Submitted:
06 December 2023
Posted:
07 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Mutant U2OS cell line establishment
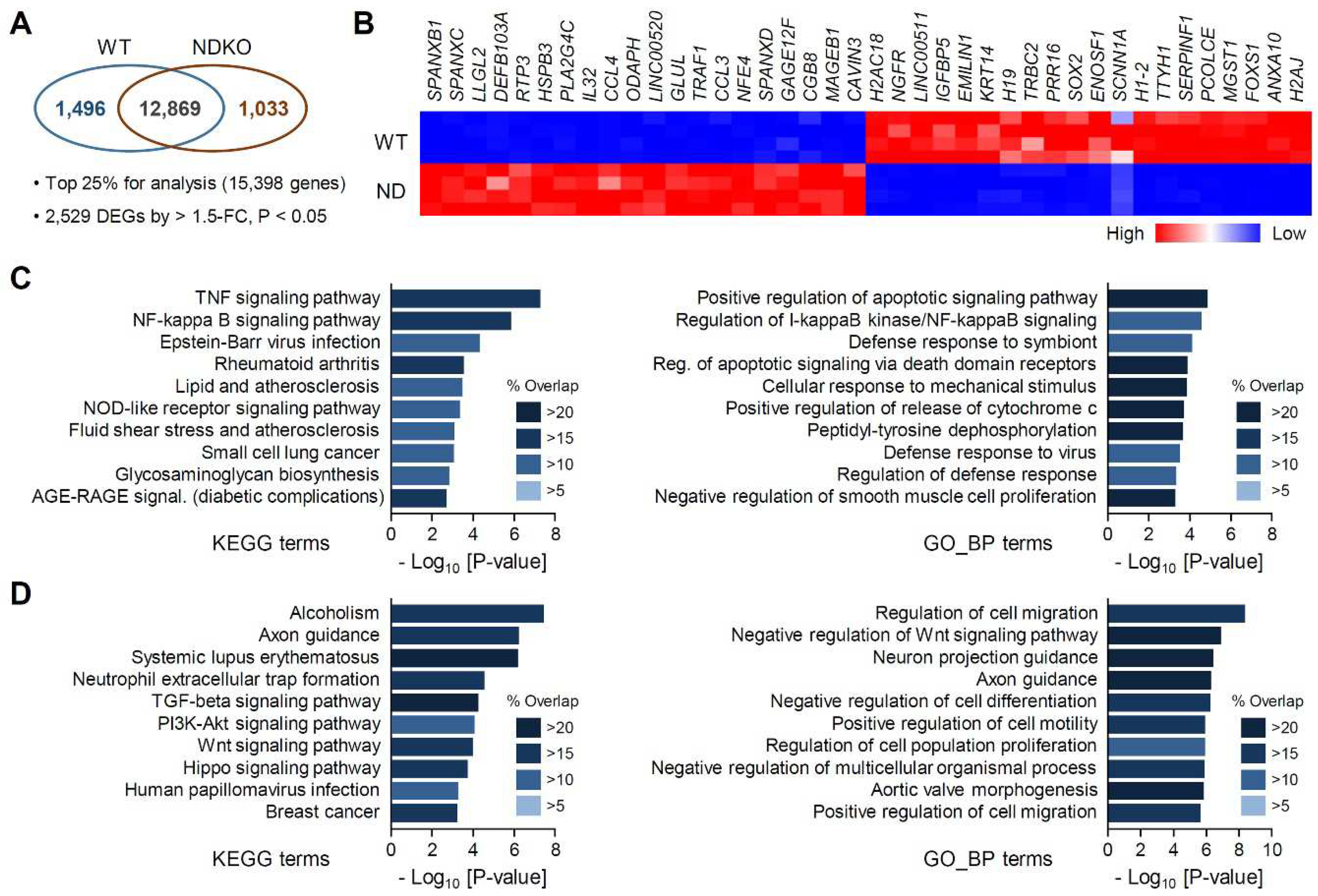
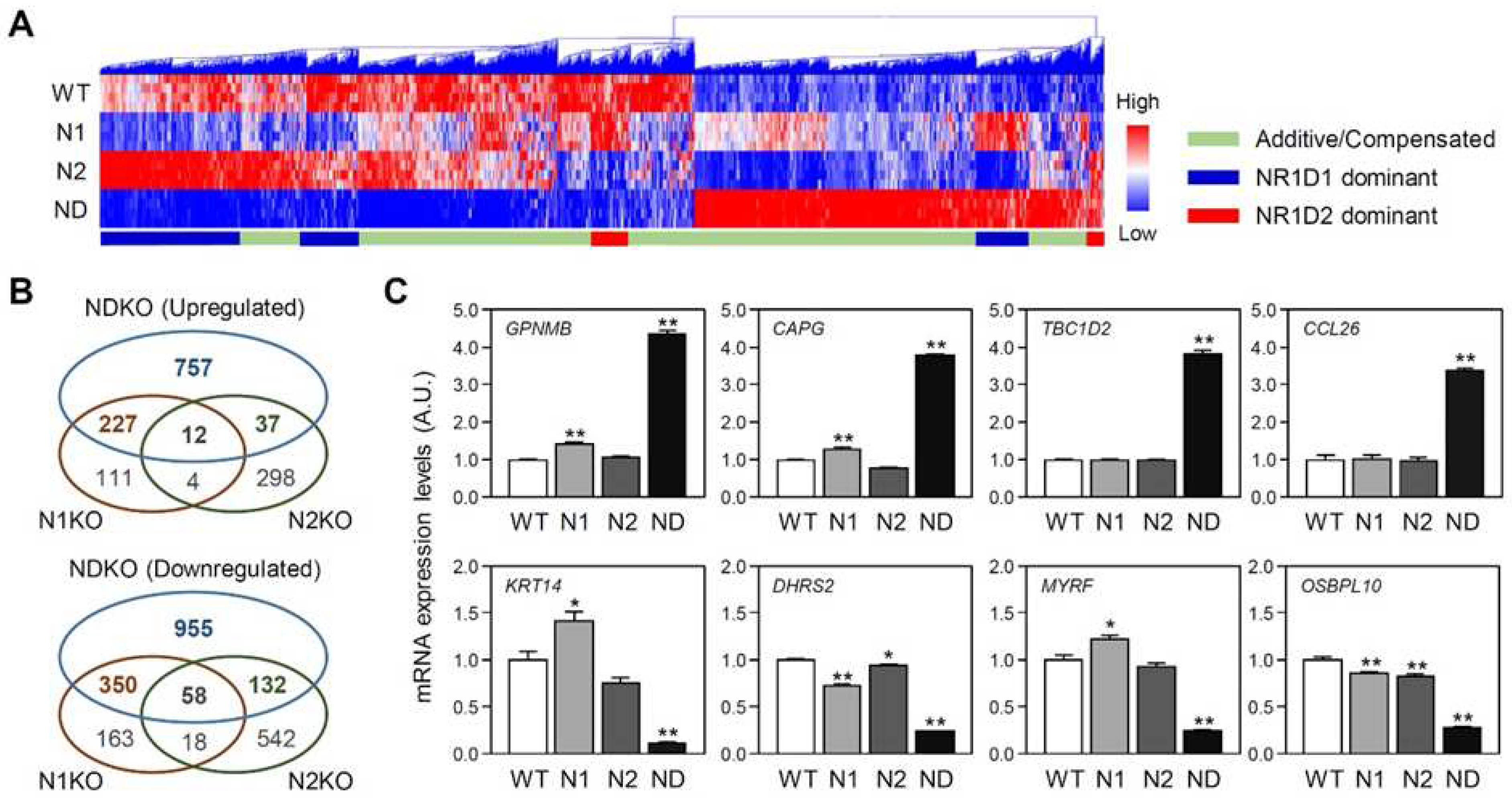
2.2. Differentially expressed gene characterization in NDKO cells
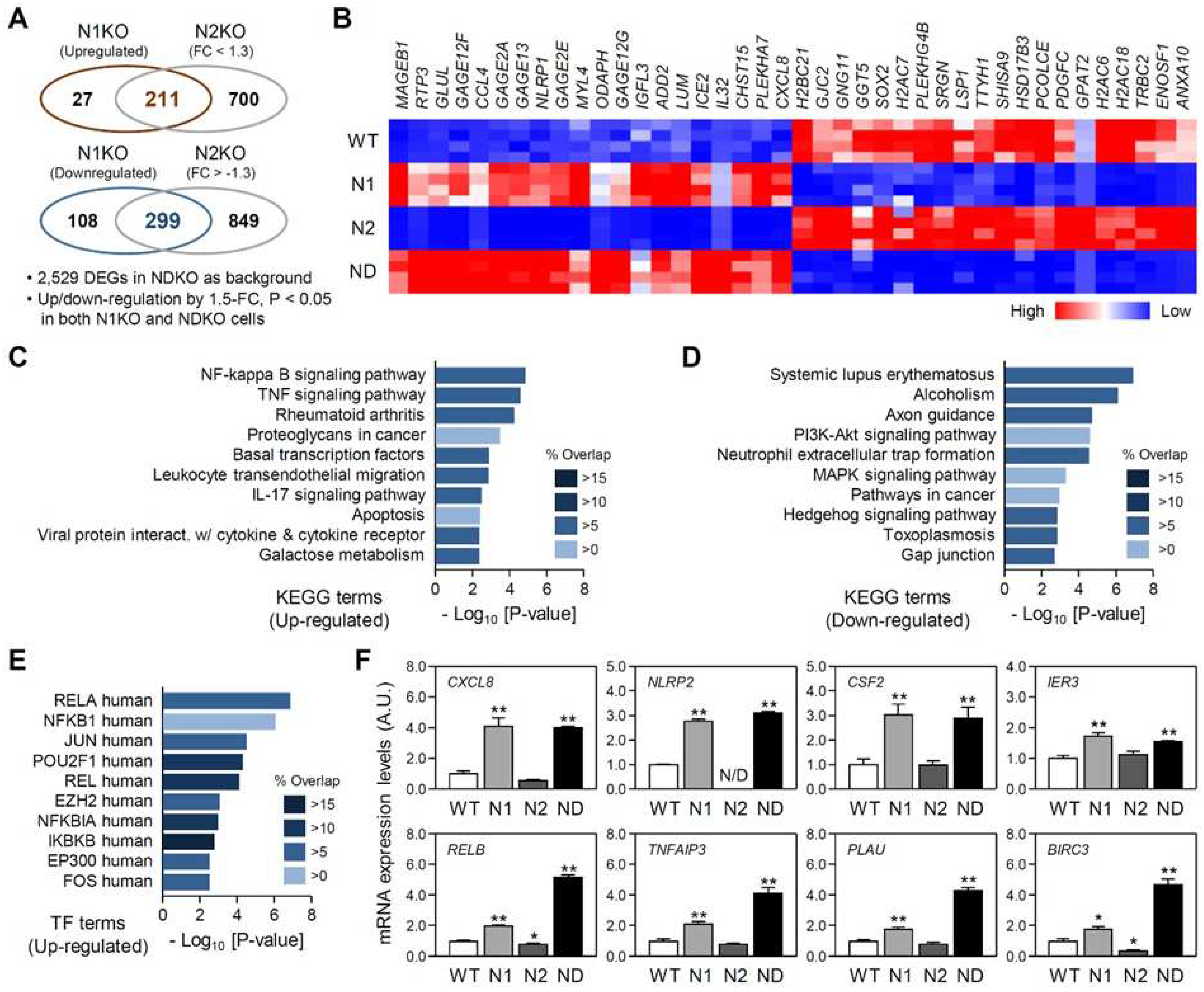
2.3. Isoform-specific target genes among the NDKO DEGs
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Cell cultures and generation of mutant cell lines
4.3. Total RNA sample preparation and reverse transcription-quantitative PCR (RT-qPCR)
4.4. Real-time bioluminescence monitoring and luciferase reporter assay
4.5. RNA sequencing (RNA-seq) and gene enrichment analyses
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed]
- Kojetin, D.J.; Burris, T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug. Discov. 2014, 13, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, F.; Lin, Y.; Wu, B. Targeting REV-ERBα for therapeutic purposes: promises and challenges. Theranostics 2020, 10, 4168. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.C.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Welsh, D.K.; Kay, S.A. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008, 4, e1000023. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.-W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Stratmann, M.; Schibler, U. REV-ERBs: more than the sum of the individual parts. Cell Metab. 2012, 15, 791–793. [Google Scholar] [CrossRef]
- Raghuram, S.; Stayrook, K.R.; Huang, P.; Rogers, P.M.; Nosie, A.K.; McClure, D.B.; Burris, L.L.; Khorasanizadeh, S.; Burris, T.P.; Rastinejad, F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 2007, 14, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.I.; Xu, X.; Reinking, J.; Schuetz, A.; Dong, A.; Liu, S.; Zhang, R.; Tiefenbach, J.; Lajoie, G.; Plotnikov, A.N. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. PLoS Biol. 2009, 7, e1000043. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Yin, L.; Collins, J.L.; Parks, D.J.; Orband-Miller, L.A.; Wisely, G.B.; Joshi, S.; Lazar, M.A.; Willson, T.M.; Zuercher, W.J. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem. Biol. 2010, 5, 925–932. [Google Scholar] [CrossRef]
- Kojetin, D.; Wang, Y.; Kamenecka, T.M.; Burris, T.P. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem. Biol. 2011, 6, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Solt, L.A.; Wang, Y.; Banerjee, S.; Hughes, T.; Kojetin, D.J.; Lundasen, T.; Shin, Y.; Liu, J.; Cameron, M.D.; Noel, R. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012, 485, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Wang, Y.; Solt, L.A.; Griffett, K.; Kazantzis, M.; Amador, A.; El-Gendy, B.M.; Huitron-Resendiz, S.; Roberts, A.J.; Shin, Y. Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat. Commun. 2014, 5, 5759. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Chaudhari, S.; Wang, R.; Campbell, S.; Mosure, S.A.; Chopp, L.B.; Lu, Q.; Shang, J.; Pelletier, O.B.; He, Y. REV-ERBα regulates TH17 cell development and autoimmunity. Cell Rep. 2018, 25, 3733–3749.e3738. [Google Scholar] [CrossRef]
- Hering, Y.; Berthier, A.; Duez, H.; Lefebvre, P.; Deprez, B.; Gribbon, P.; Wolf, M.; Reinshagen, J.; Halley, F.; Hannemann, J. Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα. J. Biol. Methods 2018, 5, e94. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhou, C.; Xu, H.; Zhang, T.; Wu, B. Chronopharmacological targeting of Rev-erbα by puerarin alleviates hyperhomocysteinemia in mice. Biomed. Pharmacother. 2020, 125, 109936. [Google Scholar] [CrossRef]
- Chung, S.; Lee, E.J.; Yun, S.; Choe, H.K.; Park, S.-B.; Son, H.J.; Kim, K.-S.; Dluzen, D.E.; Lee, I.; Hwang, O. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 2014, 157, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, B.; Emmett, M.J.; Damle, M.; Sun, Z.; Feng, D.; Armour, S.M.; Remsberg, J.R.; Jager, J.; Soccio, R.E. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 2015, 348, 1488–1492. [Google Scholar] [CrossRef]
- Hirota, T.; Lee, J.W.; Lewis, W.G.; Zhang, E.E.; Breton, G.; Liu, X.; Garcia, M.; Peters, E.C.; Etchegaray, J.-P.; Traver, D. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 2010, 8, e1000559. [Google Scholar] [CrossRef]
- Crumbley, C.; Wang, Y.; Kojetin, D.J.; Burris, T.P. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J. Biol. Chem. 2010, 285, 35386–35392. [Google Scholar] [CrossRef]
- Crumbley, C.; Burris, T.P. Direct regulation of CLOCK expression by REV-ERB. PloS One 2011, 6, e17290. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics 2013, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Lazar, M.A. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol. Cell. Biol. 1995, 15, 4791–4802. [Google Scholar] [CrossRef] [PubMed]
- Adlanmerini, M.; Lazar, M.A. The REV-ERB nuclear receptors: Timekeepers for the core clock period and metabolism. Endocrinology 2023, 164, bqad069. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Liu, T.; Sun, Z.; Bugge, A.; Mullican, S.E.; Alenghat, T.; Liu, X.S.; Lazar, M.A. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011, 331, 1315–1319. [Google Scholar] [CrossRef] [PubMed]
- Bugge, A.; Feng, D.; Everett, L.J.; Briggs, E.R.; Mullican, S.E.; Wang, F.; Jager, J.; Lazar, M.A. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012, 26, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Amador, A.; Campbell, S.; Kazantzis, M.; Lan, G.; Burris, T.P.; Solt, L.A. Distinct roles for REV-ERBα and REV-ERBβ in oxidative capacity and mitochondrial biogenesis in skeletal muscle. PLoS One 2018, 13, e0196787. [Google Scholar] [CrossRef]
- De Mei, C.; Ercolani, L.; Parodi, C.; Veronesi, M.; Vecchio, C.L.; Bottegoni, G.; Torrente, E.; Scarpelli, R.; Marotta, R.; Ruffili, R. Dual inhibition of REV-ERBβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 2015, 34, 2597–2608. [Google Scholar] [CrossRef]
- Le Martelot, G.; Claudel, T.; Gatfield, D.; Schaad, O.; Kornmann, B.; Sasso, G.L.; Moschetta, A.; Schibler, U. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009, 7, e1000181. [Google Scholar] [CrossRef]
- Dierickx, P.; Zhu, K.; Carpenter, B.J.; Jiang, C.; Vermunt, M.W.; Xiao, Y.; Luongo, T.S.; Yamamoto, T.; Martí-Pàmies, Í.; Mia, S. Circadian REV-ERBs repress E4bp4 to activate NAMPT-dependent NAD+ biosynthesis and sustain cardiac function. Nat. Cardiovasc. Res. 2022, 1, 45–58. [Google Scholar] [CrossRef]
- Negoro, H.; Kanematsu, A.; Doi, M.; Suadicani, S.O.; Matsuo, M.; Imamura, M.; Okinami, T.; Nishikawa, N.; Oura, T.; Matsui, S. Involvement of urinary bladder Connexin43 and the circadian clock in coordination of diurnal micturition rhythm. Nat. Commun. 2012, 3, 809. [Google Scholar] [CrossRef] [PubMed]
- Negoro, H.; Okinami, T.; Kanematsu, A.; Imamura, M.; Tabata, Y.; Ogawa, O. Role of Rev-erbα domains for transactivation of the connexin43 promoter with Sp1. FEBS Lett. 2013, 587, 98–103. [Google Scholar] [CrossRef]
- Yang, Z.; Tsuchiya, H.; Zhang, Y.; Lee, S.; Liu, C.; Huang, Y.; Vargas, G.M.; Wang, L. REV-ERBα activates C/EBP homologous protein to control small heterodimer partner–mediated oscillation of alcoholic fatty liver. Am. J. Pathol. 2016, 186, 2909–2920. [Google Scholar] [CrossRef] [PubMed]
- Son, G.H.; Chung, S.; Choe, H.K.; Kim, H.-D.; Baik, S.-M.; Lee, H.; Lee, H.-W.; Choi, S.; Sun, W.; Kim, H. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc. Natl. Acad. Sci. 2008, 105, 20970–20975. [Google Scholar] [CrossRef]
- Kim, S.H.; Son, G.H.; Seok, J.Y.; Chun, S.K.; Yun, H.; Jang, J.; Suh, Y.-G.; Kim, K.; Jung, J.-W.; Chung, S. Identification of a novel class of cortisol biosynthesis inhibitors and its implications in a therapeutic strategy for hypercortisolism. Life Sci. 2023, 325, 121744. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




