Submitted:
08 December 2023
Posted:
12 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
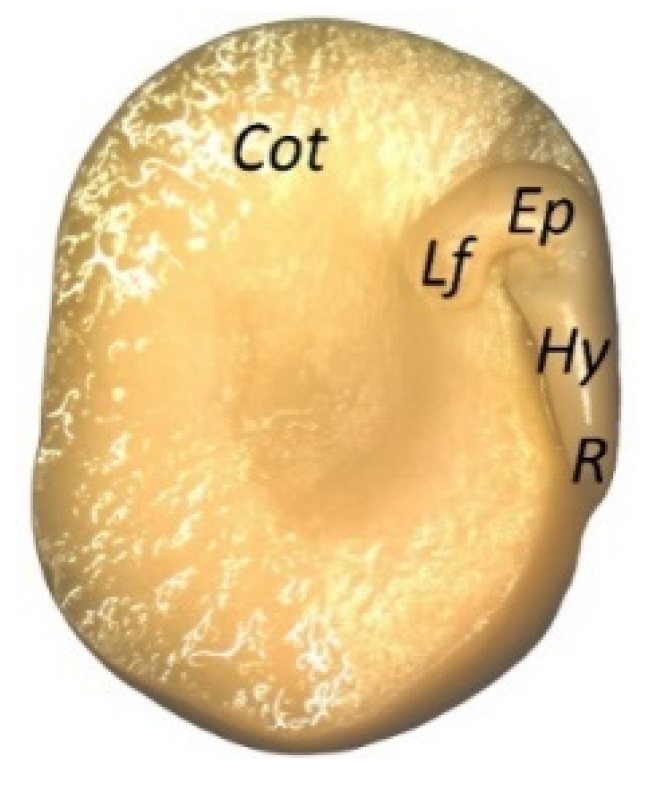
2.1. Plant Material
2.2. Quantitation of ABA and ABA-related metabolites
2.3. Annotation of ABA-Associated Genes
2.4. DNA Extraction and Sodium Bisulfite Treatment
2.5. Primer Design and in silico Analysis
2.6. PCR, Electrophoretic Analysis, Extraction, and Purification
2.7. Cloning and Sequencing the Amplified PCR Fragments
2.8. Statistical Analyses
3. Results
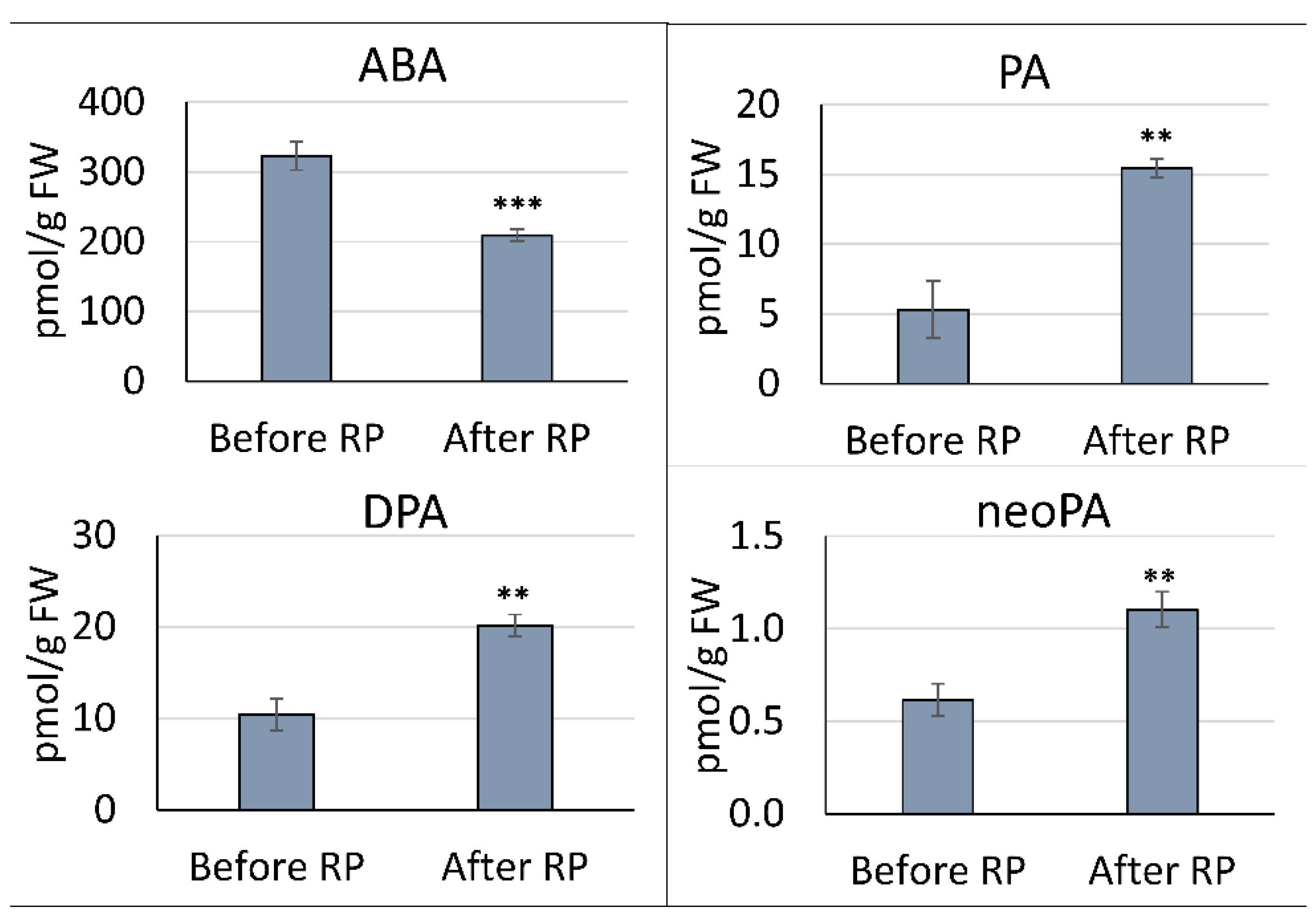
3.1. Quantitation of ABA and ABA-related metabolites in Pea Embryonic Axes Before and After Radicle Protrusion
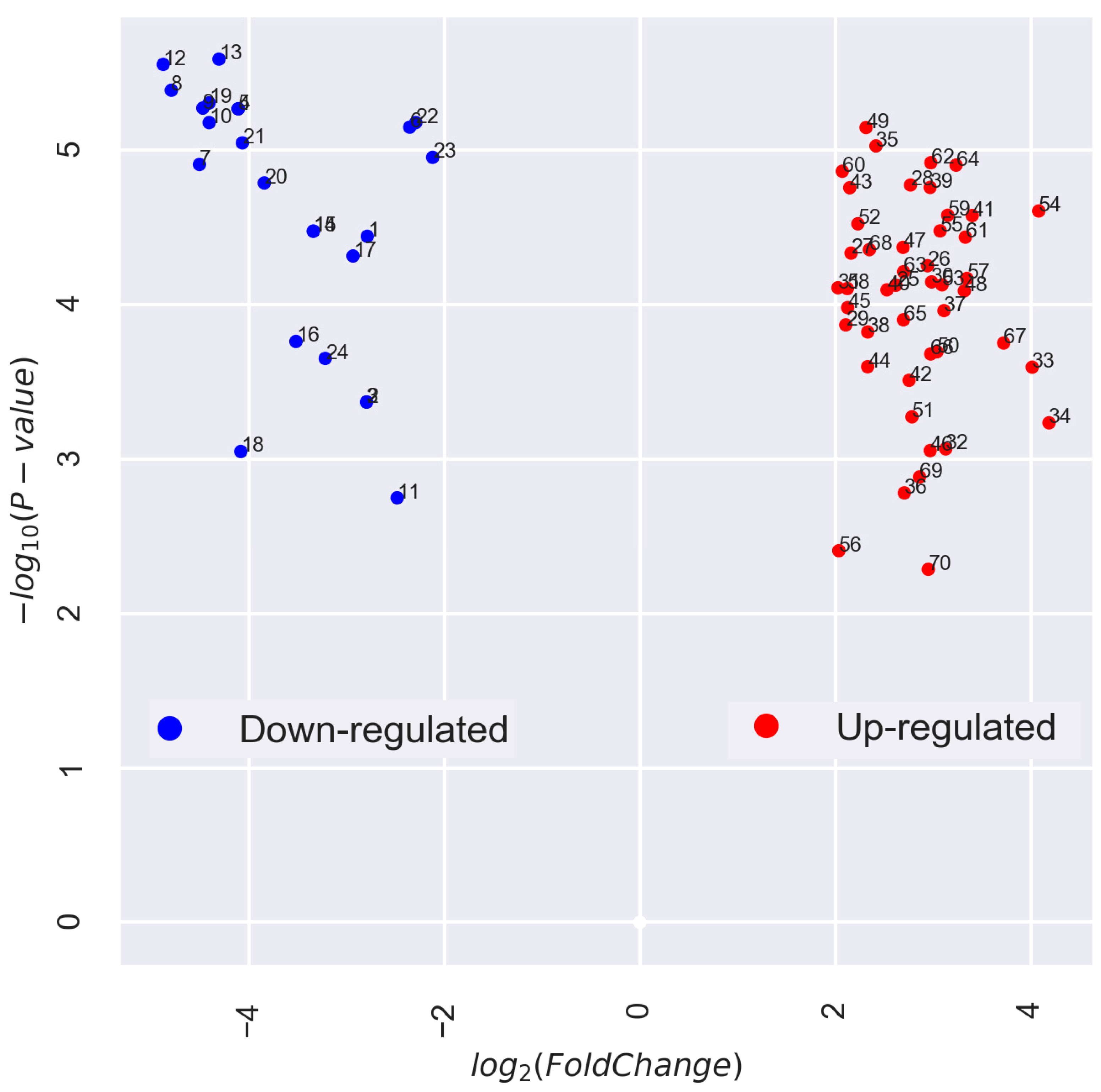
3.2. Categorization and Functional Annotation of ABA-Associated DEGs in Pea Embryonic Axes Before and After Radicle Protrusion
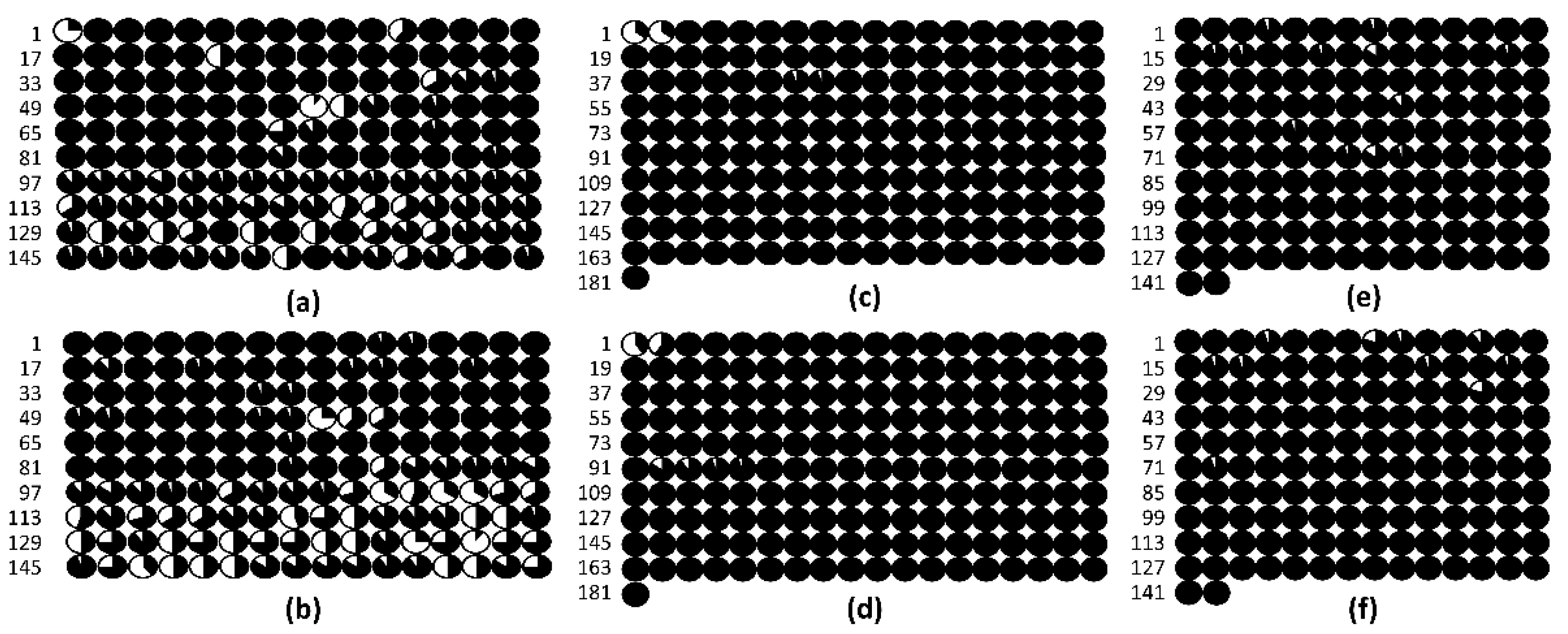
3.3. DNA Methylation in the Promoters of the PsABI3, PsABI4, and PsABI5 Genes
4. Discussion
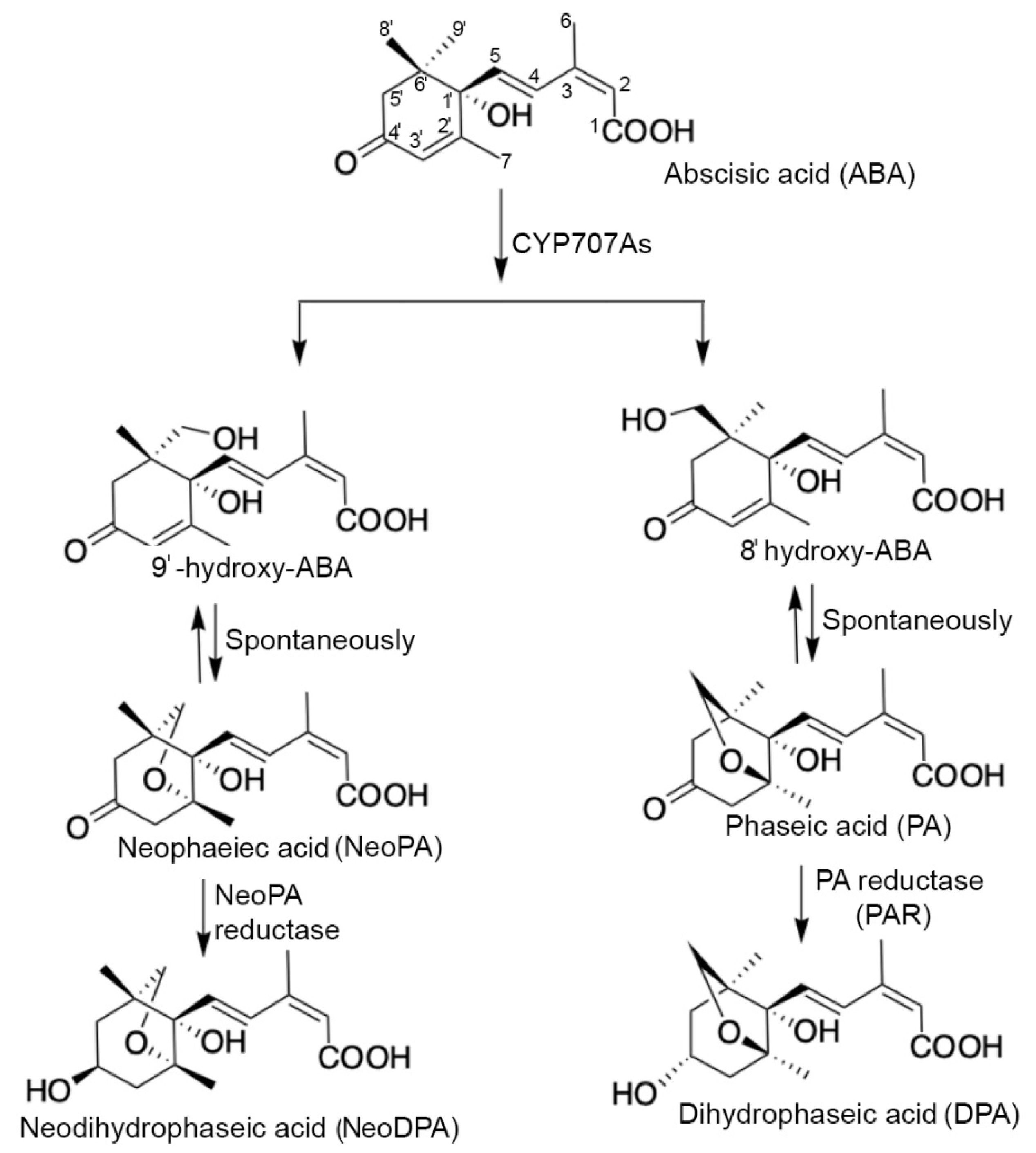
4.1. ABA catabolism
4.2. Annotation of ABA-associated DEGs
4.3. Epigenetic regulation of PsABI3, PsABI4, and PsABI5 genes based on DNA promoter methylation.
Supplementary Materials
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: a temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2017, 68, 857–869. [Google Scholar] [CrossRef]
- Yan, A.; Chen, Z. The control of seed dormancy and germination by temperature, light and nitrate. Bot. Rev. 2020, 86, 39–75. [Google Scholar] [CrossRef]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. B. 2008, 6, e0119. [Google Scholar] [CrossRef]
- Smolikova, G.; Strygina, K.; Krylova, E.; Vikhorev, A.; Bilova, T.; Frolov, A.; Khlestkina, E.; Medvedev, S. Seed-to-seedling transition in Pisum sativum L.: a transcriptomic approach. Plants 2022, 11, 1686. [Google Scholar] [CrossRef]
- Smolikova, G.; Strygina, K.; Krylova, E.; Leonova, T.; Frolov, A.; Khlestkina, E.; Medvedev, S. Transition from seeds to seedlings: hormonal and epigenetic aspects. Plants 2021, 10, 1884. [Google Scholar] [CrossRef]
- Silva, A.T.; Ligterink, W.; Hilhorst, H.W.M. Metabolite profiling and associated gene expression reveal two metabolic shifts during the seed-to-seedling transition in Arabidopsis thaliana. Plant Mol. Biol. 2017, 95, 481–496. [Google Scholar] [CrossRef]
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An updated overview on the regulation of seed germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef]
- Luján-Soto, E.; Dinkova, T.D. Time to wake up: Epigenetic and small-RNA-mediated regulation during seed germination. Plants 2021, 10, 236. [Google Scholar] [CrossRef]
- Smolikova, G.; Medvedev, S. Seed-to-seedling transition : Novel aspects. Plants 2022, 11, 1988. [Google Scholar] [CrossRef]
- Finkelstein, R.R. The role of hormones during seed development and germination. In Plant Hormones; Davies, P.J., Ed.; Springer Netherlands: Dordrecht, 2010; pp. 549–573. ISBN 978-1-4020-2684-3. [Google Scholar]
- Shu, K.; Meng, Y.J.; Shuai, H.W.; Liu, W.G.; Du, J.B.; Liu, J.; Yang, W.Y. Dormancy and germination: How does the crop seed decide? Plant Biol. 2015, 17, 1104–1112. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2022, 35, 199–214. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z. Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Steber, C.M. GA signaling is essential for the embryo-to-seedling transition during Arabidopsis seed germination, a ghost story. Plant Signal. Behav. 2020, 15, 1705028. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA metabolism and homeostasis in seed dormancy and germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic acid synthesis and response. Arab. B. 2013, 11, e0166. [Google Scholar] [CrossRef]
- Nambara, E.; Okamoto, M.; Tatematsu, K.; Yano, R.; Seo, M.; Kamiya, Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55. [Google Scholar] [CrossRef]
- Gutierrez, L.; Van Wuytswinkel, O.; Castelain, M.; Bellini, C. Combined networks regulating seed maturation. Trends Plant Sci. 2007, 12, 294–300. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Bai, Y.L.; Yin, X.; Xiong, C.F.; Cai, B.D.; Wu, Y.; Zhang, X.Y.; Wei, Z.; Ye, T.; Feng, Y.Q. Neophaseic acid catabolism in the 9′-hydroxylation pathway of abscisic acid in Arabidopsis thaliana. Plant Commun. 2022, 3, 100340. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef]
- Carbonero, P.; Iglesias-Fernández, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J. Exp. Bot. 2016, 68, erw458. [Google Scholar] [CrossRef]
- Han, J.-D.; Li, X.; Jiang, C.-K.; Wong, G.K.-S.; Rothfels, C.J.; Rao, G.-Y. Evolutionary analysis of the LAFL genes involved in the land plant seed maturation program. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Roscoe, T.T.; Guilleminot, J.; Bessoule, J.J.J.-J.; Berger, F.; Devic, M. Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in arabidopsis. Plant Cell Physiol. 2015, 56, 1215–1228. [Google Scholar] [CrossRef]
- Cagliari, A.; Turchetto-Zolet, A.C.; Korbes, A.P.; Maraschin, F. dos S.; Margis, R.; Margis-Pinheiro, M. New insights on the evolution of Leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 2014, 103, 380–387. [CrossRef]
- Jia, H.; Suzuki, M.; McCarty, D.R. Regulation of the seed to seedling developmental phase transition by the <scp>LAFL</scp> and <scp>VAL</scp> transcription factor networks. WIREs Dev. Biol. 2014, 3, 135–145. [Google Scholar] [CrossRef]
- Lepiniec, L.; Devic, M.; Roscoe, T.J.; Bouyer, D.; Zhou, D.-X.; Boulard, C.; Baud, S.; Dubreucq, B. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 2018, 31, 291–307. [Google Scholar] [CrossRef]
- Faria, J.M.R.; Buitink, J.; van Lammeren, A.A.M.; Hilhorst, H.W.M. Changes in DNA and microtubules during loss and re-establishment of desiccation tolerance in germinating Medicago truncatula seeds. J. Exp. Bot. 2005, 56, 2119–2130. [Google Scholar] [CrossRef]
- Dekkers, B.J.W.; Costa, M.C.D.; Maia, J.; Bentsink, L.; Ligterink, W.; Hilhorst, H.W.M. Acquisition and loss of desiccation tolerance in seeds: from experimental model to biological relevance. Planta 2015, 241, 563–577. [Google Scholar] [CrossRef]
- Smolikova, G.; Leonova, T.; Vashurina, N.; Frolov, A.; Medvedev, S. Desiccation tolerance as the basis of long-term seed viability. Int. J. Mol. Sci. 2021, 22, 101. [Google Scholar] [CrossRef]
- Sano, N.; Lounifi, I.; Cueff, G.; Collet, B.; Clément, G.; Balzergue, S.; Huguet, S.; Valot, B.; Galland, M.; Rajjou, L. Multi-omics approaches unravel specific features of embryo and endosperm in rice seed germination. Front. Plant Sci. 2022, 13, 1–20. [Google Scholar] [CrossRef]
- Buitink, J.; Ly Vu, B.; Satour, P.; Leprince, O. The re-establishment of desiccation tolerance in germinated radicles of Medicago truncatula Gaertn. seeds. Seed Sci. Res. 2003, 13, 273–286. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Saijo, T.; Shibata, D.; Morikami, A.; Nakamura, K. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2005, 138, 675–685. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Morikami, A.; Nakamura, K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. 2007, 104, 2543–2547. [Google Scholar] [CrossRef]
- Zinsmeister, J.; Lalanne, D.; Ly Vu, B.; Schoefs, B.; Marchand, J.; Dang, T.T.; Buitink, J.; Leprince, O. ABSCISIC ACID INSENSITIVE4 coordinates eoplast formation to ensure acquisition of seed longevity during maturation in Medicago truncatula. Plant J. 2023, 113, 934–953. [Google Scholar] [CrossRef]
- Zinsmeister, J.; Lalanne, D.; Terrasson, E.; Chatelain, E.; Vandecasteele, C.; Vu, B.L.; Dubois-Laurent, C.; Geoffriau, E.; Signor, C. Le; Dalmais, M.; et al. ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell 2016, 28, 2735–2754. [Google Scholar] [CrossRef]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.-H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef]
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef]
- Ding, X.; Jia, X.; Xiang, Y.; Jiang, W. Histone modification and chromatin remodeling during the seed life cycle. Front. Plant Sci. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Xing, M.Q.; Zhang, Y.J.; Zhou, S.R.; Hu, W.Y.; Wu, X.T.; Ye, Y.J.; Wu, X.X.; Xiao, Y.P.; Li, X.; Xue, H.W. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015, 168, 1417–1432. [Google Scholar] [CrossRef]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y. V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, D.; Kramdi, A.; Kassam, M.; Heese, M.; Schnittger, A.; Roudier, F.; Colot, V. DNA methylation dynamics during early plant life. Genome Biol. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Park, K.; Shin, S.-Y.; Frost, J.M.; Hsieh, P.-H.; Shin, C.; Fischer, R.L.; Hsieh, T.-F.; Choi, Y. Distinct regulatory pathways contribute to dynamic CHH methylation patterns in transposable elements throughout Arabidopsis embryogenesis. Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef]
- Li, W.-Y.; Chen, B.-X.; Chen, Z.-J.; Gao, Y.-T.; Chen, Z.; Liu, J. Reactive oxygen species generated by NADPHoxidases promote radicle protrusion and root elongation during rice seed germination. Int. J. Mol. Sci. 2017, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cabellos, S.; Toorop, P.E.; Cañal, M.J.; Iannetta, P.P.M.; Fernández-Pascual, E.; Pritchard, H.W.; Visscher, A.M. Global DNA methylation and cellular 5-methylcytosine and H4 acetylated patterns in primary and secondary dormant seeds of Capsella bursa-pastoris (L.) Medik. (shepherd’s purse). Protoplasma 2022, 259, 595–614. [Google Scholar] [CrossRef]
- Dew-Budd, K.J.; Chow, H.T.; Kendall, T.; David, B.C.; Rozelle, J.A.; Mosher, R.A.; Beilstein, M.A. RNA-directed DNA Methylation is required for seed development in outbreeding species. bioRxiv 2023, 1–20. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.; Pearson, J.; Hsieh, T.-F.; An, Y.-Q.; et al. Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [PubMed]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed]
- Kreplak, J.; Madoui, M.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, F.; Luo, X.; Dai, Y.; Yang, Y.; Zheng, C.; Yang, W.; Shu, K. A matter of life and death: Molecular, physiological, and environmental regulation of seed longevity. Plant Cell Environ. 2020, 43, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J.J. Molecular mechanisms of seed dormancy. Plant. Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Jikumaru, Y.; Hanada, A.; Nambara, E.; Abrams, S.R.; Kamiya, Y.; Seo, M. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707A s encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oOxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Milborrow, B. V; Vaughan, G.T. Characterization of dihydrophaseic acid 4′-O-β-D-glucopyranoside as a major metabolite of abscisic acid. Funct. Plant Biol. 1982, 9, 361–372. [Google Scholar] [CrossRef]
- Cai, W.-J.; Zeng, C.; Zhang, X.-Y.; Ye, T.; Feng, Y.-Q. A structure–guided screening strategy for the discovery and identification of potential gibberellins from plant samples using liquid chromatography–mass spectrometry assisted by chemical isotope labeling. Anal. Chim. Acta 2021, 1163, 338505. [Google Scholar] [CrossRef]
- Zeevaart, J.A.D.; Milborrow, B. V Metabolism of abscisic acid and the occurrence of epi-dihydrophaseic acid in Phaseolus vulgaris. Phytochemistry 1976, 15, 493–500. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Raschke, K. Effects of phaseic acid and dihydrophaseic acid on stomata and the photosynthetic apparatus. Plant Physiol. 1980, 65, 291–297. [Google Scholar] [CrossRef]
- Walker-Simmons, M.K.; Holappa, L.D.; Abrams, G.D.; Abrams, S.R. ABA metabolites induce group 3 LEA mRNA and inhibit germination in wheat. Physiol. Plant. 1997, 100, 474–480. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef]
- Liu, W.; Thapa, P.; Park, S.-W. RD29A and RD29B rearrange genetic and epigenetic markers in priming systemic defense responses against drought and salinity. Plant Sci. 2023, 337, 111895. [Google Scholar] [CrossRef]
- Medvedev, S.S. Principles of calcium signal generation and transduction in plant cells. Russ. J. Plant Physiol. 2018, 65, 771–783. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y. How do plants maintain pH and ion homeostasis under saline-alkali stress? Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef]
- Dyla, M.; Basse Hansen, S.; Nissen, P.; Kjaergaard, M. Structural dynamics of P-type ATPase ion pumps. Biochem. Soc. Trans. 2019, 47, 1247–1257. [Google Scholar] [CrossRef]
- Sim, S.I.; Park, E. P5-ATPases: Structure, substrate specificities, and transport mechanisms. Curr. Opin. Struct. Biol. 2023, 79, 102531. [Google Scholar] [CrossRef]
- Aguilar-Martínez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef]
- Ferrero, L. V.; Gastaldi, V.; Ariel, F.D.; Viola, I.L.; Gonzalez, D.H. Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 2021, 105, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Alem, A.L.; Ariel, F.D.; Cho, Y.; Hong, J.C.; Gonzalez, D.H.; Viola, I.L. TCP15 interacts with GOLDEN2-LIKE1 to control cotyledon opening in Arabidopsis. Plant J. 2022, 110, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Reeves, W.M.; Lynch, T.J.; Mobin, R.; Finkelstein, R.R. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 2011, 75, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Chen, Y.; Wang, C.; Kong, Y.; Wu, W.; Chen, Y. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, M.; Qu, L.; Yang, J.; Liu, X.; Zhao, Q.; Liu, X.; Zhao, X. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Wei, J.; Li, X.; Song, P.; Wang, Y.; Ma, J. Studies on the interactions of AFPs and bZIP transcription factor ABI5. Biochem. Biophys. Res. Commun. 2022, 590, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef]
- Maymon, T.; Eisner, N.; Bar-Zvi, D. The ABCISIC ACID INSENSITIVE (ABI) 4 transcription factor is stabilized by stress, ABA and phosphorylation. Plants 2022, 11, 2179. [Google Scholar] [CrossRef]
- Gregorio, J.; Hernández-Bernal, A.F.; Cordoba, E.; León, P. Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA-INSENSITIVE (ABI) 4 transcription factor. Mol. Plant 2014, 7, 422–436. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Wang, F.; Zheng, Q.; Niza, V.M.A.G.E.; Downie, A.B.; Perry, S.E. Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 2020, 103, 1679–1694. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, X.; Wang, K.; Li, Z.; Jia, Q.; Zhao, C.; Zhang, M. ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2022, 73, 2077–2092. [Google Scholar] [CrossRef]
- Parcy, F.; Valon, C.; Kohara, A.; Misera, S.; Giraudat, J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, AND LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 1997, 9, 1265–1277. [Google Scholar]
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxidants Redox Signal. 2011. [CrossRef] [PubMed]
- Chen, H.; Chu, P.; Zhou, Y.; Ding, Y.; Li, Y.; Liu, J.; Jiang, L.; Huang, S. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 2016, 88, 608–619. [Google Scholar] [CrossRef]
- Chen, H.; Ruan, J.; Chu, P.; Fu, W.; Liang, Z.; Li, Y.; Tong, J.; Xiao, L.; Liu, J.; Li, C.; et al. AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 2020, 101, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Haslekås, C.; Viken, M.K.; Grini, P.E.; Nygaard, V.; Nordgard, S.H.; Meza, T.J.; Aalen, R.B. Seed 1-cysteine peroxiredoxin antioxidants are not involved in dormancy, but contribute to inhibition of germination during stress. Plant Physiol. 2003, 133, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant group II LEA proteins: intrinsically disordered structure for multiple functions in response to environmental stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Qi, S.; Li, H.; Liu, P.; Li, P.; Wu, C.; Zheng, C.; Huang, J. Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014, 454, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Ecker, J.R. Diversity and dynamics of dna methylation: Epigenomic resources and tools for crop breeding. Breed. Sci. 2019, 69, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Plitta-Michalak, B.P.; Suszka, J.; Naskręt-Barciszewska, M.Z.; Kotlarski, S.; Barciszewski, J.; Chmielarz, P. Identification of DNA methylation changes in European beech seeds during desiccation and storage. Int. J. Mol. Sci. 2023, 24, 3557. [Google Scholar] [CrossRef] [PubMed]
- An, Y.Q.C.; Goettel, W.; Han, Q.; Bartels, A.; Liu, Z.; Xiao, W. Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Le, B.H.; Chen, M.; Henry, K.F.; Hur, J.; Hsieh, T.-F.; Chen, P.-Y.; Pelletier, J.M.; Pellegrini, M.; Fischer, R.L.; et al. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. 2017, 114, E9730–E9739. [Google Scholar] [CrossRef]
- Weng, J.-K.; Ye, M.; Li, B.; Noel, J.P. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 2016, 166, 881–893. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




