1. Introduction
The incidence of type 1 diabetes mellitus (T1DM) is increasing and because the genetic transmission of this disease remains the same, other culprits have been proposed, like pollutants, endocrine disruptors, vitamin D deficit and a pathologic colonization of the gastrointestinal tract. The gastrointestinal microbiota is comprised of all the bacteria, viruses and fungi that colonize this environment. An imbalance of this microbiota (dysbiosis) with the predominance of proinflammatory bacteria that affect the immune system is thought to explain the global rise in T1DM especially the early onset of this disease [
1].
The human gastrointestinal tract harbors a diverse array of microbial species. Among these microorganisms, the most prevalent bacterial phyla found are
Bacteroidetes, Firmicutes, Proteobacteria, and
Actinobacteria. These phyla collectively contribute to the microbial ecosystem within the human gut, playing essential roles in various aspects of human health and physiology and offering numerous advantages to its host. These include breaking down dietary substances for easier digestion, producing essential vitamins, regulating the immune system, and safeguarding against harmful intestinal pathogens. This protection is achieved through competition for nutrients and the production of bacteriocins and other antimicrobial compounds (hydrogen peroxide, lactic acid) [
3,
4].
The delicate balance of the gut microbiota can be disrupted by various factors, leading to what is known as dysbiosis. These factors include a high fat diet, specific health conditions such as inflammatory bowel disease and cancer, antibiotics, infections, stress, and other environmental influences. However, it's important to recognize that the microbiota also plays a vital role in the development and progression of numerous diseases. One concerning health issue we are currently grappling with worldwide is the escalating incidence of T1DM, striking at younger ages, especially as it does not affect all individuals with genetic susceptibility [
4,
5,
6,
7].
Dysbiosis and T1DM
Recent data strongly supports the crucial role of external triggers in the rapid increase of T1DM incidence. The alterations in the microbiota and the chronic inflammation, due to environmental influences, are being suggested as explanations for the shifts in the prevalence of this disease [
4,
8,
9,
10].
Several animal studies have provided evidence of a potential link between the microbiota and T1DM. For instance, experiments involving the administration of single antibiotics (such as vancomycin) or combinations of antibiotics in non-obese diabetic (NOD) mice resulted in microbiota alterations, which, in turn, either accelerated or delayed disease progression. These findings offer valuable insights into the intricate relationship between the gut microbiota and T1DM [
4,
11,
12,
13]. In the context of T1DM, changes in the gut microbiota have been observed prior to the emergence of systemic signs of islet autoimmunity. This shift in the microbiota could be attributed to the fact that previous studies primarily identified these modifications through gene analysis of 16S rRNA, which might not capture specific structural and functional characteristics potentially involved in disease progression. To address this concern, subsequent investigations employed specialized designs to control for all known factors influencing T1DM susceptibility and analyzed microbiome characteristics using longitudinal metagenomic sequencing of stool samples. Irrespective of geographical location, the environmental niche associated with T1DM was found to harbor a proinflammatory environment, with a higher abundance of Bacteroidetes and a lower abundance of Firmicutes. The reduction in Firmicutes may be detrimental to the host since this phylum includes many producers of the short-chain fatty acid (SCFA) butyrate, which plays a crucial role in intestinal homeostasis. On the other hand, the Bacteroidetes phylum encompasses, among others, strains of Bacteroides and Prevotella. Studies have consistently shown that T1DM is characterized by a dominance of Bacteroides, a taxa correlated with intestinal inflammation, while Prevotella, which is believed to be protective, is reduced [
4,
14,
15,
16,
17,
18,
19,
20].
Despite the considerable variability observed in T1DM-associated microbiota, numerous studies have consistently reported a link between
Bacteroides and T1DM development. Species within this genus can ferment glucose and lactate to produce propionate, acetate, and succinate, but they lack the ability to generate butyrate. Butyrate is a crucial metabolite for intestinal homeostasis, stimulating mucin synthesis and contributing to a decrease in gut permeability by facilitating the assembly of tight junctions (TJ). Additionally, lactate-producing bacteria, including certain probiotic strains like
Lactobacillus rhamnosus,
L. reuteri, L. johnsonii N6.2, L. plantarum, and
Bifidobacterium lactis, have the ability to synthesize butyrate, thereby reinforcing the intestinal barrier function. These findings underscore the intricate role of the gut microbiota in T1DM and suggest that targeting specific microbial components could hold potential for interventions aimed at supporting intestinal health in individuals with T1DM [
21,
22,
23,
24,
25].
2. Results
In our 31 patients the median of age was 9 +/- 0.5 SD years, the ratio male: female was 1, and the BMI was 17.48 kg/m2. The age of onset of the disease was 9 +/- 0.5 SD, the median glycaemia was 350 +/- 98 mg/dl. The ratio normal: caesarian birth was 0.36, representing 42 % (13/31) had a family history of autoimmune disease and 33% (10/31) of T1DM.
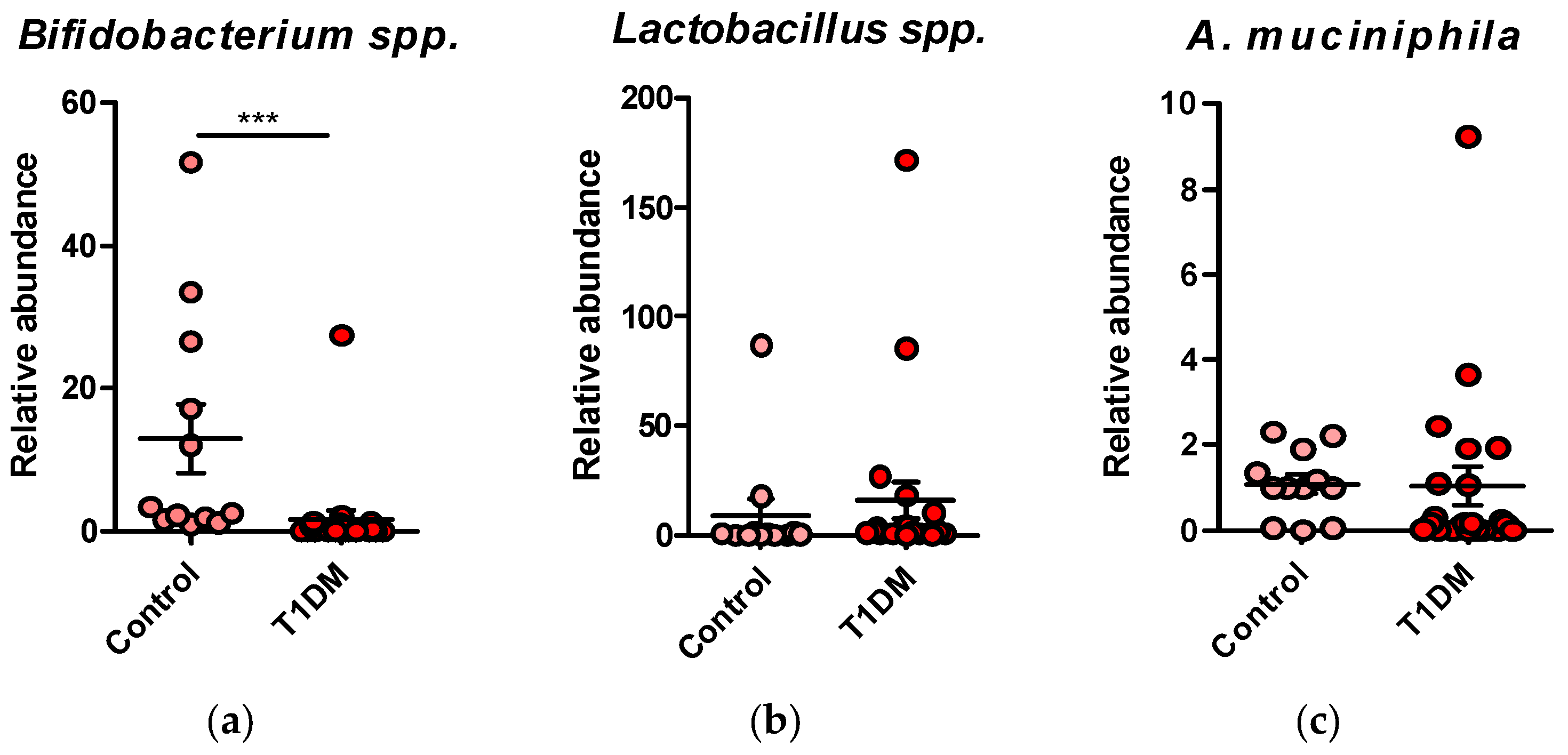
We observed a dysbiosis in the T1DM, with a predominance of detrimental bacteria (Clostridium coccoides, Faecalibacterium, Bacterioides, Enterobacteriaceae) and fungi versus the healthy control group. Also, reduced Bifidobacterium spp. in T1DM in the beneficial taxa was noticed.
We observed a dysbiosis in the T1DM, with a predominance of detrimental bacteria (Clostridium coccoides, Faecalibacterium, Bacterioides, Enterobacteriaceae) and fungi versus the healthy control group. Also, reduced Bifidobacterium spp. in T1DM in the beneficial taxa was noticed.
Figure 1.
(a) Relative abundance of Bifidobacterium spp. in T1DM patients compared to healthy controls; (b) Relative abundance of Lactobacillus spp. in T1DM patients compared to healthy controls; (c) Relative abundance of A. muciniphila in T1DM patients compared to healthy controls.
Figure 1.
(a) Relative abundance of Bifidobacterium spp. in T1DM patients compared to healthy controls; (b) Relative abundance of Lactobacillus spp. in T1DM patients compared to healthy controls; (c) Relative abundance of A. muciniphila in T1DM patients compared to healthy controls.
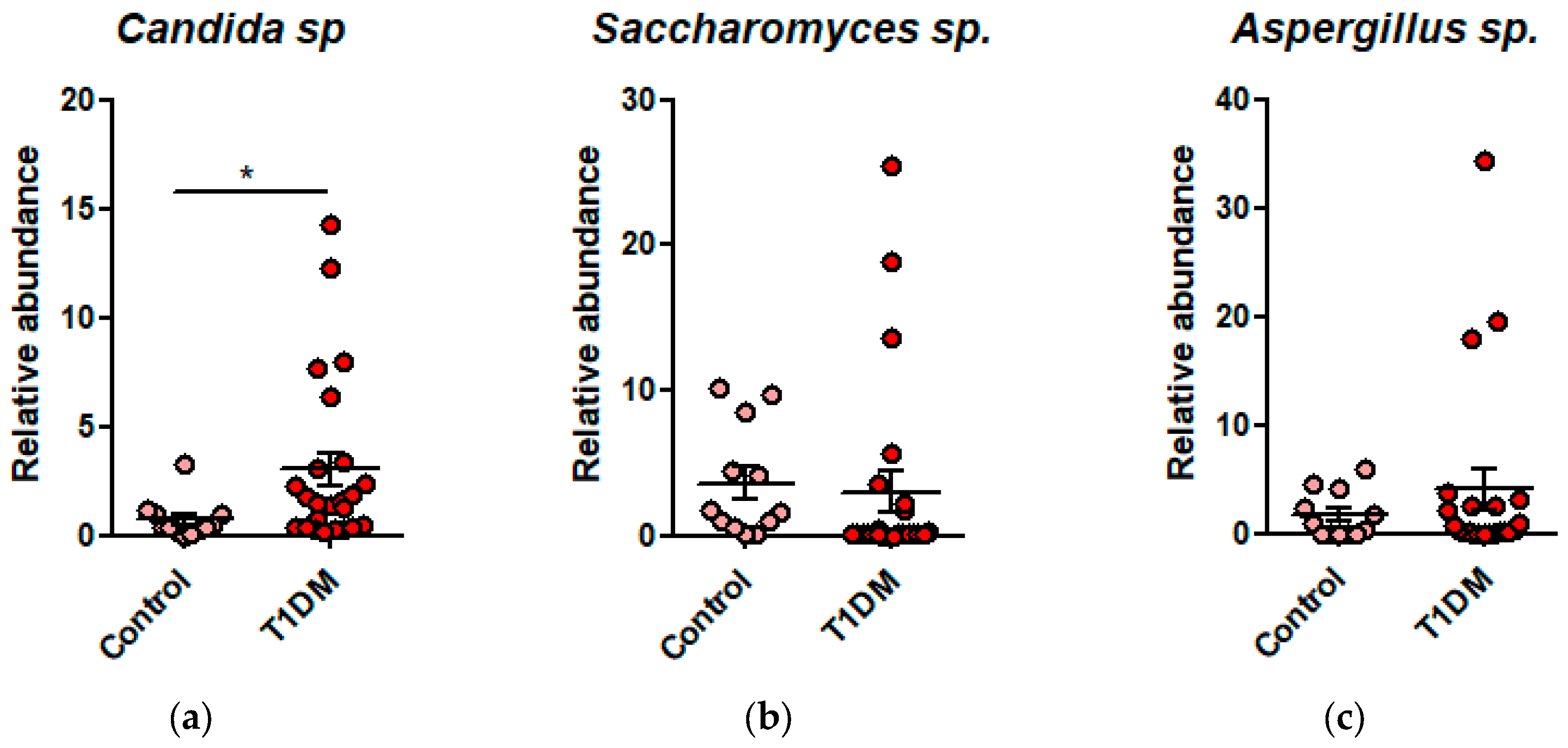
Figure 2.
(a) Dysbiosis with increased abundance of Candida sp. in T1DM; (b) Dysbiosis with increased abundance of Saccharomyces sp. in T1DM; (c) Dysbiosis with increased abundance of Aspergillus sp. in T1DM.
Figure 2.
(a) Dysbiosis with increased abundance of Candida sp. in T1DM; (b) Dysbiosis with increased abundance of Saccharomyces sp. in T1DM; (c) Dysbiosis with increased abundance of Aspergillus sp. in T1DM.
Figure 3.
(a) Dysbiosis with increased abundance of Clostridium coccoides in T1DM group; (b) Dysbiosis with increased abundance of Bacteroides spp. in T1DM group; (c) A reduction of butyrate producing bacteria was seen in the T1DM group; (d) Dysbiosis with increased abundance of Enterobacteriaceae in T1DM group.
Figure 3.
(a) Dysbiosis with increased abundance of Clostridium coccoides in T1DM group; (b) Dysbiosis with increased abundance of Bacteroides spp. in T1DM group; (c) A reduction of butyrate producing bacteria was seen in the T1DM group; (d) Dysbiosis with increased abundance of Enterobacteriaceae in T1DM group.
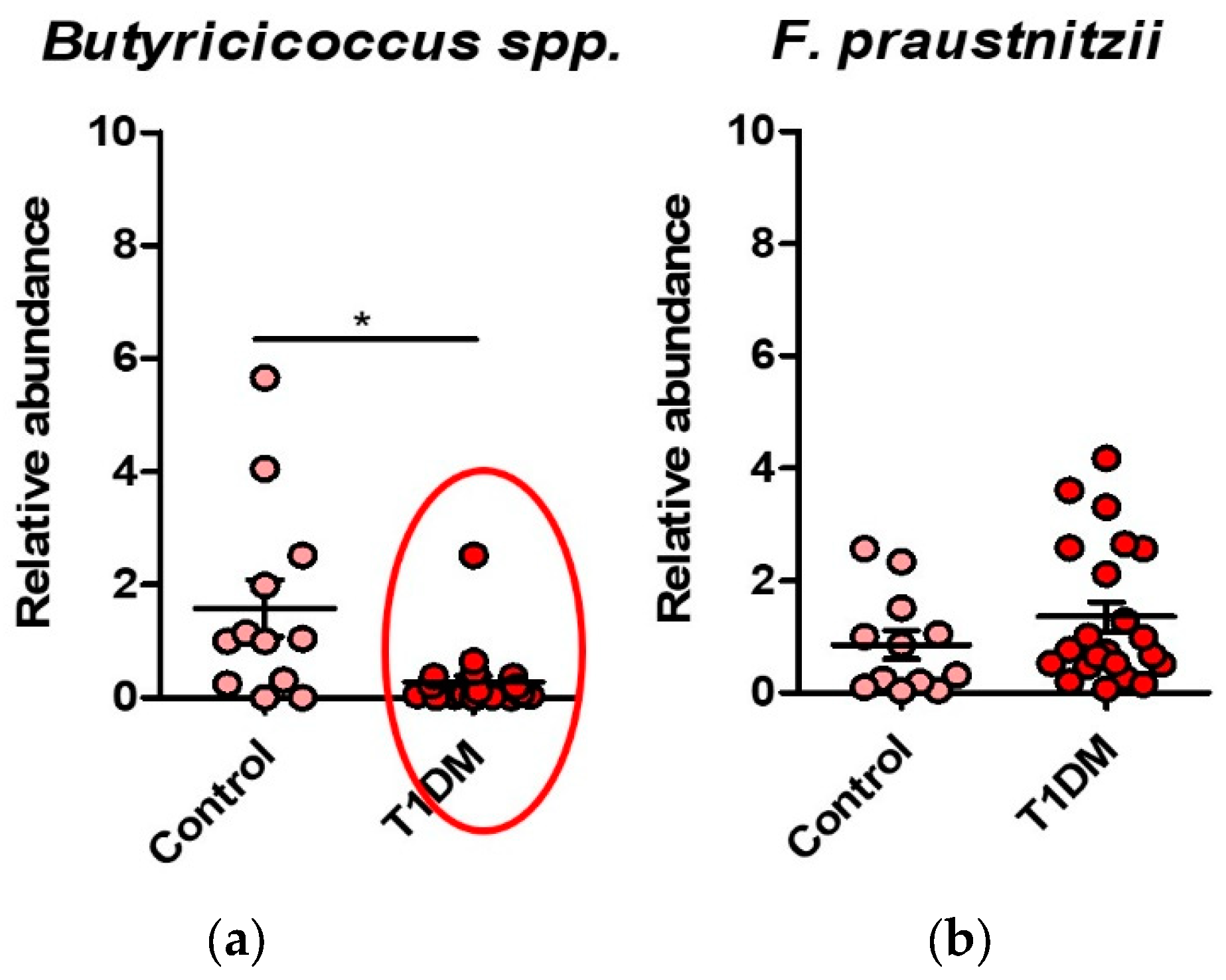
Figure 4.
(a) Dysbiosis with a reduction of Butyricicoccus spp. in T1DM group; (b) Dysbiosis with a reduction of F. Praustnitzii in T1DM group.
Figure 4.
(a) Dysbiosis with a reduction of Butyricicoccus spp. in T1DM group; (b) Dysbiosis with a reduction of F. Praustnitzii in T1DM group.
The Spearman correlation showed a positive correlation between the presence of Bacteroides sp. (p=0.0442), Butyricicoccus sp (p=0.0164), and Clostridium leptum (p=0.0023) with the recent onset of diabetes. The younger age onset correlated with the presence of Butyricicoccus sp.(p=0.019).
Antibiotic use in the last month before admission is associated with the presence of fungi in the microbiota, especially Candida spp. (p=0.0442).
High values of blood glucose at diagnosis negatively correlate to the presence of Bacteroides and the presence of diabetic ketoacidosis positively correlate to the detrimental taxa-Enterobacteriaceae and Aspergillus, but not significantly.
High insulin doses in the treatment of patients positively correlated with Bacteroides (p= 0.0351) and Candida sp. (p= 0.0427) while high CRP was correlated with the presence of Enterobacteriaceae (p=0.0022).
In the calcium-phosphorous metabolism, calcium levels positively correlate with Aspergillus, IGF-1 with Lactobacillus (p= 0.0337), phosphorus with Lactobacilli (p= 0.0226) and vitamin D with Bacteroides levels (p= 0.0376).
The positive anti-thyroglobulin antibodies correlated with Candida (p= 0.0388) and low fT4 with Bacteroides levels (p= 0.0427). The anti-transglutaminase antibodies positive correlated with Enterobacteriaceae, but not significantly.
Figure 5.
Spearman correlations between microbiota patterns and clinical parameters; blue=negative correlation, red=positive correlation.
Figure 5.
Spearman correlations between microbiota patterns and clinical parameters; blue=negative correlation, red=positive correlation.
3. Discussion
Our study reveals a dysbiosis characterized by an overabundance of proinflammatory bacteria,
Clostridium coccoides, Faecalibacterium, Bacterioides, Enterobacteriaceae) and fungi versus the healthy control group in the pediatric romanian T1DM cohort. The literature cites
Bifidobacterium and
Ruminococcus and the risk of this disease [
1,
2,
3,
4,
5,
33].
The positive correlations found between specific proinflammatory bacterial species – mainly
Bacteroides but also
Butyricicoccus sp and the disease onset, especially with early onset correlates with the data found in the literature [
21,
22,
23,
24,
25].
The presence of antibiotic use during acute infections in the last 2 months in the cohort of children with T1DM perturbs microbiota with the overabundance of fungi.
Diabetic ketoacidosis positively, but not significantly, correlates to the detrimental taxa-
Enterobacteriaceae and
Aspergillus. As children that received antibiotics during the last two weeks were excluded from the microbiota study, the microbiota detection prior to antibiotic administration in diabetic ketoacidosis would help asses if in T1DM patients as in type 2 diabetic patients from Romania these pathological taxa correlate with inflammation and a more severe onset of the disease [
32].
The presence of pro inflammatory bacteria and fungi (Bacteroides and Candida) correlate with higher insulin dosses to achieve euglycemia and high CRP with the presence of Enterobacteriaceae showing the importance of this dysbiosis and the inflammation that aggravate the evolution and control of T1DM.
The role of microbiota in the onset of other autoimmune diseases is showed by the positive correlation between anti-thyroglobulin antibodies and the presence of Candida sp. and the anti-transglutaminase antibodies and
Enterobacteriaceae, but not with Bifidobacterium as in other studies [
33].
The calcium-phosphorous metabolism is also influenced by the microbiota, as calcium levels positively correlate with
Aspergillus, IGF-1 and phosphorus with
Lactobacilli and vitamin D with
Bacteroides levels. As impaired bone health in type 2 diabetes was found to be linked to dysbiosis and higher abundance of
Lactobacilli the determination of growth and also of bone density is important to be assessed in these children [
34].
The opportunity of this study is that it’s one of very few researches of the microbiota in recently diagnosed T1DM patients in Romania, especially in the group with early onset. Also, the correlation between this dysbiosis and other autoimmune diseases like Hashimoto’s thyroiditis and Coeliac disease that was not determined in this region is very important to assess the associations between autoimmune diseases and the gut bacterial and fungi population.
The limitations of this study are the fact that it was a heterogeneous study group, the study was a retrospective one with a small number of patients, with mostly urban predominance and the exclusion of the patients that had antibiotics after infections (mostly with diabetic ketoacidosis).
3. Future research
In future studies proteomics and metabolomics profiling technologies are of interest for the assessment of functional changes in the microbiome. Identifying unique biological signatures (SCFA) and their delivery to a recipient host might allow personalized treatments in T1DM.
This type of treatment with probiotics, prebiotics, and dietary factors that normalize the dysbiosis and help control the autoimmunity might be a solution for the genetic predisposed siblings of T1DM patients.
Also, the associations between dysbiosis and other autoimmune diseases –Hashimoto’s thyroiditis, celiac disease need to be investigated more attentively regarding the presence of antibodies but also the clinical control of this diseases.
Tools that estimate the risk of developing T1DM are becoming more advanced. These tools take into account factors like specific genetic traits (HLA haplotypes), the presence of autoantibodies, and a family history of the disease. By considering these aspects, these risk calculators can potentially predict the likelihood of someone getting diabetes even before they actually develop the condition. This early prediction could open up opportunities for people to try out new treatments that aim to extend the period before the disease becomes apparent. Intervening early, when there’s still a reasonable amount of the insulin-producing β-cells in the pancreas, might offer valuable help to these kids by modulating their immune responses. This could potentially slow down the progression of the disease and provide better outcomes [
26,
27].
4. Materials and Methods
Using 16S rRNA qRT-PCR, we analyzed phyla abundance as well as the relative abundance of specific bacterial and fungal groups.
Study population
This was a retrospective case control study of 31 T1DM pediatric patients 4-18 years of age recently diagnosed (within the last six months) according to the American Diabetes Association criteria and their 7 first-degree T1DM family members.
All the participants were of Caucasian origin, as defined by Gorodezky, et al. [
26,
27,
28].
The exclusion criteria were antibiotic/probiotic administration in the last 2 weeks prior to the admission to the department, T1DM more than 6 months from diagnosis and other types of diabetes mellitus. The newly diagnosed T1DM patients were matched by age, gender, BMI, same type of environment (urban) to normal (without any chronic disease) children.
The study was conducted at the Pediatric Endocrinology and Diabetes Department of the Elias Emergency and University Hospital, in Bucharest, Romania, between 2019-2021 with the hospital approval by the Etic Commission for Scientific Research.
All participants or their parents were asked to sign an informed consent. From all study subjects, a complete medical history was taken (age, sex, hereditary-collateral background of autoimmune diseases/diabetes, personal physiological/pathological history including autoimmune, type of birth, antibiotic therapy, breastfeeding, weaning, cesarean/vaginal birth, history of breastfeeding, viral infections, antibiotic therapy history, parasite history, date of T1DM diagnosis, number of hospitalizations, blood glucose control, ketoacidosis events, emergency hospitalizations due to hypo/hyperglycemia, type of treatment - presence of sensor and pump).
Current dietary survey (focusing on intake of soluble/insoluble fibers, saturated/unsaturated fats, fast/slow carbohydrates, red meat, sugars, and anthropometric measurements (age, weight, height, body mass index [BMI], blood pressure, Tanner Stage, waist to hip ratio) were taken. A 10 cc peripheral blood sample was obtained eight hours after the last meal for a complete blood count and blood chemistry.
Laboratory procedures
Complete blood count and blood chemistry were performed, including glucose, creatinine, uric acid, total cholesterol, HDL-cholesterol, LDL-cholesterol, triglycerides, glycated hemoglobin (HbA1c), hepatic function, β-cell function tests (C-peptide), and β-cell antibodies (anti-GAD). Anti-thyroglobulin and anti-thyroperoxidase antibodies (autoimmune thyroid disorders) and anti-tissue transglutaminase antibodies (for celiac disease) were dosed, to assess autoimmune diseases. Thyroid function was assessed by TSH and free T4. IGF1 and phospho-calcium metabolism were evaluated.
Microbiota analysis
Stool samples were gathered during hospitalization or at home using a standardized procedure that involved antiseptic handling, collection in sterile tubes (without culture media), and immediate freezing at −20 °C. Subsequently, fecal DNA was extracted utilizing the PureLink Microbiome Purification Kit (Invitrogen) following the manufacturer's instructions. The concentration of DNA was assessed with a Qubit 4 fluorometer (Thermo Scientific). For qPCR analysis, DNA samples were diluted in DNase-free water to a concentration of 3 ng/μl. qRT-PCR was employed to determine the relative abundance of intestinal microorganisms in stool DNA isolated from both MetSyn patients and healthy controls, using a ViiA7© Fast Real-Time instrument (Applied Biosystems). Bacterial or fungal group-specific primers (16S rDNA and 18S rDNA, respectively) were utilized at their designated annealing temperatures, with primer sequences selected from literature [55–57] and listed in
Table 1. Each PCR reaction comprised 2.5 nM of forward and reverse primers, 9 ng of DNA, and 2x SYBR Green Master Mix (Applied Biosystems). Negative controls were included, consisting of samples without DNA templates. The samples underwent incubation at 95ºC for 5 min, followed by amplification through 40 cycles of 95ºC for 10 s, 60ºC for 30 s, and 72ºC for 1 s.
Statistical analysis
Our study data are presented as mean ±SEM and were graphed using the GraphPad Prism 9.0 software. Power analysis was initially performed with a set power (1) of 0.90 and of 0.05 for two groups (Control, MetSyn) tested using difference in means and standard deviation as parameters. The * p < 0.05 was considered as statistically significant. Statistical significance levels were * p < 0.05; ** p < 0.01; *** p < 0.001. Standardized statistical test methods were used to analyze the results of demography and laboratory tests (biochemistry tests and metabolite levels). Continuous variables were expressed as means SD. The analysis of differences between groups was performed by a normality test; a p-value 0.05 was considered to be normal and homogeneous, followed by parametric testing (t-test); a p-value < 0.05 was considered to be statistically significant.
A Spearman correlation analysis evaluating the association between the risk haplotype and early onset T1DM, and high-level onset glycaemia and ATPO antibodies was performed using SPSS Windows v. 17.0 (SPSS, Inc). Also, a binary logistic regression analysis was used to study the association between HLA alleles, ketoacidosis, anti-thyroglobulin antibodies and vitamin D deficit.
For all tests, p < 0.05 was considered statistically significant.
5. Conclusions
In our T1D children cohort, there is a dysbiosis similar to what is found in the literature, which correlates with inflammatory pathogens. Thus, it may be a reasonable explanation for the more frequent severe onset of T1D, taking into account the fact that genetics are not to be changed.
Author Contributions
Conceptualization, A.A.I., S.F. and G.G.P.; methodology, S.I.; software, T.P.; validation, A.A.I, G.G.P; formal analysis, T.P.; investigation, A.A.I and G.G.P; resources, G.G.P; data curation, A.A.I. and ; writing—original draft preparation, A.A.I.; writing—review and editing, A.A.I., G.G.P; visualization, A.A.I; supervision, G.G.P; project administration, S.F..; funding acquisition, S.F and G.G.P.All authors have read and agreed to the published version of the manu-script.”.
Funding
This research was funded by a UEFISCDI grant, project ID PN-III-P1-1.1-PD-2019-0499, Nr 224/2021.
Institutional Review Board Statement
The study was conducted in accordance with the Decla-ration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of THE ELIAS HOSPITAL (protocol code 1695, 12.03.2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Acknowledgments
The entire departments of endocrinology, diabetes and metabolic diseases and pediatric endocrinology and diabetes.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Durazzo, M.; Ferro, A.; Gruden, G. Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art. J Clin Med. 2019 Nov 2;8(11):1843. [CrossRef]
- Dedrick, S.; Sundaresh, B.; Huang, Q.; Brady, C.; Yoo, T.; Cronin, C.; Rudnicki, C.; Flood, M.; Momeni, B.; Ludvigsson, J.; Altindis, E. The Role of Gut Microbiota and Environmental Factors in Type 1 Diabetes Pathogenesis. Front Endocrinol (Lausanne) 2020, 11, 78. [CrossRef]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; Van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521. [CrossRef]
- Gradisteanu Pircalabioru G.; Corcionivoschi N.; Gundogdu O.; Chifiriuc M.C.; Marutescu L.G.; Ispas B.; Savu O. Dysbiosis in the Development of Type I Diabetes and Associated Complications: From Mechanisms to Targeted Gut Microbes Manipulation Therapies. Int J Mol Sci. 2021 Mar 9;22(5):2763. [CrossRef]
- Patterson, C.; Guariguata, L.; Dahlquist, G.; Soltész, G.; Ogle, G.; Silink, M. Diabetes in the young—A global view and worldwide estimates of numbers of children with type 1 diabetes. Diabetes Res. Clin. Pract. 2014, 103, 161–175. [CrossRef]
- Mayer-Davis, E.J.; Kahkoska, A.R.; Jefferies, C.; Dabelea, D.; Balde, N.; Gong, C.X.; Aschner, P.; Craig, M.E. ISPAD clinical practice consensus guidelines 2018: Definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr. Diabetes 2018, 19, 7–19. [CrossRef]
- Katsarou, A.; Gudbjörnsdottir, S.; Rawshani, A.; Dabelea, D.; Bonifacio, E.; Anderson, B.J.; Jacobsen, L.M.; Schatz, D.A.; Lernmark. Type 1 diabetes mellitus. Nat. Rev. Dis. Primers 2017, 3. [CrossRef]
- Hooper, L.V.; Littman, D.R.; MacPherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336,1268–1273. [CrossRef]
- Umoh, F.I.; Kato, I.; Ren, J.;Wachowiak, P.L.; Ruffin, M.T.; Turgeon, D.K.; Sen, A.; Brenner, D.E.; Djuric, Z. Markers of systemic exposures to products of intestinal bacteria in a dietary intervention study. Eur. J. Nutr. 2016, 55, 793–798. [CrossRef]
- Quercia, S.; Candela, M.; Giuliani, C.; Turroni, S.; Luiselli, D.; Rampelli, S.; Brigidi, P.; Franceschi, C.; Bacalini, M.G.; Garagnani, P.; et al. From lifetime to evolution: Timescales of human gut microbiota adaptation. Front. Microbiol. 2014, 5, 587. [CrossRef]
- Candon, S.; Perez-Arroyo, A.; Marquet, C.; Valette, F.; Foray, A.-P.; Pelletier, B.; Milani, C.; Ventura, M.; Bach, J.-F.; Chatenoud, L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 2015, 10, e0125448. [CrossRef]
- Hu, Y.; Jin, P.; Peng, J.; Zhang, X.; Wong, F.S.; Wen, L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J. Autoimmun. 2016, 72, 47–56. [CrossRef]
- Hansen, C.H.F.; Krych, L.; Nielsen, D.S.; Vogensen, F.K.; Hansen, L.H.; Sørensen, S.J.; Buschard, K.; Hansen, A.K. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012, 55, 2285–2294. [CrossRef]
- Maffeis, C.; Martina, A.; Corradi, M.; Quarella, S.; Nori, N.; Torriani, S.; Plebani, M.; Contreas, G.; Felis, G.E. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab. Res. Rev. 2016, 32, 700–709. [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [CrossRef]
- 16. Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian,W.A.; Rewers, M.J.; She, J.-X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nat. Cell Biol. 2018, 562, 589–594. [CrossRef]
- Hagopian, W.A.; Erlich, H.; Lernmark, Å.; Rewers, M.; Ziegler, A.G.; Simell, O.; Akolkar, B.; Vogt, R.; Blair, A.; Ilonen, J.; et al. The environmental determinants of diabetes in the young (TEDDY): Genetic criteria and international diabetes risk screening of 421,000 infants. Pediatr. Diabetes 2011, 12, 733–743. [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [CrossRef]
- Durazzo, M.; Ferro, A.; Gruden, G. Gastrointestinal microbiota and type 1 diabetes mellitus: The state of art. J. Clin. Med. 2019, 8,1843. [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [CrossRef]
- Hague, A.; Butt, A.J.; Paraskeva, C. The role of butyrate in human colonic epithelial cells: An energy source or inducer of differentiation and apoptosis? Proc. Nutr. Soc. 1996, 55, 937–943. [CrossRef]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [CrossRef]
- Rosenfeldt, V.; Benfeldt, E.; Valerius, N.H.; Pærregaard, A.; Michaelsen, K.F. Effect of probiotics on gastrointestinal symptoms and small intestinal permeability in children with atopic dermatitis. J. Pediatr. 2004, 145, 612–616. [CrossRef]
- Valladares, R.; Sankar, D.; Li, N.; Williams, E.; Lai, K.-K.; Abdelgeliel, A.S.; Gonzalez, C.F.; Wasserfall, C.H.; Larkin, J.; Schatz, D.; et al. Lactobacillus johnsonii N6.2 Mitigates the development of type 1 diabetes in BB-DP rats. PLoS ONE 2010, 5, e10507. [CrossRef]
- Karczewski, J.; Troost, F.J.; Konings, I.; Dekker, J.; Kleerebezem, M.; Brummer, R.-J.M.; Wells, J.M. Regulation of human epithelial tight junction proteins by lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Liver Physiol. 2010, 298, G851–G859. [CrossRef]
- Steenkiste A.; Valdes A.M.; Feolo M.; Hoffman D.; Concannon P.; Noble J.; Schoch G.; Hansen J.; Helmberg W.; Dorman J.S.; Thomson G.; Pugliese A. 13th IHWS 1 Diabetes Component participating investigators. 14th International HLA and Immunogenetics Workshop: report on the HLA component of type 1 diabetes. Tissue Antigens. 2007 Apr;69 Suppl 1:214-25. [CrossRef]
- Pociot F.; McDermott M.F. Genetics of type 1 diabetes mellitus. Genes Immun. 2002;3(5):235-49. [CrossRef]
- ZThomson G.; Valdes A.M.; Noble J.A.; Kockum I.; Grote M.N.; Gorodezky C. et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens 2007, 70:110–127. [CrossRef]
- Ziegler A.G.; Hoffmann G.F.; Hasford J.; Larsson H.E.; Danne T.; Berner R.; Penno M.; Koralova A.; Dunne J.; Bonifacio E. Screening for asymptomatic β-cell autoimmunity in young children. Lancet Child Adolesc Health 2019 May;3(5):288-290. [CrossRef]
- Vaarala O. Gut microbiota and type 1 diabetes. Rev Diabet Stud. 2012 Winter;9(4):251-9. [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012 Jun 13;486(7402):207-14. [CrossRef]
- Gradisteanu Pircalabioru G.; Ilie I.; Oprea L.; Picu A.; Petcu L.M.; Burlibasa L.; Chifiriuc M.C.; Musat M. Microbiome, Mycobiome and Related Metabolites Alterations in Patients with Metabolic Syndrome-A Pilot Study. Metabolites 2022 Feb 28;12(3):218. [CrossRef]
- Xu Q.; Ni J.J.; Han B.X.; Yan S.S.; Wei X.T.; Feng G.J.; Zhang H.; Zhang L.; Li B.; Pei YF. Causal Relationship Between Gut Microbiota and Autoimmune Diseases: A Two-Sample Mendelian Randomization Study. Front Immunol. 2022 Jan 24;12:746998. [CrossRef]
- Knudsen J.K.; Leutscher P.; Sørensen S. Gut Microbiota in Bone Health and Diabetes. Curr Osteoporos Rep. 2021 Aug;19(4):462-479. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).