDTA solution. Cell aliquots containing equal cell amount (500 000 cells) were stained for 30 min in a CO2 incubator in the full incubation medium (DMEM/F12) with 10 nM 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)) to measure mitochondrial membrane potential or with 50 nM MitoTracker Deep Red to assess mitochondrial mass. Experimental data obtained using BD Accuri C6 flow cytometer (BD Bioscience, USA) were processed using BD Accuri C6 CFlow software (BD Bioscience, USA) and resulting histograms were plotted using FlowJo v10 software (BD Bioscience, USA).
4.6. Membrane potential measurement in intact cells using fluorescence microscopy
SUM159 cells were plated on 0.13 mm thick 25-mm round coverslips (Menzel-Gläser, Germany) in 35 mm Petri dishes at a density of 10,000 cells/cm2 and treated with 150 nM TR-57 for 24, 48 and 72 hours. Following TR-57 treatment coverslips with cells were incubated in 1 ml of complete incubation medium containing 20 mM HEPES (PanEco) and 20 nM TMRM for 30 minutes in a cell culture incubator (95/5% air/CO2). After staining, the incubation media was replaced with a similar one containing 20 nM TMRM, and the cells were transferred to a microscopic stage. A Leica DMI6000 B fluorescence microscope (Leica microsystems, Germany) equipped with Hamamatsu EM-CCD digital camera C9100 (Hamamatsu, Japan), the OSRAM HXP R 120W/45C UV mercury lamp, HC PL APO 20x/0.70 water immersion lens, green excitation filter (530 nm), red 640 mm emission filter were used to evaluate TMRM fluorescence. Exposure time was set to 100 ms. 2x2 camera binning, gain 1100, an additional magnification lens 1.6x and 50% neutral density filter were used to decrease excitation light intensity. Time-lapse images were acquired frame by frame with an interval of 1 frame per minute. The baseline (initial TMRM fluorescence) was taken for 5 minutes, then 2 μM oligomycin was added to the cells and the change in TMRM fluorescence was recorded for 5 minutes, after that 5 μM antimycin A and 1 μM CCCP uncoupler were added and the images were acquired for 5 minutes each. Obtained stacks were analyzed using FiJi software as described [
24]. The level of mitochondrial membrane potential was determined as intensity of TMRM fluorescence averaged from 5 min of baseline. For tracking of changes in mitochondrial membrane potential in each series, 10 regions of cell mitochondria and 10 corresponding regions of nuclei and 1 region of the extracellular environment were selected, and the fluorescence of TMRM was measured. The relative level of membrane potential (Δ𝜓) was calculated using the following formula:
Δψ = (Fmt – Fbackground)/(Fnucleus – Fbackground), where Fmt is the average fluorescence level of the mitochondrial region of the cell, Fnucleus is the average fluorescence level of the cell nucleus area, Fbackground is the average fluorescence level of the extracellular area. Obtained values of membrane potential at each point were normalized to baseline level (referred as 100%) and CCCP (referred as 0%).
4.7. Western blot analysis of mitochondrial proteins
Intact and TR-57-treated cells (4×106) were trypsinized, washed twice in PBS, pelleted at 300×g for 5 min and lysed in RIPA Lysing Buffer System with 1 mM Na3VO4, 2 mM PMSF, and a complete protease inhibitor cocktail (Santa Cruz, CA, USA). Samples were solubilized in Laemmli loading buffer (BioRad, Hercules, CA, USA) and boiled at 95°C for 5 min (except samples for Total OXPHOS detection heated at 37°C for 5 min). The concentration of protein in samples was measured by Bradford method and 30 μg of each sample were subjected to PAAG electrophoresis followed by transfer to nitrocellulose membrane. Then membranes were blocked with 5% dry milk in PBST for 1 h at room temperature and incubated overnight at 4°C with primary antibodies against target proteins. After incubation with corresponding secondary antibodies conjugated with HRP for 1h at room temperature target proteins were visualized with ClarityWestern ECL substrate (Bio-Rad, Hercules, CA, USA). Chemiluminescence was detected with Chemidoc Touch Imaging System (Bio-Rad, Hercules, CA, USA) and Image Lab software was used for the processing and quantification of obtained results.
4.8. Statistical analysis
All experiment performed at least in triplicates were analyzed using the ANOVA one-way variance analysis with a post hoc Bonferroni test [
21] and values are presented as mean ± SD and statistical significance of p<0.05.
Author Contributions
Conceptualization, E.H. and I.O.; methodology, A.M.,E.M., M.K., E.H.; software, A.M., A.B..; formal analysis, A.M., I.O.; investigation, A.M., E.M., A.V.B., M.K., Y.L., I.O..; resources, A.B., I.O.; writing—original draft preparation, A.M., I.O.; writing—review and editing, A.V.B., E.H., I.O.; visualization, A.M., M.K., Y.L., I.O; supervision, E.H., I.O.; project administration, E.H. and I.O.; funding acquisition, I.O. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Effect of TR-57 on the number of mitochondrial nucleoids, mitochondrial morphology and size and the expression of key mitochondrial regulatory proteins of SUM159 cells. (a) representative confocal fluorescent images of intact (0 h) and treated with 150 nM TR-57 for 72 hours (72 h) SUM159 cells and loaded with Hoechst 33342 (Hoechst, blue), SYBR Green I (SYBR, green) and MitoTracker Deep Red (MTDR, red); Scale bar – 20 µm. Number of mitochondrial nucleoids (b), average mitochondrial size (c) and average intensity of MitoTracker Deep Red fluorescence (d) in SUM159 cells treated with 150 nM TR-57 for 0 (intact), 24, 48 and 72 hours measured by confocal fluorescent microscopy (CM); (e) average intensity of MTDR fluorescence measured by flow cytometry (FC); (f) Western-blot analysis of mitochondrial proteins TUFM, TFAM and Tom20 in SUM159 cells treated with 150 nM TR-57 for 0, 24, 48 and 72 hours. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. * – p < 0.05, ns – p > 0.05.
Figure 1.
Effect of TR-57 on the number of mitochondrial nucleoids, mitochondrial morphology and size and the expression of key mitochondrial regulatory proteins of SUM159 cells. (a) representative confocal fluorescent images of intact (0 h) and treated with 150 nM TR-57 for 72 hours (72 h) SUM159 cells and loaded with Hoechst 33342 (Hoechst, blue), SYBR Green I (SYBR, green) and MitoTracker Deep Red (MTDR, red); Scale bar – 20 µm. Number of mitochondrial nucleoids (b), average mitochondrial size (c) and average intensity of MitoTracker Deep Red fluorescence (d) in SUM159 cells treated with 150 nM TR-57 for 0 (intact), 24, 48 and 72 hours measured by confocal fluorescent microscopy (CM); (e) average intensity of MTDR fluorescence measured by flow cytometry (FC); (f) Western-blot analysis of mitochondrial proteins TUFM, TFAM and Tom20 in SUM159 cells treated with 150 nM TR-57 for 0, 24, 48 and 72 hours. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. * – p < 0.05, ns – p > 0.05.
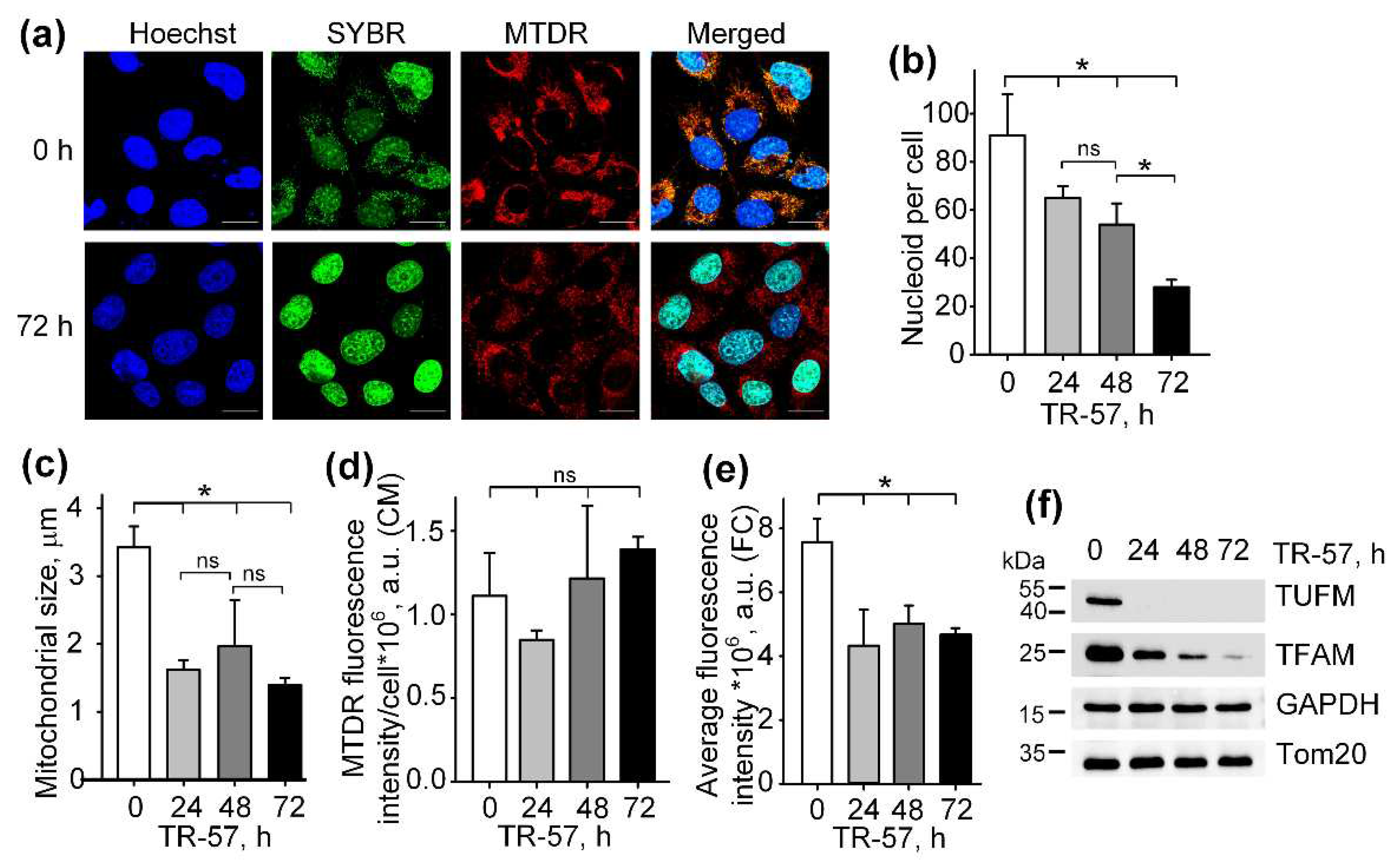
Figure 2.
TR-57 induces gradual decrease in mitochondrial respiratory chain activity and electron transport chain (ETC) proteins content. (a-b) representative curves and oxygen consumption rates SUM159 cells, control (white bars) and treated with 150 nM TR-57 for 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars), oxidizing Complex I substrates; (c-d) representative curves and oxygen consumption rates of SUM159 cells oxidizing Complex II substrates; Arrows on curves indicate additions of 0.003% digitonin, 2 mM ADP, 5 μM carboxyatractyloside (CATR), 30 μM 2,4-dinitrophenol (DNP) and 1 mM NaN3; (e) representative Western blot of ETC complex proteins; (f) quantitative analysis of ETC proteins normalized to GAPDH level. The ratio of protein level to GAPDH under control conditions was taken as 100%. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, # p > 0.01, “ns” – non significant.
Figure 2.
TR-57 induces gradual decrease in mitochondrial respiratory chain activity and electron transport chain (ETC) proteins content. (a-b) representative curves and oxygen consumption rates SUM159 cells, control (white bars) and treated with 150 nM TR-57 for 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars), oxidizing Complex I substrates; (c-d) representative curves and oxygen consumption rates of SUM159 cells oxidizing Complex II substrates; Arrows on curves indicate additions of 0.003% digitonin, 2 mM ADP, 5 μM carboxyatractyloside (CATR), 30 μM 2,4-dinitrophenol (DNP) and 1 mM NaN3; (e) representative Western blot of ETC complex proteins; (f) quantitative analysis of ETC proteins normalized to GAPDH level. The ratio of protein level to GAPDH under control conditions was taken as 100%. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, # p > 0.01, “ns” – non significant.
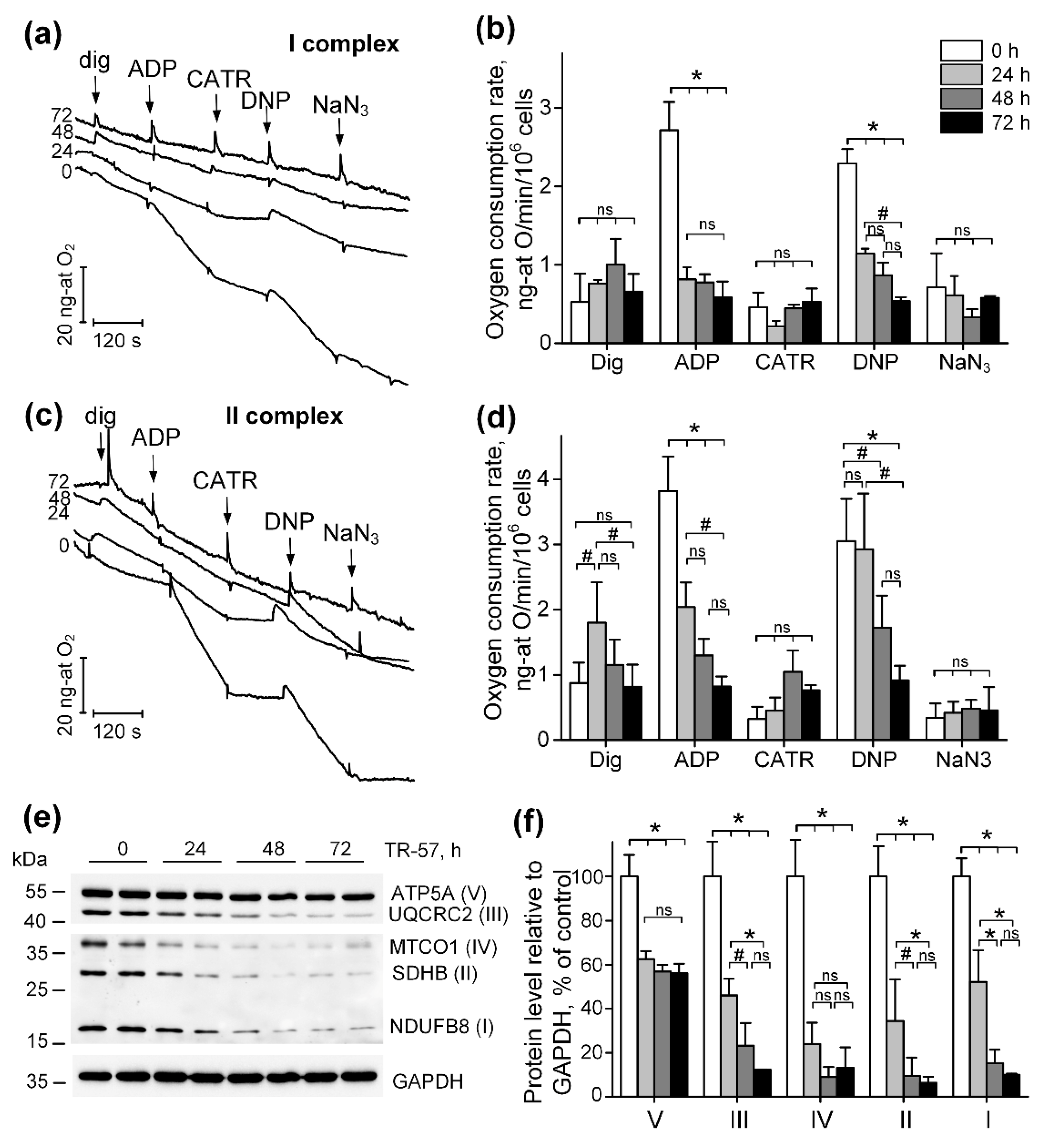
Figure 3.
Effect of TR-57 treatment on mitochondrial membrane potential of SUM159 cells. (a) Representative confocal fluorescent images of SUM159 cells treated with 150 nM TR-57 for 0 (control), 24, 48 and 72 hours and stained with 20 nM TMRM; (b) Average intensity of TMRM in mitochondria of SUM159 from panel (a); (c-d) Mitochondrial membrane potential of control (0) and TR-59 treated SUM159 cells measured with DiOC3(6) fluorescent dye by flow cytometry; (e) Representative confocal fluorescent images of SUM159 cells treated with 150 nM TR-57 for 0 (control) and 72 hours, stained with 20 nM TMRM and sequentially exposed to 2 µM Oligomycin (Oligo), 5 µM Antimycin A (AntA) and 1 µM CCCP; (f) Relative mitochondrial membrane potential of SUM159 from panel E during sequential addition of Oligo, AntA and CCCP, fluorescence of TMRM was normalized to 100% baseline and 0% after CCCP addition. (g-h) Representative Western blots and quantitative analysis of FOF1-ATPase proteins of SUM159 cells following treatment with 150 nM TR-57 for 0 (white bars), 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars). Tom20 was used as loading control. The ratio of protein level to Tom20 under control conditions was taken as 100%. Scale bar is 20 µm. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, # p > 0.01, “ns” – non significant.
Figure 3.
Effect of TR-57 treatment on mitochondrial membrane potential of SUM159 cells. (a) Representative confocal fluorescent images of SUM159 cells treated with 150 nM TR-57 for 0 (control), 24, 48 and 72 hours and stained with 20 nM TMRM; (b) Average intensity of TMRM in mitochondria of SUM159 from panel (a); (c-d) Mitochondrial membrane potential of control (0) and TR-59 treated SUM159 cells measured with DiOC3(6) fluorescent dye by flow cytometry; (e) Representative confocal fluorescent images of SUM159 cells treated with 150 nM TR-57 for 0 (control) and 72 hours, stained with 20 nM TMRM and sequentially exposed to 2 µM Oligomycin (Oligo), 5 µM Antimycin A (AntA) and 1 µM CCCP; (f) Relative mitochondrial membrane potential of SUM159 from panel E during sequential addition of Oligo, AntA and CCCP, fluorescence of TMRM was normalized to 100% baseline and 0% after CCCP addition. (g-h) Representative Western blots and quantitative analysis of FOF1-ATPase proteins of SUM159 cells following treatment with 150 nM TR-57 for 0 (white bars), 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars). Tom20 was used as loading control. The ratio of protein level to Tom20 under control conditions was taken as 100%. Scale bar is 20 µm. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, # p > 0.01, “ns” – non significant.
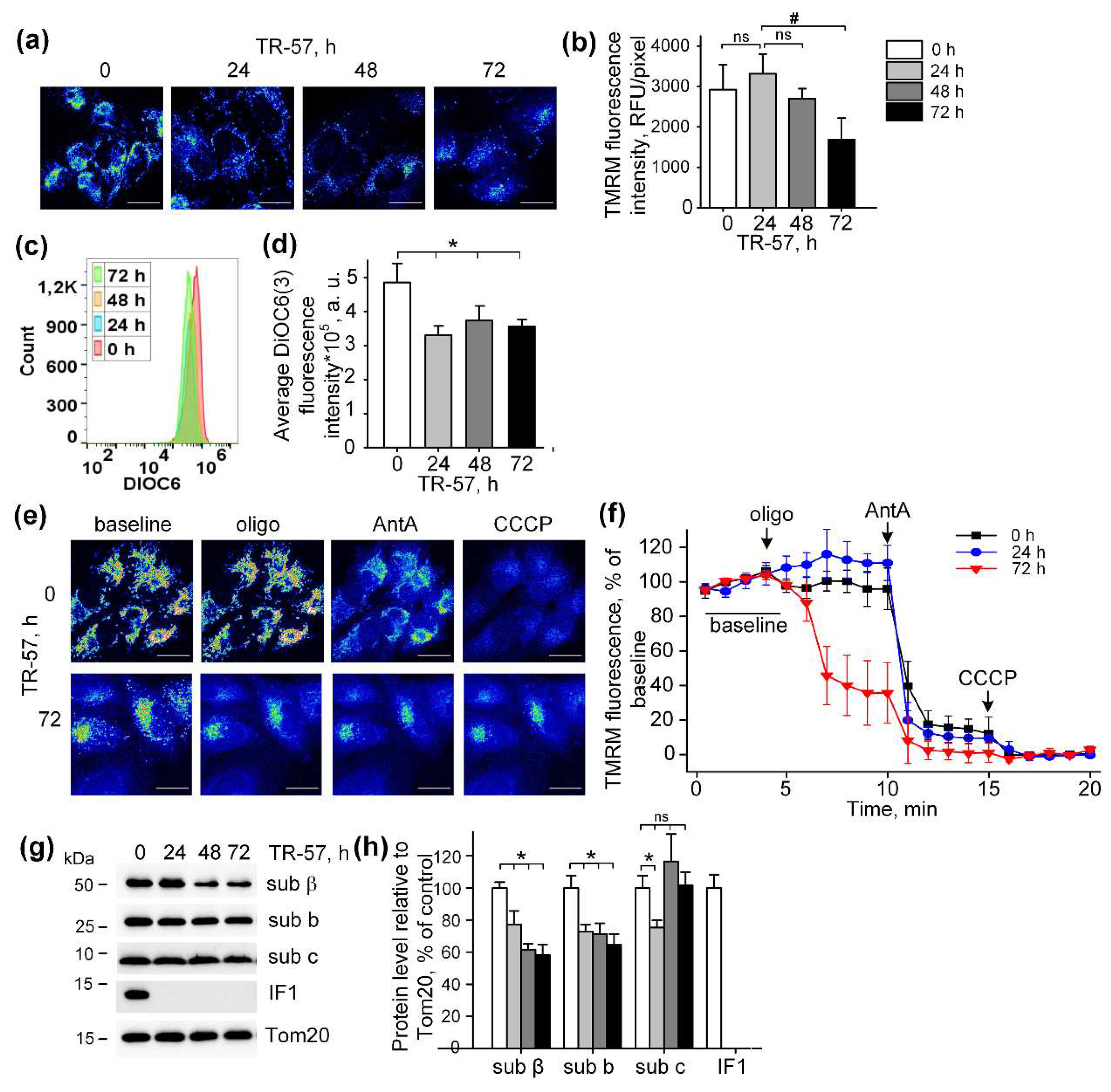
Figure 4.
Effect of TR-57 on mitochondrial proteins responsible for ATP and phosphate ion transport. (a) Representative Western blots of proteins of control (white bars) SUM159 cells and cells treated with 150 nM TR-57 for 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars); (b) – quantitative analysis of target protein normalized to Tom20. The ratio of protein level to Tom20 under control conditions was taken as 100%. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, “ns” – non significant.
Figure 4.
Effect of TR-57 on mitochondrial proteins responsible for ATP and phosphate ion transport. (a) Representative Western blots of proteins of control (white bars) SUM159 cells and cells treated with 150 nM TR-57 for 24 h (light gray bars), 48 h (dark gray bars) and 72 h (black bars); (b) – quantitative analysis of target protein normalized to Tom20. The ratio of protein level to Tom20 under control conditions was taken as 100%. The data are presented as the means ± SD of at least three independent experiments. The data were analyzed using a one-way ANOVA with post-hoc Bonferroni test. *p > 0.001, “ns” – non significant.