Submitted:
04 December 2023
Posted:
08 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA extraction
2.3. SSR Markers Genotyping
2.3.1. SSR markers and Polymerase Chain Reaction (PCR)
2.3.2. Gel electrophoresis
2.3.3. Band Scoring
2.4. Analysis of Genetic Diversity
2.5. Analysis of Genetic Structure
3. Results
3.1. Genetic Diversity Parameters
| Markers | MaF | AnL | He | Ho | FIT | FIS | FST | PIC |
|---|---|---|---|---|---|---|---|---|
| SSRY4 | 0.35 | 5 | 0.74 | 0.65 | 0.05 | 0.03 | 0.02 | 0.69 |
| SSRY9 | 0.67 | 4 | 0.51 | 0.53 | -0.01 | -0.05 | 0.04 | 0.48 |
| SSRY12 | 0.51 | 3 | 0.58 | 0.98 | -0.70 | -0.74 | 0.02 | 0.49 |
| SSRY19 | 0.58 | 4 | 0.58 | 0.75 | -0.24 | -0.31 | 0.05 | 0.51 |
| SSRY20 | 0.50 | 6 | 0.68 | 0.77 | -0.16 | -0.20 | 0.04 | 0.65 |
| SSRY21 | 0.72 | 4 | 0.43 | 0.54 | -0.25 | -0.28 | 0.02 | 0.39 |
| SSRY34 | 0.94 | 2 | 0.12 | 0.11 | 0.20 | 0.19 | 0.01 | 0.11 |
| SSRY38 | 0.98 | 3 | 0.03 | 0.03 | -0.02 | -0.08 | 0.06 | 0.03 |
| SSRY51 | 0.38 | 3 | 0.66 | 0.90 | -0.35 | -0.35 | 0.00 | 0.59 |
| SSRY59 | 0.46 | 4 | 0.62 | 0.98 | -0.57 | -0.58 | 0.01 | 0.54 |
| SSRY63 | 0.60 | 3 | 0.55 | 0.81 | -0.46 | -0.50 | 0.02 | 0.48 |
| SSRY64 | 0.70 | 2 | 0.42 | 0.25 | 0.39 | 0.33 | 0.09 | 0.33 |
| SSRY69 | 0.50 | 5 | 0.68 | 0.62 | 0.13 | 0.12 | 0.01 | 0.64 |
| SSRY82 | 0.40 | 4 | 0.68 | 0.78 | -0.12 | -0.15 | 0.03 | 0.62 |
| SSRY100 | 0.36 | 5 | 0.69 | 0.63 | 0.02 | 0.02 | 0.00 | 0.63 |
| SSRY102 | 0.78 | 2 | 0.35 | 0.45 | -0.26 | -0.26 | 0.00 | 0.29 |
| SSRY103 | 0.58 | 3 | 0.57 | 0.60 | -0.03 | -0.05 | 0.01 | 0.51 |
| SSRY105 | 0.93 | 3 | 0.14 | 0.13 | 0.18 | 0.16 | 0.03 | 0.13 |
| SSRY106 | 0.72 | 3 | 0.43 | 0.47 | -0.11 | -0.15 | 0.04 | 0.37 |
| SSRY108 | 0.76 | 4 | 0.39 | 0.48 | -0.23 | -0.22 | -0.01 | 0.36 |
| SSRY110 | 0.97 | 2 | 0.06 | 0.06 | -0.02 | -0.02 | 0.00 | 0.06 |
| SSRY135 | 0.69 | 3 | 0.47 | 0.40 | 0.09 | 0.04 | 0.05 | 0.43 |
| SSRY147 | 0.61 | 2 | 0.48 | 0.66 | -0.36 | -0.37 | 0.01 | 0.36 |
| SSRY148 | 0.95 | 2 | 0.09 | 0.09 | -0.05 | -0.09 | 0.03 | 0.08 |
| SSRY151 | 0.32 | 5 | 0.73 | 0.87 | -0.17 | -0.16 | -0.01 | 0.68 |
| SSRY155 | 0.58 | 2 | 0.49 | 0.26 | 0.47 | 0.45 | 0.02 | 0.37 |
| SSRY161 | 0.97 | 2 | 0.06 | 0.06 | -0.03 | -0.04 | 0.01 | 0.06 |
| SSRY169 | 0.96 | 2 | 0.08 | 0.08 | -0.04 | -0.06 | 0.02 | 0.08 |
| SSRY177 | 0.47 | 4 | 0.67 | 0.46 | 0.28 | 0.27 | 0.00 | 0.61 |
| SSRY179 | 0.63 | 3 | 0.52 | 0.21 | 0.47 | 0.43 | 0.06 | 0.44 |
| SSRY180 | 0.84 | 2 | 0.27 | 0.28 | -0.13 | -0.17 | 0.03 | 0.23 |
| SSRY182 | 0.45 | 4 | 0.65 | 0.53 | 0.17 | 0.12 | 0.06 | 0.58 |
| Mean | 0.65 | 3.3 | 0.45 | 0.48 | -0.07 | -0.10 | 0.03 | 0.40 |
3.2. Population Structure and Genetic Relationships
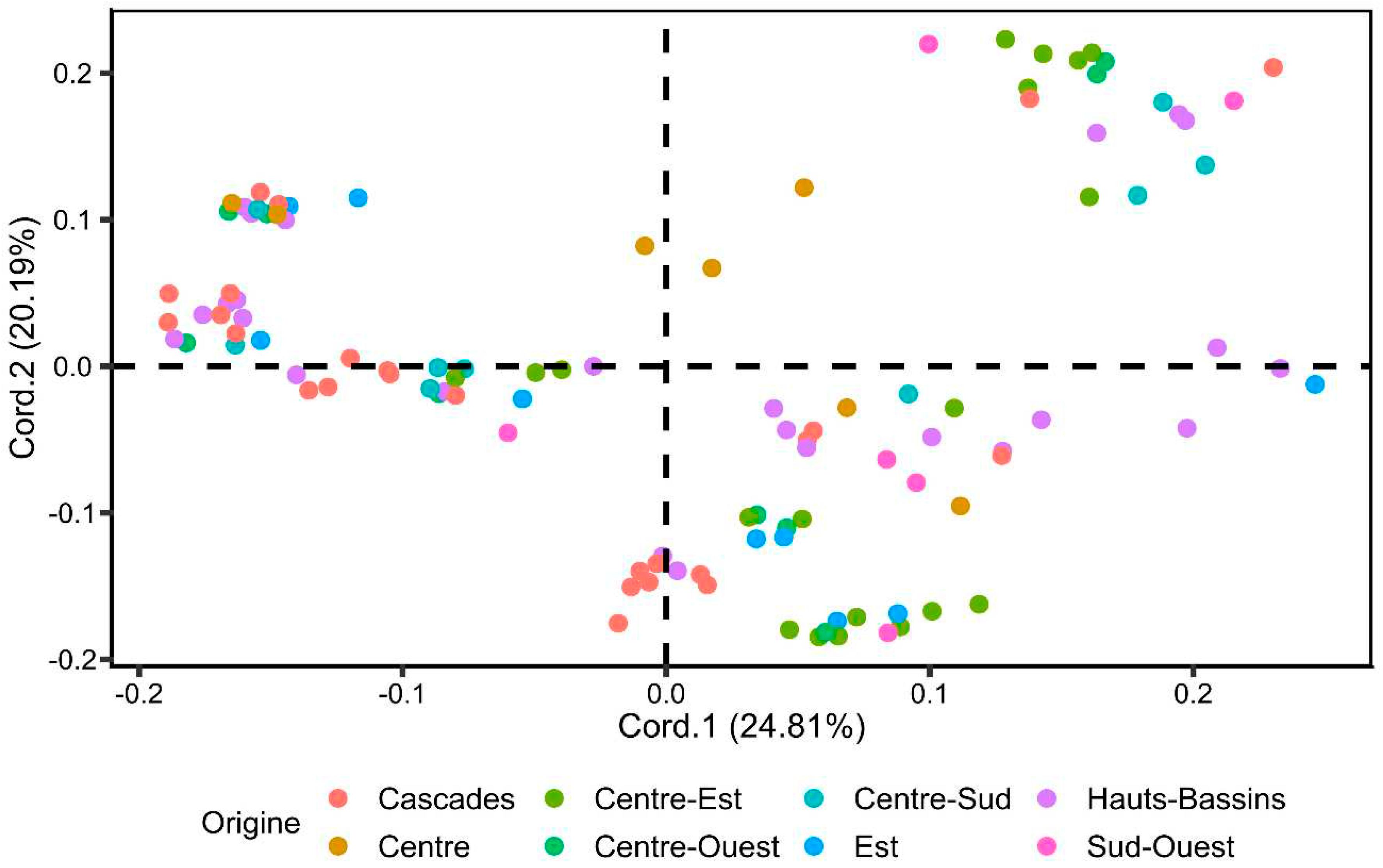
3.2.1. Principal Coordinates Analysis (PCoA)
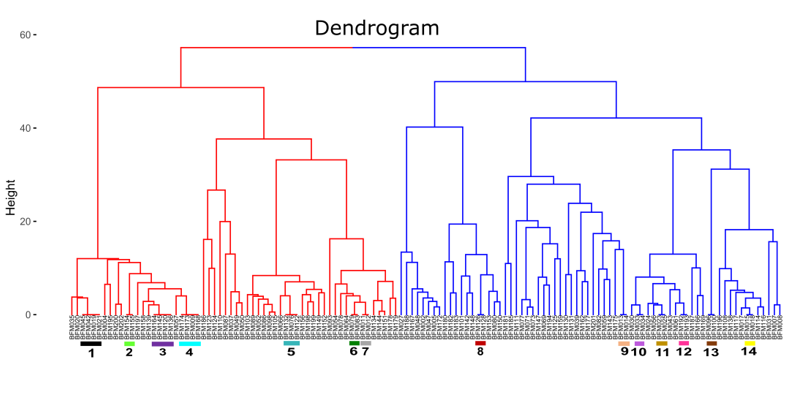
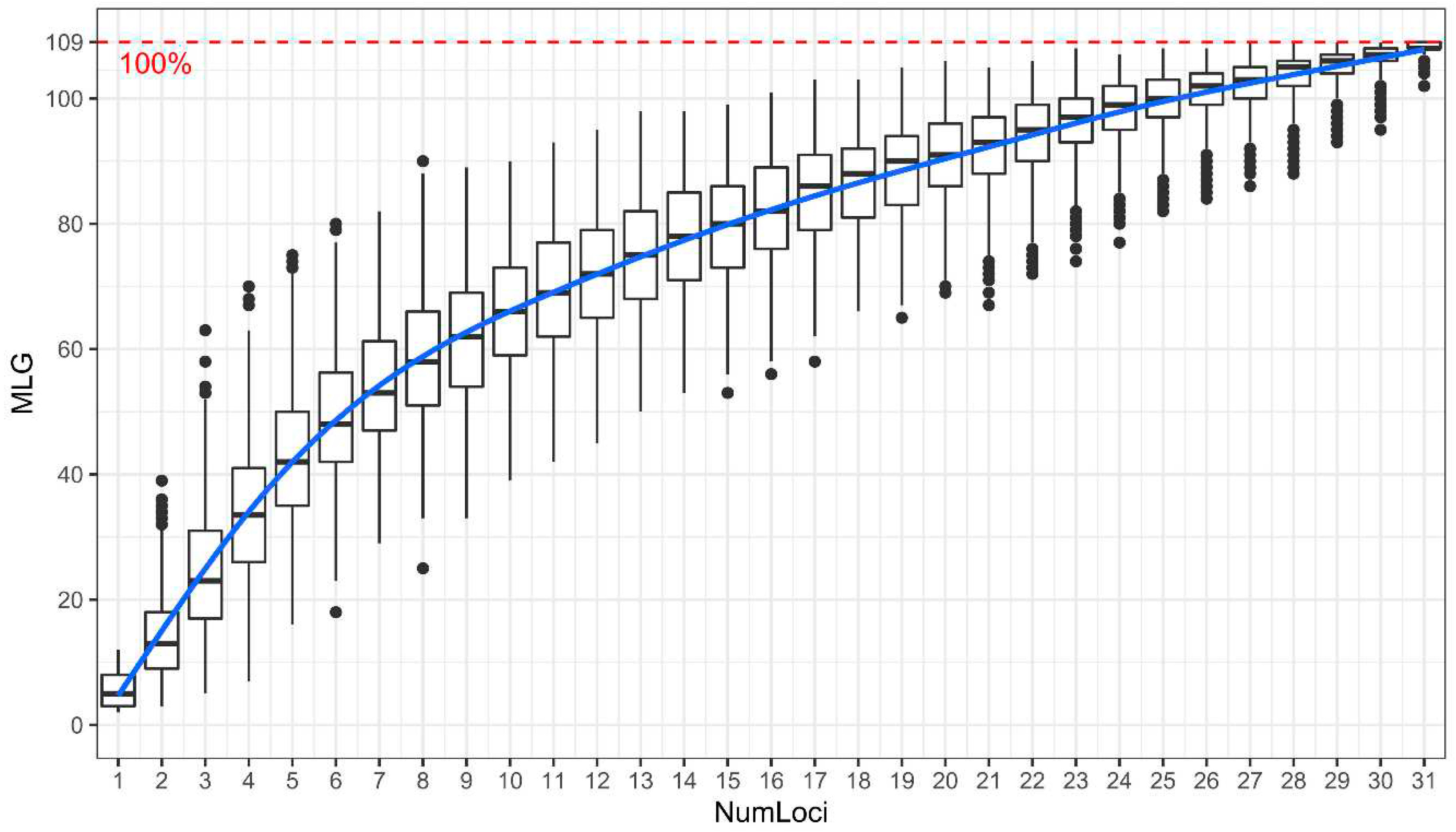
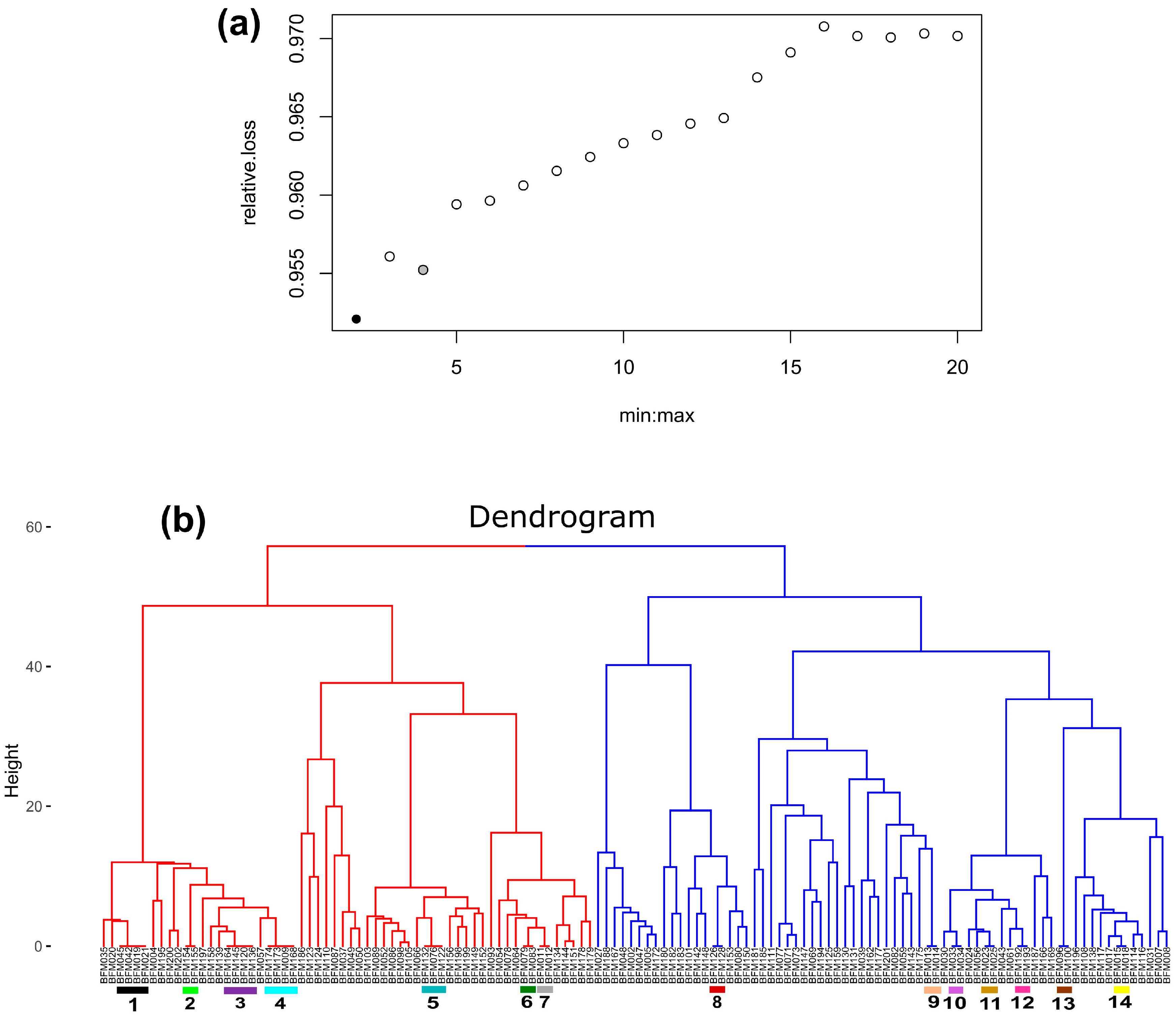
3.2.2. Hierarchical Clustering Analysis and Identification of Duplicate Accessions
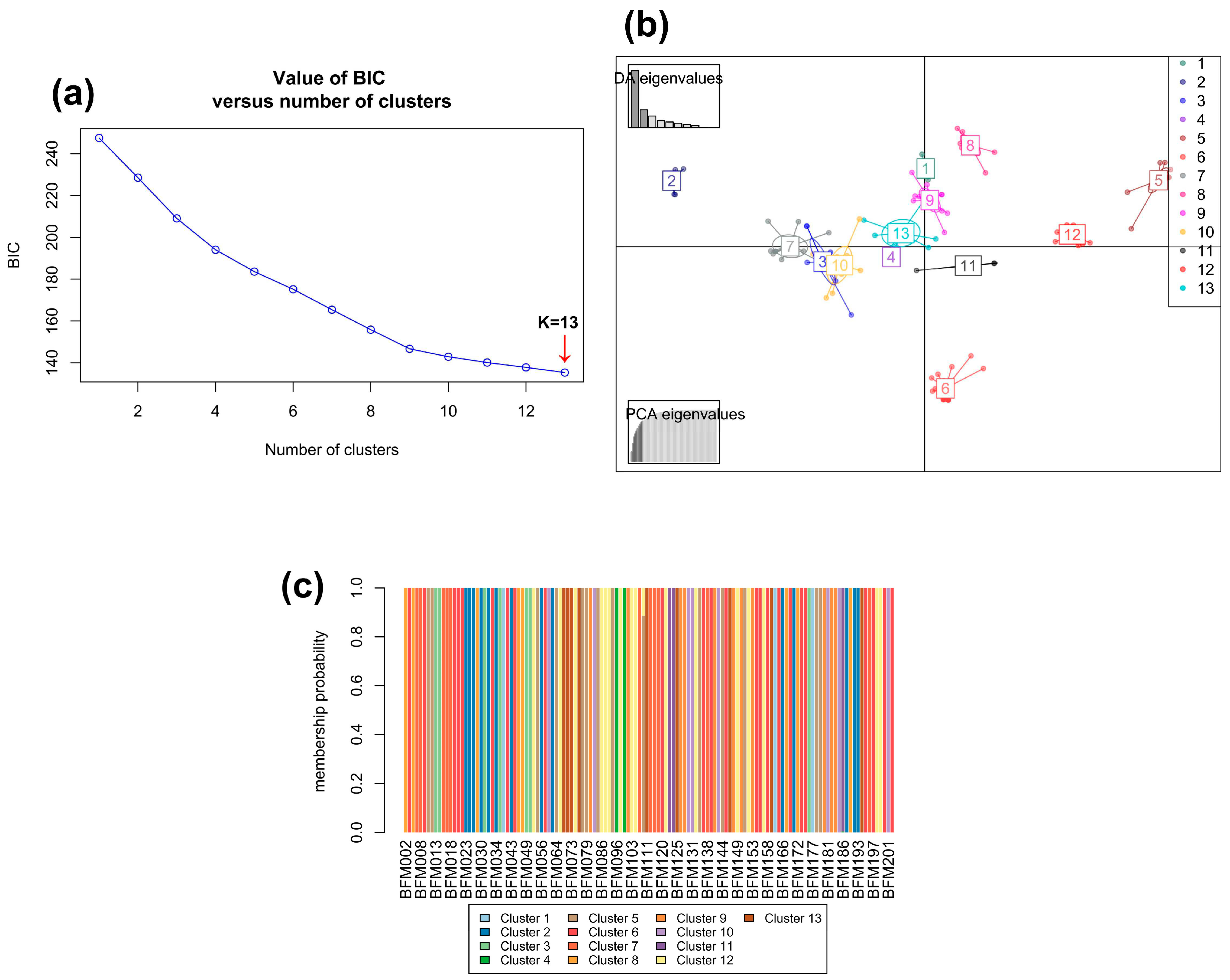
3.2.3. Bayesian analysis
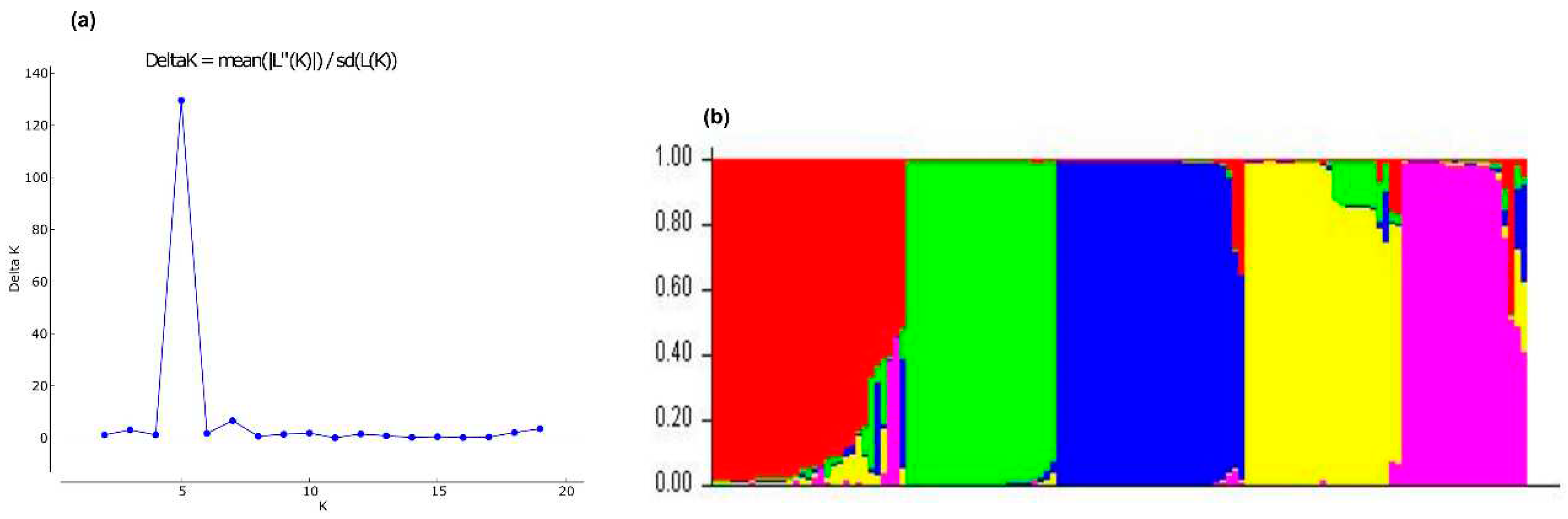
3.2.4. Discriminant Analysis of Principal Components (DAPC)
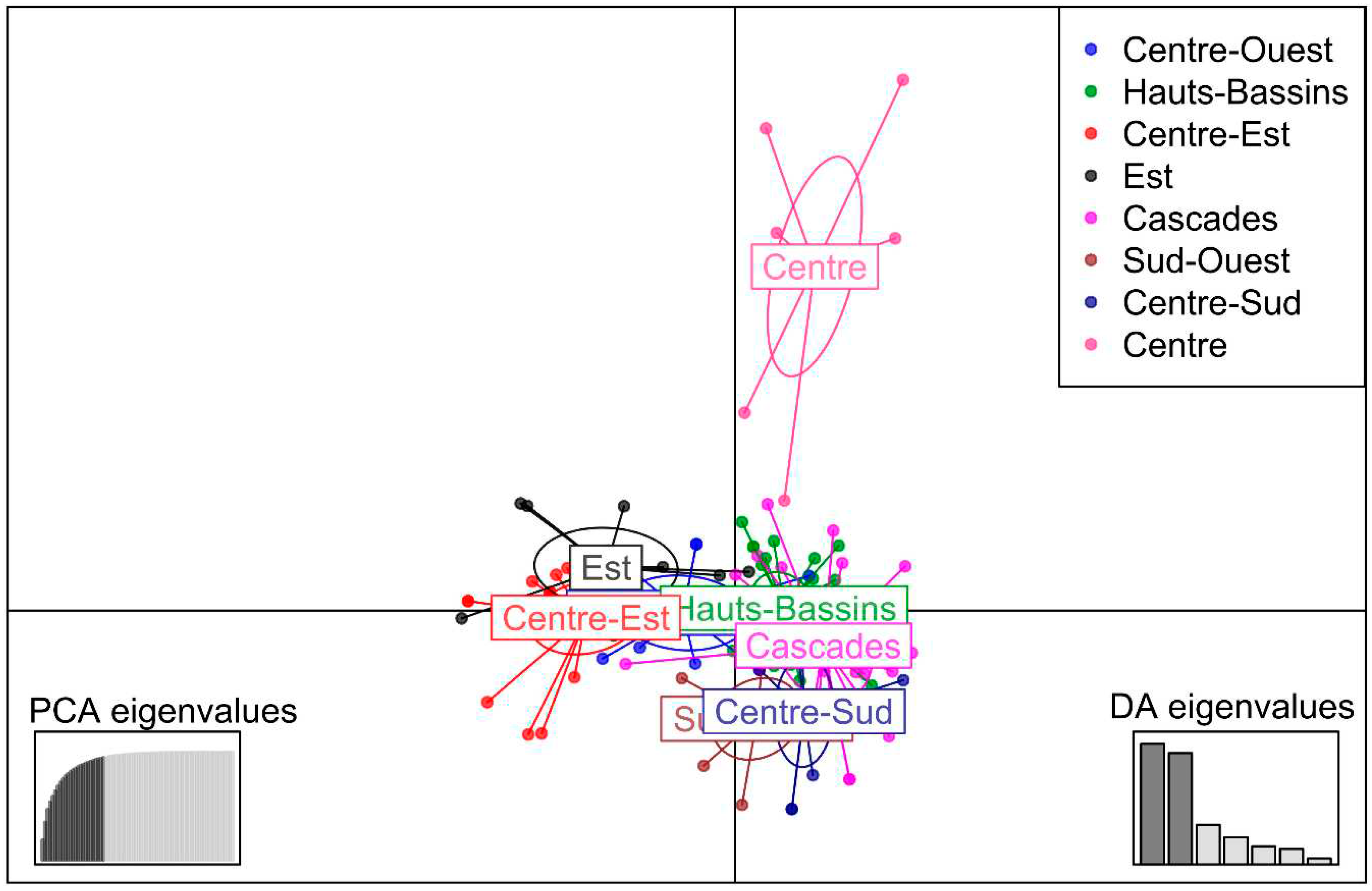
3.2.4. Analysis of Molecular Variance (AMOVA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adjebeng-Danquah, J.; Manu-Aduening, J.; Asante, I.K.; Agyare, R.Y.; Gracen, V.; Offei, S.K. Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon. 2020, 6, 1–9. [CrossRef]
- Montagnac, J.A.; Davis, C.R.; Tanumihardjo, S.A. Nutritional value of cassava for use as a staple food and recent advances for improvement. Compr Rev Food Sci Food Saf. 2009, 8, 181–194. [CrossRef]
- Salvador, E.M.; Steenkamp, V.; McCrindle, C.M.E. Production, consumption and nutritional value of cassava (Manihot esculenta, Crantz) in Mozambique: An overview. J Agric Biotechnol Sustain Dev. 2014, 6, 29–38. [CrossRef]
- Latif, S.; Müller, J. Potential of cassava leaves in human nutrition: A review. Trends Food Sci Technol. 2015, 44, 147–158. [CrossRef]
- Ally, H.M.; Hamss, H.; Simiand, C.; Maruthi, M.N.; Colvin, J.; Omongo, C.A.; Delatte, H. What has changed in the outbreaking populations of the severe crop pest whitefly species in cassava in two decades? Sci Rep. 2019, 9, 1–13. [CrossRef]
- FAOSTAT. Food and agriculture organization o the united nations statistics division. Https://www.fao.org/faostat/en/#data/QCL. 2023, 30/10/2023,.
- El-Sharkawy, M.A. Physiological characteristics of cassava tolerance to prolonged drought in the tropics: Implications for breeding cultivars adapted to seasonally dry and semiarid environments. Brazilian J Plant Physiol. 2007, 19, 257–286. [CrossRef]
- El-Sharkawy, M.A. Cassava biology and physiology. Plant Mol Biol. 2004, 56, 481–501. [CrossRef]
- Oliveira, E.J. de.; Ferreira, C.F.; Silva, S.D.V.; Jesus, O.D.N.; Oliveira, G.A.F.; Silva, M.D.S. Potential of SNP markers for the characterization of Brazilian cassava germplasm. Theor Appl Genet. 2014, 127, 1423–1440. [CrossRef]
- Elias, M.; Penet, L.; Vindry, P.; McKey, D.; Panaud, O.; Robert, T. Unmanaged sexual reproduction and the dynamics of genetic diversity of a vegetatively propagated crop plant, cassava (Manihot esculenta Crantz), in a traditional farming system. Mol Ecol. 2001, 10, 1895–1907. [CrossRef]
- Park, Y.; Dixit, A.; Ma, K.; Kang, J.; Rao, V.R.; Cho, E. On-farm Conservation Strategy to Ensure Crop Genetic Diversity in Changing Agro-ecosystems in the Republic of Korea. J Agron Crop Sci. 2005, 191, 401–410.
- Rao, S.A.; Bounphanousay, C.; Schiller, J.M.; Alcantara, A.P.; Jackson, M.T. Naming of traditional rice varieties by farmers in the Lao PDR. Genet Resour Crop Evol. 2002, 49, 83–88.
- Ferguson, M.E.; Shah, T.; Kulakow, P.; Ceballos, H. A global overview of cassava genetic diversity. PLoS One. 2019, 14, 1–16. Available: . [CrossRef]
- Kawuki, R.S.; Ferguson, M.; Labuschagne, M.T.; Herselman, L.; Orone, J.; Ralimanana, I.; Bidiaka, M.; Lukombo, S.; Kanyange, M.C.; Gashaka, G.; et al. Variation in qualitative and quantitative traits of cassava germplasm from selected national breeding programmes in sub-Saharan Africa. F Crop Res. 2011, 122, 151–156. [CrossRef]
- Kamanda, I.; Blay, E.T.; Asante, I.K.; Danquah, A.; Ifie, B.E.; Parkes, E.; Kulakow, P.; Rabbi, I.; Conteh, A.; Kamara, J.S.; et al. Genetic diversity of provitamin-A cassava (Manihot esculenta Crantz) in Sierra Leone. Genet Resour Crop Evol. 2020, 67, 1193–1208. [CrossRef]
- Gonçalves, T.M.; Filho, P.S.V.; Vidigal, M.C.G.; Ferreira, R.C.U.; Rocha, V.P.C.; Ortiz, A.H.T.; Moiana, L.D.; Kvitschal, M.V. Genetic diversity and population structure of traditional sweet cassava accessions from Southern of Minas Gerais State , Brazil , using microsatellite markers. African J Biotechnol. 2017, 16, 346–358. [CrossRef]
- Albuquerque, H.D.Y.G.; Oliveira, E.D.J.; Brito, A.C.; Andrade, L.D.R.B.; Carmo, C.D.D.; Morgante, C.V.; Vieira, E.A.; Moura, E.F.; Faleiro, F.G. Identification of duplicates in cassava germplasm banks based on single-nucleotide polymorphisms (SNPs). Sci Agric. 2019, 76, 328–336. [CrossRef]
- Prempeh, R.N.A.; Manu-Aduening, J.A.; Quain, M.D.; Asante, I.K.; Offei, S.K.; Danquah, E.Y. Assessment of genetic diversity among cassava landraces using single nucleotide polymorphic markers. African J Biotechnol. 2020, 19, 383–391. [CrossRef]
- Asare, P.A.; Galyuon, I.K.A.; Sarfo, J.K.; Tetteh, J.P. Morphological and molecular based diversity studies of some cassava (Manihot esculenta crantz) germplasm in Ghana. African J Biotechnol. 2011, 10, 13900–13908. [CrossRef]
- Mezette, T.F.; Blumer, C.G.; Veasey, E.A. Morphological and molecular diversity among cassava genotypes. Pesqui Agropecuária Bras. 2013, 48, 510–518. [CrossRef]
- Fregene, M.; Angel, F.; Gómez, R.; Rodriguez, F.; Chavarriaga, P.; Roca, W.; Tohme, J.; Bonierbale, M. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor Appl Genet. 1997, 95, 431–441.
- Elias, M.; Panaud, O.; Robert, T. Assessment of genetic variability in a traditional cassava (Manihot esculenta Crantz) farming system, using AFLP markers. Heredity. 2000, 85, 219–230. [CrossRef]
- Kawuki, R.S.; Herselman, L.; Labuschagne, M.T.; Nzuki, I.; Ralimanana, I.; Bidiaka, M.; Kanyange, M.C.; Gashaka, G.; Masumba, E.; Mkamilo, G.; et al. Genetic diversity of cassava (Manihot esculenta Crantz) landraces and cultivars from southern, eastern and central Africa. Plant Genet Resour. 2013, 11, 170–181. [CrossRef]
- Albuquerque, H.D.Y.G.; Carmo, C.D.D.; Brito, A.C.; Oliveira, E.D.J. Genetic diversity of Manihot esculenta Crantz germplasm based on single-nucleotide polymorphism markers. Ann Appl Biol. 2018, 173, 271–284. [CrossRef]
- Adu, B.G.; Yeboah, A.; Akromah, R.; Bobobee, E.; Amoah, S.; Kena, A.W.; Amoah, R.A. Whole genome SNPs and phenotypic characterization of cassava (Manihot esculenta Crantz) germplasm in the semi-deciduous forest ecology of Ghana. Ecol Genet Genomics. 2020, 17, 1–12. [CrossRef]
- Karim, K.Y.; Ifie, B.; Dzidzienyo, D.; Danquah, E.Y.; Blay, E.T.; Whyte, J.B.A.; Kulakow, P.; Rabbi, I.; Parkes, E.; Omoigui, L.; et al. Genetic characterization of cassava (Manihot esculenta Crantz) genotypes using agro-morphological and single nucleotide polymorphism markers. Physiol Mol Biol Plants. 2020, 26, 317–330. [CrossRef]
- Nelimor, C.; Badu-Apraku, B.; Garcia-Oliveira, A.L.; Tetteh, A.; Paterne, A.; N’guetta, A.S.-P.; Gedil, M. Genomic Analysis of Selected Maize Landraces from Sahel and Coastal West Africa Reveals Their. Genes. 2020, 11, 1–14.
- Pierre, N.; Wamalwa, L.N.; Muiru, W.M.; Simon, B.; Kanju, E.; Ferguson, M.E.; Ndavi, M.M.; Tumwegamire, S. Genetic diversity of local and introduced cassava germplasm in Burundi using DArTseq molecular analyses.2022, 1–19. [CrossRef]
- Li, X.; Qiao, L.; Chen, B.; Zheng, Y.; Zhi, C.; Zhang, S.; Pan, Y. Plant Diversity SSR markers development and their application in genetic diversity evaluation of garlic ( Allium sativum ) germplasm. Plant Divers. 2022, 44, 481–491. [CrossRef]
- Luo, Z.; Yao, Z.; Yang, Y.; Wang, Z.; Zou, H.; Zhang, X.; Chen, J. Genetic fingerprint construction and genetic diversity analysis of sweet potato ( Ipomoea batatas ) germplasm resources. BMC Plant Biol. 2023, 1–14. [CrossRef]
- Suvi, W.T.; Shimelis, H.; Laing, M.; Mathew, I.; Titus, W.; Shimelis, H.; Laing, M.; Mathew, I. Assessment of the genetic diversity and population structure of rice genotypes using SSR markers.2020, 4710,. [CrossRef]
- Soro, M.; Pita, J.S.; Somé, K.; Otron, D.H.; Yéo, E.; Mutuku, J.M.; Néya, J.B.; Tiendrébéogo, F.; Koné, D. Genomic analysis and identification of potential duplicate accessions in Burkina Faso cassava germplasm based on single nucleotide polymorphism. Front Sustain Food Syst. 2023, 7, 1–15. [CrossRef]
- Singh, N.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Singh, N.K.; Singh, R. Comparison of SSR and SNP Markers in Estimation of Genetic Diversity and Population Structure of Indian Rice Varieties.2013, 8, 1–14. [CrossRef]
- Permingeat, H.R.; Romagnoli, M. V.; Juliana, I.; Vallejos, R.H. A Simple Method for Isolating DNA of High Yield and Quality from Cotton (Gossypium hirsutum L.) Leaves. Plant Mol Biol Report. 1998, 16, 89.
- Mba, R.E.C.; Stephenson, P.; Edwards, K.; Melzer, S.; Nkumbira, J.; Gullberg, U.; Apel, K.; Gale, M.; Tohme, J.; Fregene, M. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: Towards an SSR-based molecular genetic map of cassava. Theor Appl Genet. 2001, 102, 21–31. [CrossRef]
- Twumasi, P.; Acquah, E.W.; Quain, M.D.; Parkes, E.Y. Use of simple sequence repeat ( SSR ) markers to establish genetic relationships among cassava cultivars released by different research groups in Ghanaian. Int J Genet Mol Biol. 2014, 6, 29–36. [CrossRef]
- Beovides, Y.; Fregene, M.; Gutiérrez, J.P.; Milián, M.D.; Coto, O.; Buitrago, C.; Cruz, J.A.; Ruiz, E.; Basail, M.; Rayas, A.; et al. Molecular diversity of Cuban cassava (Manihot esculenta Crantz) cultivars assessed by simple sequences repeats (SSR). Biotechnol Agron Soc Environ. 2015, 19, 364–377. Available: http://www.scopus.com/inward/record.url?eid=2-s2.0-84948747783&partnerID=40&md5=0c80b85a06bead1492b9c6b70300cd52.
- Acquah, W.E.; Quain, D.M.; Twumasi, P. Genetic relationships between some released and elite Ghanaian cassava cultivars based on distance matrices. African J Biotechnol. 2011, 10, 913–921. Available: http://www.academicjournals.org/AJB.
- Grünwald, N.J.; Everhart, S.E.; Kamvar, Z.N. Best Practices for Population Genetic Analyses. Pap Plant Pathol. 2017, 421. Available: http://digitalcommons.unl.edu/plantpathpapers/421.
- Arnaud-Haond, S.; Duarte, C.M.; Alberto, F.; Serrao, E.A. Standardizing methods to address clonality in population studies. Mol Ecol. 2007, 16, 5115–5139. [CrossRef]
- Liu, K.; Muse, S. V. PowerMaker: An integrated analysis environment for genetic maker analysis. Bioinforma Appl Note. 2005, 21, 2128–2129. [CrossRef]
- Goudet, J.; Jombart, T.; Kamvar, Z.N.; Archer, E.; Hardy, O. Estimation and Tests of Hierarchical F-Statistics.2020. pp. 1–68.
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R package version 2.5-7.2020.
- Villanueva, R.A.M.; Chen, Z.J. ggplot2 : Elegant Graphics for Data Analysis (2nd ed.). Meas Interdiscip Res Perspect. 2019, 17, 160–167. [CrossRef]
- Larmarange, J. JLutils: Collection of R functions. R package version 1.22.0.2021. Available: https://github.com/larmarange/JLutils.
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000, 155, 945–959. [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005, 14, 2611–2620. [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012, 4, 359–361. [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 1–15. [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica. 2005, 142, 169–196. [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018, 32, 261–285. [CrossRef]
- Anne, C. Choosing the right molecular genetic markers for studying biodiversity: From molecular evolution to practical aspects. Genetica. 2006, 127, 101–120. [CrossRef]
- Amiteye, S. Basic Concepts And Methodologies Of Dna Marker Systems In Plant Molecular Breeding. Heliyon. 2021, 7, e08093. [CrossRef]
- Moyib, O.K.; Odunola, O.A.; Dixon, A.G.O. SSR markers reveal genetic variation between improved cassava cultivars and landraces within a collection of Nigerian cassava germplasm. African J Biotechnol. 2007, 6, 2666–2674.
- Turyagyenda, L.F.; Kizito, E.B.; Ferguson, M.E.; Baguma, Y.; Harvey, J.W.; Gibson, P.; Wanjala, B.W.; Osiru, D.S.O. Genetic diversity among farmer-preferred cassava landraces in uganda. African Crop Sci J. 2012, 20, 15–30.
- Pedri, E.C.M.; Hoogerheide, E.S.S.; Tiago, A.V.; Cardoso, E.S.; Pinto, J.M.A.; Santos, L.L.; Yamashita, O.M.; Rossi, A.A.B. Genetic diversity of cassava landraces cultivated in northern Mato Grosso State , Brazil , using microsatellite markers. Genet Mol Res. 2019, 18, 1–11. [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Gr̈unwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014, 2, 1–14. [CrossRef]
- Rabbi, I.Y.; Kulakow, P.A.; Manu-aduening, J.A.; Dankyi, A.A.; Asibuo, J.Y.; Parkes, E.Y.; Abdoulaye, T.; Girma, G.; Gedil, M.A.; Ramu, P.; et al. Tracking crop varieties using genotyping- by-sequencing markers : a case study using cassava (Manihot esculenta Crantz). BMC Genet. 2015, 16, 1–11. [CrossRef]
- Yang, X.; Xu, Y.; Shah, T. Comparison of SSRs and SNPs in assessment of genetic relatedness in maize.2011, 1045–1054. [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape.2013, 1–17.
| Markers | Forward primer | Reverse primer | PS (pb) | AT (°C) | |
|---|---|---|---|---|---|
| SSRY4 | ATAGAGCAGAAGTGCAGGCG | CTAACGCACACGACTACGGA | 287 | 55 | |
| SSRY9 | ACAATTCATCATGAGTCATCAACT | CCGTTATTGTTCCTGGTCCT | 278 | 55 | |
| SSRY12 | AACTGTCAAACCATTCTACTTGC | GCCAGCAAGGTTTGCTACAT | 266 | 55 | |
| SSRY19 | TGTAAGGCATTCCAAGAATTATCA | TCTCCTGTGAAAAGTGCATGA | 214 | 55 | |
| SSRY20 | CATTGGACTTCCTACAAATATGAAT | TGATGGAAAGTGGTTATGTCCTT | 143 | 55 | |
| SSRY21 | CCTGCCACAATATTGAAATGG | CAACAATTGGACTAAGCAGCA | 192 | 55 | |
| SSRY34 | TTCCAGACCTGTTCCACCAT | ATTGCAGGGATTATTGCTCG | 279 | 55 | |
| SSRY38 | GGCTGTTCGTGATCCTTATTAAC | GTAGTTGAGAAAACTTTGCATGAG | 122 | 55 | |
| SSRY51 | AGGTTGGATGCTTGAAGGAA | GGATGCAGGAGTGCTCAACT | 298 | 55 | |
| SSRY59 | GCAATGCAGTGAACCATCTTT | CGTTTGTCCTTTCTGATGTTC | 158 | 55 | |
| SSRY63 | TCAGAATCATCTACCTTGGCA | AAGACAATCATTTTGTGCTCCA | 290 | 55 | |
| SSRY64 | CGACAAGTCGTATATGTAGTATTC | GCAGAGGTGGCTAACGAGAC | 194 | 55 | |
| SSRY69 | CGATCTCAGTCGATACCCAAG | CACTCCGTTGCAGGCATTA | 239 | 55 | |
| SSRY82 | TGTGACAATTTTCAGATAGCTTCA | CACCATCGGCATTAAACTTTG | 211 | 55 | |
| SSRY100 | ATCCTTGCCTGACATTTTGC | TTCGCAGAGTCCAATTGTTG | 210 | 55 | |
| SSRY102 | TTGGCTGCTTTCACTAATGC | TTGAACACGTTGAACAACCA | 179 | 55 | |
| SSRY103 | TGAGAAGGAAACTGCTTGCAC | CAGCAAGACCATCACCAGTTT | 272 | 55 | |
| SSRY105 | CAAACATCTGCACTTTTGGC | TCGAGTGGCTTCTGGTCTTC | 225 | 55 | |
| SSRY106 | GGAAACTGCTTGCACAAAGA | CAGCAAGACCATCACCAGTTT | 270 | 55 | |
| SSRY108 | ACGCTATGATGTCCAAAGGC | CATGCCACATAGTTCGTGCT | 203 | 55 | |
| SSRY110 | TTGAGTGGTGAATGCGAAAG | AGTGCCACCTTGAAAGAGCA | 247 | 55 | |
| SSRY127 | GCTGAACTGCTTTGCCAACT | CTTCGGCCTCTACAAAAGGA | 130 | 45 | |
| SSRY132 | CTTTTTGCCAGTCTTCCTGC | TGTCCAATGTCTTCCTTTCCTT | 196 | 55 | |
| SSRY135 | CCAGAAACTGAAATGCATCG | AACATGTGCGACAGTGATTG | 253 | 45 | |
| SSRY147 | GTACATCACCACCAACGGGC | AGAGCGGTGGGGCGAAGAGC | 113 | 55 | |
| SSRY148 | GGCTTCATCATGGAAAAACC | CAATGCTTTACGGAAGAGCC | 114 | 55 | |
| SSRY151 | AGTGGAAATAAGCCATGTGATG | CCCATAATTGATGCCAGGTT | 182 | 55 | |
| SSRY155 | CGTTGATAAAGTGGAAAGAGCA | ACTCCACTCCCGATGCTCGC | 158 | 55 | |
| SSRY161 | AAGGAACACCTCTCCTAGAATCA | CCAGCTGTATGTTGAGTGAGC | 220 | 55 | |
| SSRY169 | TCAAACAAGAATTAGCAGAACTGG | TGAGATTTCGTAATATTCATTTCACTT | 187 | 45 | |
| SSRY171 | ACAGCTCTAAAAACTGCAGCC | AACGTAGGCCCTAACTAACCC | 100 | 55 | |
| SSRY177 | ACTGTGCCAAAATAGCCAAATAGT | TCATGAGTGTGGGATGTTTTTATG | 291 | 55 | |
| SSRY179 | ACCACAAACATAGGCACGAG | CACCCAATTCACCAATTACCA | 268 | 45 | |
| SSRY180 | CAGGCTCAGGTGAAGTAAAGG | GCGAAAGTAAGTCTACAACTTTTCTAA | 226 | 55 | |
| SSRY181 | CCTTGGCAGAGATGAATTAGAG | GGGGCATTCTACATGATCAATAA | 163 | 55 | |
| SSRY182 | GGTAGATCTGGATCGAGGAGG | CAATCGAAACCGACGATACA | 199 | 55 | |
| Source of variation | Geographical origin | Source of variation | Breeding patterns | |||||
|---|---|---|---|---|---|---|---|---|
| df | Mean Sq | % of variation | df | Mean Sq | % of variation | |||
| Between clusters | 7 | 13.04 | 6.31 | Between groups | 1 | 14.83 | 3.33 | |
| Within individuals | 122 | 6.40 | 93.69 | Within individuals | 128 | 6.70 | 96.67 | |
| Total | 129 | 6.76 | 100.00 | Total | 129 | 6.78 | 100.00 | |
| Source of variation | DAPC | Source of variation | Bayesian approach | |||||
| Df | Mean Sq | % of variation | df | Mean Sq | % of variation | |||
| Between groups | 12 | 51.55 | 70.09 | Between groups | 4 | 94.06 | 46.73 | |
| Within individuals | 117 | 2.17 | 29.91 | Within individuals | 125 | 3.97 | 53.27 | |
| Total | 129 | 6.76 | 100.00 | Total | 129 | 6.76 | 100.00 | |
| SNP markers | |
|---|---|
| Type of clustering | FST |
| Geographical origin | 0.025 |
| Breeding patterns | 0.008 |
| DAPC clusters | 0.307 |
| Bayesian clusters | 0.192 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





