1. Introduction
Antibiotics are substances employed to address infections stemming from bacterial sources. They exhibit diverse modes of action, including hindering bacterial growth, eradicating bacteria, or impeding their reproductive processes. Within the realm of medicine, antibiotics find extensive use in treating a broad spectrum of bacterial infections[
1,
2,
3,
4].
It's crucial to recognize that antibiotics lack efficacy against viral infections, such as the common cold or influenza. Inappropriate antibiotic utilization, such as unnecessary intake or failure to complete the prescribed regimen, can contribute to the emergence of antibiotic-resistant bacteria, constituting a burgeoning concern in public health. Antibiotics are categorized based on their mechanism of action and their target bacteria. Examples encompass penicillins, cephalosporins, tetracyclines, macrolides, quinolones, and sulfonamides[
1,
2,
3,
4].
Antibiotics are generally grouped into classes based on their chemical structure. It is important to note that antibiotics of the same class can affect the body differently and can be effective on different bacteria. Let's review the main antibiotic classes based on their mechanism of action[
1,
2].
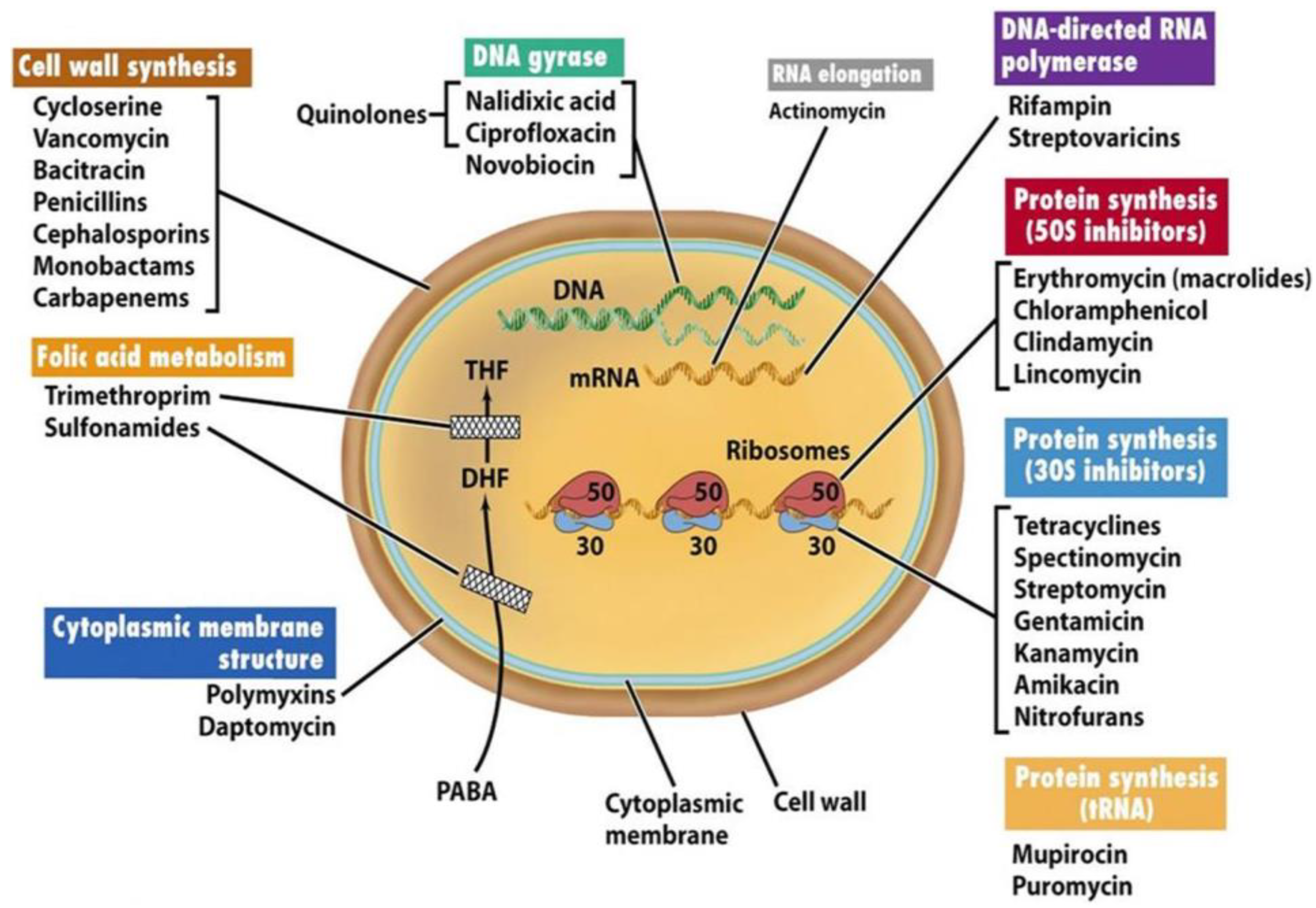
Figure 1 shows the mechanical actions of several antibiotics.
This work present a brief overview of some main antibiotic classes and their general mechanisms of action[
5,
6,
7]:
Mechanism: Inhibit bacterial cell wall synthesis by binding to enzymes (penicillin-binding proteins) involved in cell wall formation.
Example: Amoxicillin, Ampicillin.
- -
Cephalosporins:
Mechanism: Similar to penicillins, they also target bacterial cell wall synthesis.
Example: Cephalexin, Ceftriaxone.
- -
Tetracyclines:
Mechanism: Inhibit bacterial protein synthesis by binding to the bacterial ribosome.
Example: Doxycycline, Tetracycline.
- -
Macrolides:
Mechanism: Interfere with bacterial protein synthesis by binding to the ribosome.
Example: Azithromycin, Erythromycin.
- -
Quinolones/Fluoroquinolones:
Mechanism: Inhibit DNA gyrase, an enzyme involved in bacterial DNA replication.
Example: Ciprofloxacin, Levofloxacin.
- -
Sulfonamides:
Mechanism: Inhibit bacterial folic acid synthesis, a crucial component for DNA and RNA synthesis.
Example: Trimethoprim-Sulfamethoxazole (TMP-SMX).
- -
Aminoglycosides:
Mechanism: Disrupt bacterial protein synthesis by binding to the bacterial ribosome.
Example: Gentamicin, Amikacin.
- -
Glycopeptides:
Mechanism: Inhibit bacterial cell wall synthesis.
Example: Vancomycin.
- -
Oxazolidinones:
Mechanism: Inhibit bacterial protein synthesis.
Example: Linezolid.
Figure 1.
Target sites of antibiotics (Madigan and Martinko, 2006) - "Antibiotics: Classification and mechanisms of action with a focus on molecular perspectives." Figure was reproduced according to Ref [
7].
Figure 1.
Target sites of antibiotics (Madigan and Martinko, 2006) - "Antibiotics: Classification and mechanisms of action with a focus on molecular perspectives." Figure was reproduced according to Ref [
7].
The objective of this succinct scientific investigation is to evaluate different antibiotics based on various parameters related to their mechanisms of action and subsequently use the pkCSM Database[
8] to pinpoint the least detrimental among them. several toxicity parameters were performed by PkCSM Database[
8] (Preclinical Knowledge-Based Consensus Models) for antibiotics.
These parameters include:
AMES Toxicity: Assessing mutagenicity using the AMES test.
Max. Tolerated Dose (Human): Determining the maximum dose tolerated by humans.
Oral Rat Acute Toxicity (LD50): Identifying the lethal dose required to cause mortality in 50% of rats after oral administration.
Oral Rat Chronic Toxicity (LOAEL): Establishing the Lowest Observable Adverse Effect Level in rats after prolonged oral exposure.
T. Pyriformis Toxicity: Assessing toxicity specifically related to T. pyriformis (a ciliated protozoan often used in toxicity testing).
Minnow Toxicity: Evaluating toxicity with respect to minnows, a type of small freshwater fish.
These parameters provide a comprehensive view of antibiotic toxicity, encompassing mutagenicity, dose tolerance in humans, acute and chronic effects in rats, as well as specific impacts on aquatic organisms. The use of PkCSM, which relies on these parameters, helps in identifying antibiotics with potentially lower toxicity profiles.
2. Material and Methods
Toxicity parameters thorugh PkCSM [
8] investigated are:
-AMES Toxicity>> Positive Test: Indicates that the compound exhibits mutagenic properties
-Oral Rat Acute Toxicity (LD50) [(mol/kg)]>>
-Oral Rat Chronic Toxicity (LOAEL) [(log mg/kg_bw/day)]>>
-T. Pyriformis Toxicity [(log ug/L)]>> T. pyriformis, measured by pIGC50 (the concentration required to inhibit 50% growth), is evaluated for a given compound. If the pIGC50 value is greater than -0.5 log µL/L, it is considered toxic.
-Minnow Toxicity (LC50)[(log mM)]>> LC50 represents the concentration of a molecule required to cause the death of 50% of the population. Furthermore, if the LC50 value is below 0.5 mM (log LC50 < -0.3), it is considered to exhibit high acute toxicity.
-Max. Tolerated Dose (Human)>> [MRTD (log mg/kg/day)]: For a given compound, if the logarithm of the dose in milligrams per kilogram per day is less than or equal to 0.477, it is considered low. If the logarithm of the dose in milligrams per kilogram per day is greater than 0.477, it is considered high.
-Hepatotoxicity
-Skin Sensitisation
3. Results and Discussion
For the first time this work described the toxicity parameters thorugh PkCSM (Preclinical Knowledge-Based Consensus Models) wich it is capable of predicting. PkCSM is a computational tool designed to predict various toxicity endpoints for chemical compounds, including antibiotics[
8]. Each of the mentioned toxicity parameters serves a specific purpose in assessing the safety and potential risks associated with the use of antibiotics:
-AMES Toxicity: Predicting mutagenicity through the AMES test helps identify substances that may cause genetic mutations.
-Max. Tolerated Dose (Human): Estimating the maximum dose tolerated by humans assists in understanding the potential for adverse effects at high doses.
-Oral Rat Acute Toxicity (LD50): Predicting the lethal dose in rats after oral administration provides insights into acute toxicity and potential harm.
-Oral Rat Chronic Toxicity (LOAEL): Determining the Lowest Observable Adverse Effect Level in rats after prolonged oral exposure helps assess chronic toxicity.
-T. Pyriformis Toxicity: Assessing toxicity specifically related to T. pyriformis aids in understanding the impact on this organism, often used in toxicity testing.
-Minnow Toxicity: Evaluating toxicity with respect to minnows provides information on the potential harm to small freshwater fish.
The predictive capabilities of pkCSM in these areas contribute to the early identification of potential risks associated with antibiotics, facilitating more informed decision-making in drug development and safety assessment.
In establishing the relative toxicity of compounds, we must take into account the presented toxicity parameters. These include measures such as AMES toxicity, Max. tolerated dose (human), Oral Rat Acute Toxicity (LD50), Oral Rat Chronic Toxicity (LOAEL), T. Pyriformis toxicity, and Minnow toxicity.It's crucial to emphasize that assessing toxicity requires a thorough analysis, and individual parameters may hold varying degrees of influence on the overall toxicity profile. Furthermore, the criteria for classifying a compound as less toxic may vary based on the specific context and the particular toxicity endpoint under consideration. From these toxicity results, based on the specified parameters: Max. Tolerated Dose (Human) (log mg/kg/day),Oral Rat Acute Toxicity (LD50) (mol/kg), Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) and Minnow Toxicity (LC50) (log mM)
Four potential antibiotics named cefotaxime,ceftriaxone,imipenem and meropenem respectively are showed lower toxicity compared to other compounds:
- -
Max. Tolerated Dose (Human) (log mg/kg/day):
cefotaxime: 1,625 log mg/kg/day
ceftriaxone: 1,489 log mg/kg/day
imipenem: 1,431 log mg/kg/day
meropenem: 1.423 log mg/kg/day
- -
Oral Rat Acute Toxicity (LD50) (mol/kg):
cefotaxime: 2,145 mol/kg
ceftriaxone: 2,352 mol/kg
imipenem: 1,641 mol/kg
meropenem: 1.946 mol/kg
- -
Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day):
cefotaxime: 2.109 log mg/kg_bw/day
ceftriaxone: 2,269 log mg/kg_bw/day
imipenem: 1.958 log mg/kg_bw/day
meropenem: 2.371 log mg/kg_bw/day
- -
Minnow Toxicity (LC50) (log mM):
cefotaxime: 4.835 log mM
ceftriaxone: 4,976 log mM
imipenem: 3,435 log mM
meropenem: 4.074 log mM
In each parameter, the values for cefotaxime, ceftriaxone, imipenem, and meropenem are either higher or within a range that indicates lower toxicity compared to other compounds. However, it's essential to consider the specific context and additional factors in a clinical scenario.
To sum Cefotaxime, ceftriaxone, imipenem, and meropenem demonstrated lower toxicity, suggesting a favorable safety profile.
However, clinical application and additional factors must be considered for a comprehensive assessment.
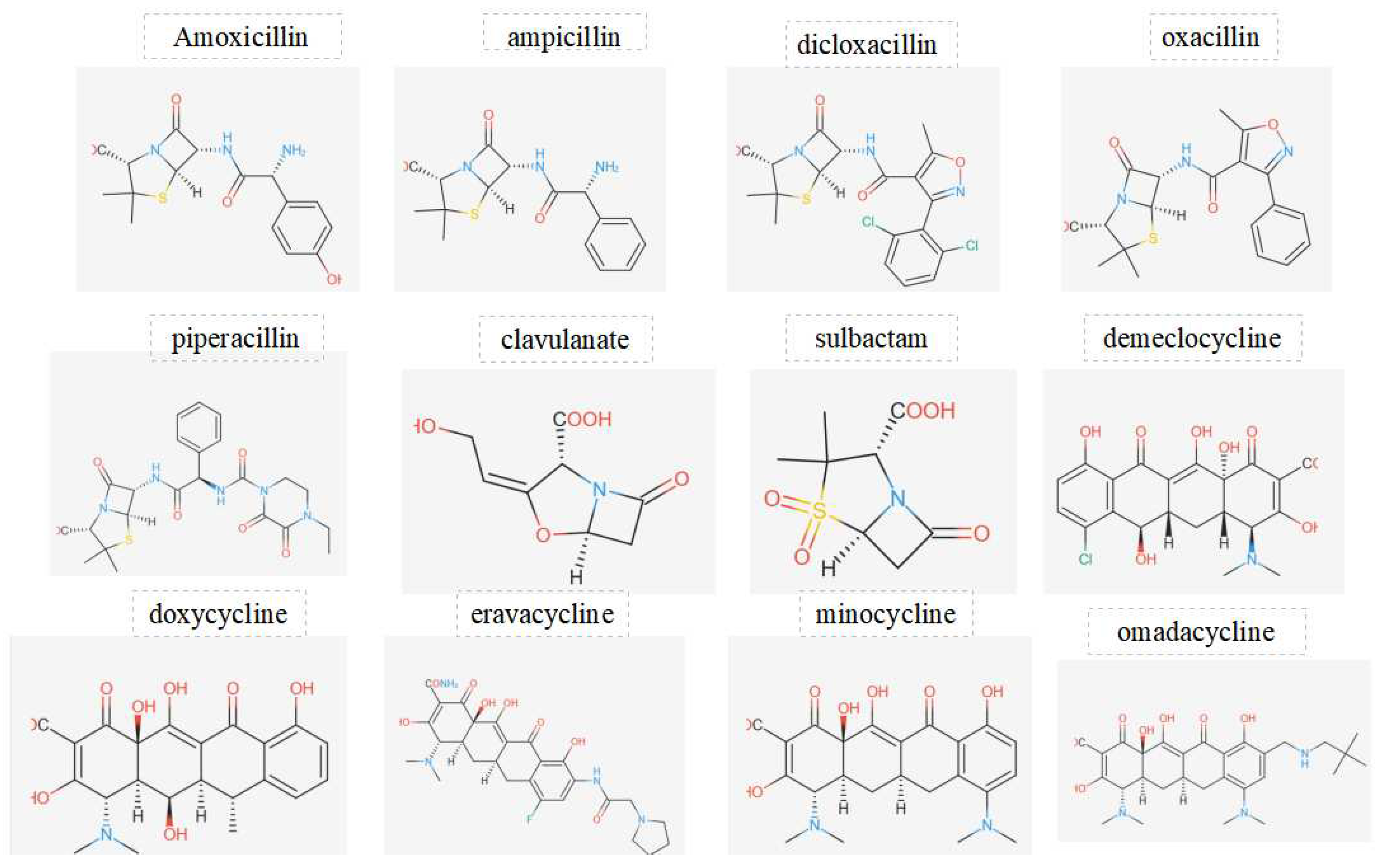
Figure 2.
Displays the chemical structures of antibiotics investigated in this work.
Figure 2.
Displays the chemical structures of antibiotics investigated in this work.
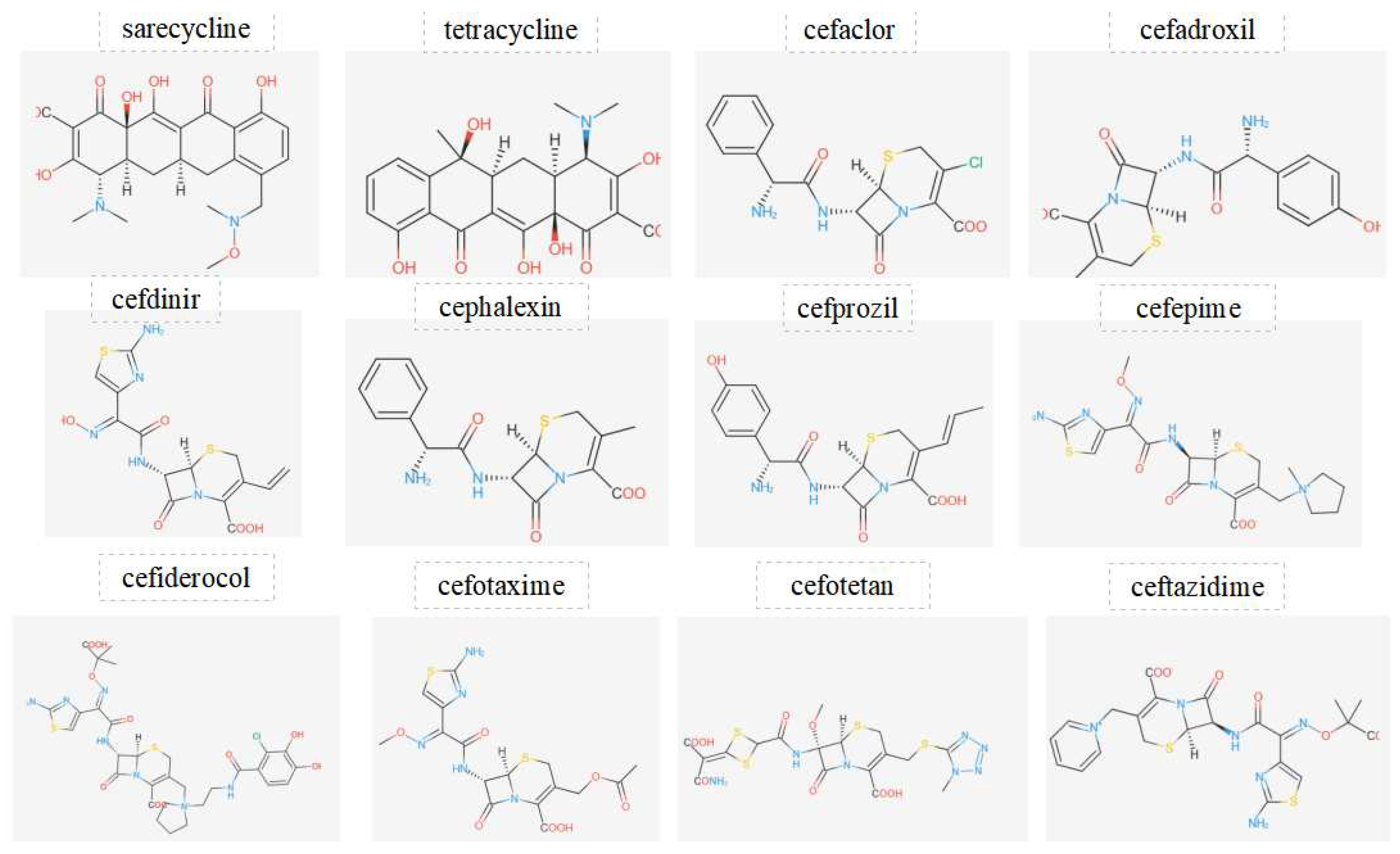
Figure 3.
Displays the chemical structures of antibiotics investigated in this work.
Figure 3.
Displays the chemical structures of antibiotics investigated in this work.
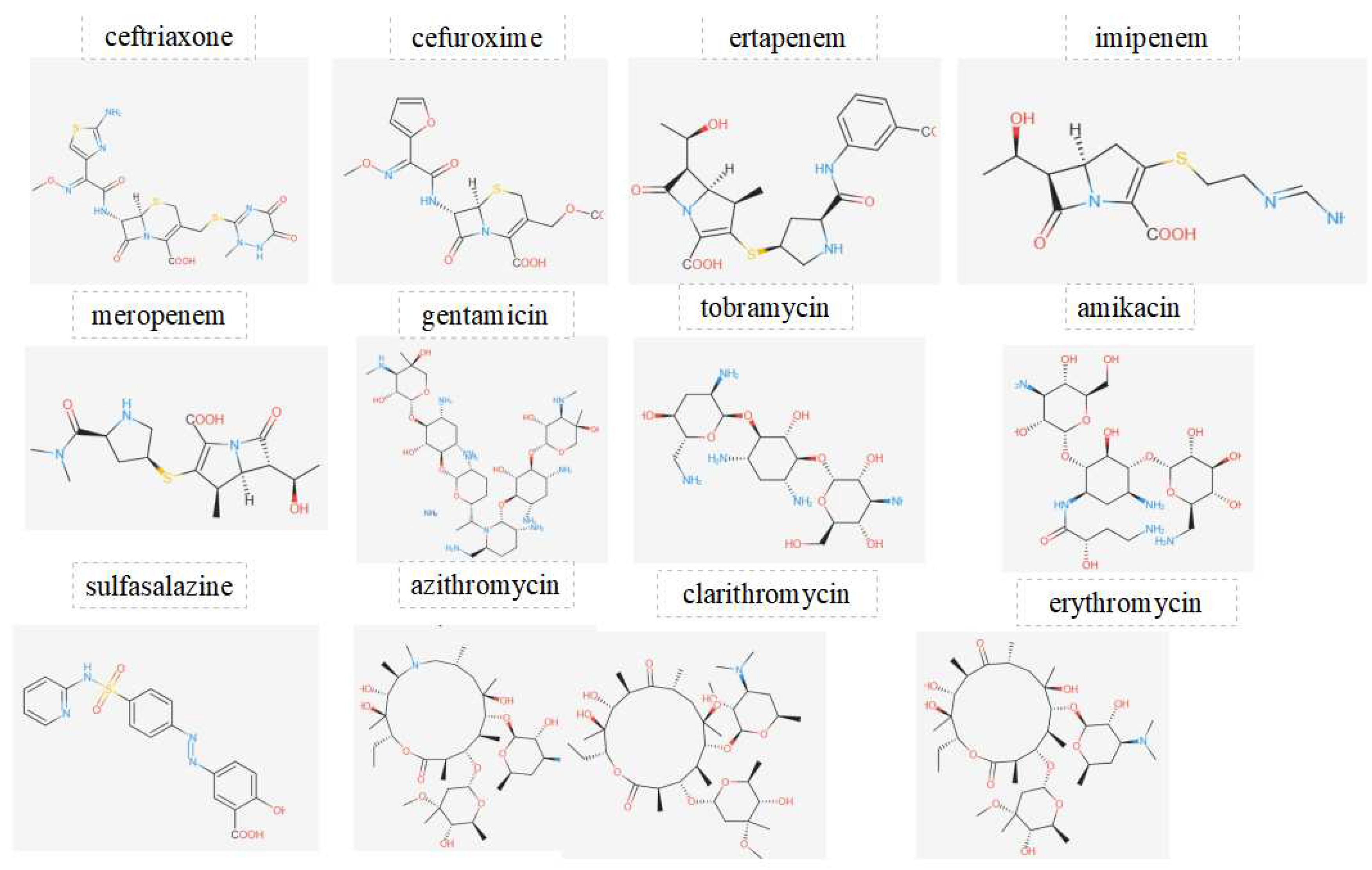
Figure 4.
Displays the chemical structures of antibiotics investigated in this work.
Figure 4.
Displays the chemical structures of antibiotics investigated in this work.
Figure 5.
Displays the chemical structures of antibiotics investigated in this work.
Figure 5.
Displays the chemical structures of antibiotics investigated in this work.
Table 1.
Displays the comparison of predicted toxicity properties of investigated antibiotics in this work.
Table 1.
Displays the comparison of predicted toxicity properties of investigated antibiotics in this work.
| Compounds |
Brand
Name |
AMES toxicity |
Max.
tolerated
dose (human) (log mg/kg/day) |
Oral Rat Acute Toxicity (LD50) (mol/kg) |
Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) |
Hepatotoxicity |
Skin
Sensitisation |
T.
Pyriformis
toxicity
(log ug/L) |
Minnow
toxicity
(log mM) |
| Amoxicillin |
Amoxil |
No |
1.04 |
1.77 |
2.638 |
yes |
no |
0.285 |
4.065 |
| ampicillin |
Unasyn |
No |
0.921 |
1.693 |
2.663 |
yes |
no |
0.285 |
3.502 |
| dicloxacillin |
/ |
No |
0.984 |
2.227 |
2.404 |
yes |
no |
0.285 |
2.842 |
| oxacillin |
/ |
No |
0.67 |
2.062 |
2.525 |
yes |
no |
0.285 |
3.286 |
| Phenoxymethylpenicillin |
Pen VK |
No |
0.751 |
2.037 |
2.81 |
yes |
no |
0.285 |
3.884 |
| piperacillin |
Pipracil |
No |
0.96 |
2.149 |
2.81 |
yes |
no |
0.285 |
5.136 |
| Clavulanic acid |
Aumetin |
No |
1.534 |
1.441 |
2.167 |
yes |
no |
0.273 |
3.766 |
| Sulbactam |
Unasyn |
No |
1.234 |
1.851 |
2.253 |
yes |
no |
0.272 |
3.689 |
| Demeclocycline |
/ |
No |
1.05 |
2.555 |
4.427 |
no |
no |
0.285 |
4.601 |
| Doxycycline |
Acticlate |
No |
1.108 |
2.401 |
5.056 |
No |
No |
0.285 |
3.518 |
| Eravacycline |
Xerava |
No |
1.19 |
2.366 |
4.509 |
No |
No |
0.285 |
4.997 |
| minocycline |
Amzeeq |
No |
0.746 |
2.235 |
4.045 |
No |
No |
0.285 |
2.615 |
| omadacycline |
Nuzyra |
No |
0.813 |
2.392 |
4.015 |
No |
No |
0.285 |
1.995 |
| sarecycline |
/ |
No |
0.843 |
2.238 |
4.055 |
No |
No |
0.285 |
2.992 |
| tetracycline |
Pylera |
No |
1.173 |
2.503 |
4.288 |
No |
No |
0.285 |
4.775 |
| cefaclor |
/ |
No |
0.928 |
1.836 |
2.643 |
yes |
No |
0.285 |
3.323 |
| cefadroxil |
/ |
No |
1.043 |
1.771 |
2.633 |
yes |
No |
0.285 |
4.061 |
| cefdinir |
Omnicef |
No |
1.671 |
2.244 |
2.342 |
yes |
No |
0.285 |
3.924 |
| cephalexin |
Keflex |
No |
0.926 |
1.695 |
2.658 |
yes |
No |
0.285 |
3.498 |
| cefprozil |
/ |
No |
0.903 |
1.836 |
2.665 |
yes |
No |
0.285 |
3.739 |
| cefepime |
/ |
No |
0.764 |
1.743 |
1.448 |
yes |
No |
0.285 |
3.332 |
| cefiderocol |
Fetroja |
No |
1.407 |
2.403 |
1.847 |
yes |
No |
0.285 |
3.459 |
| cefotaxime |
Claforan |
No |
1.625 |
2.145 |
2.109 |
yes |
No |
0.285 |
4.835 |
| cefotetan |
Cefotan |
No |
1.285 |
2.378 |
2.975 |
yes |
No |
0.285 |
3.586 |
| ceftazidime |
Avycaz |
No |
1.365 |
2.328 |
1.275 |
yes |
No |
0.285 |
3.054 |
| ceftriaxone |
Rocephin |
No |
1.489 |
2.352 |
2.269 |
yes |
No |
0.285 |
4.976 |
| cefuroxime |
Ceftin |
No |
1.264 |
1.602 |
2.516 |
yes |
No |
0.285 |
4.397 |
| ertapenem |
Invanz |
No |
0.967 |
2.209 |
2.721 |
yes |
No |
0.285 |
3.681 |
| imipenem |
Primaxin |
No |
1.431 |
1.641 |
1.958 |
yes |
No |
0.285 |
3.435 |
| meropenem |
Vabomere |
No |
1.423 |
1.946 |
2.371 |
yes |
No |
0.285 |
4.074 |
| gentamicin |
Gentak |
No |
0.45 |
2.482 |
7.124 |
No |
No |
0.285 |
11.971 |
| tobramycin |
Bethkis |
No |
1.291 |
1.877 |
4.144 |
No |
No |
0.285 |
7.987 |
| Amikacin |
Arikayce |
No |
1.11 |
2.663 |
5.724 |
No |
No |
0.285 |
11.087 |
| sulfasalazine |
Azulfidine |
No |
0.271 |
1.682 |
0.991 |
No |
No |
0.305 |
1.228 |
| azithromycin |
Azasite |
No |
1.252 |
2.446 |
1.975 |
yes |
No |
0.285 |
6.118 |
| clarithromycin |
Biaxin |
No |
1.268 |
2.703 |
1.658 |
yes |
No |
0.285 |
7.403 |
| erythromycin |
Aktipak |
No |
1.187 |
2.517 |
1.978 |
yes |
No |
0.285 |
6.92 |
| fidaxomicin |
Dificid |
No |
-0.507 |
3.465 |
3.382 |
yes |
No |
0.285 |
2.907 |
| ciprofloxacin |
Cetraxal |
yes |
-0.157 |
2.248 |
1.615 |
yes |
No |
0.485 |
1.013 |
| delafloxacin |
Baxdela |
yes |
0.746 |
2.142 |
2 |
yes |
No |
0.285 |
1.997 |
| levofloxacin |
Levaquin |
No |
0.425 |
2.218 |
1.802 |
yes |
No |
0.285 |
1.332 |
| moxifloxacin |
Avelox |
No |
0.846 |
2.166 |
1.6 |
yes |
No |
0.285 |
2.02 |
| gemifloxacin |
Factive |
No |
0.93 |
2.024 |
1.786 |
yes |
No |
0.285 |
2.256 |
| clindamycin |
Acanya |
No |
0.901 |
2.464 |
3.194 |
yes |
No |
0.285 |
1.97 |
| lincomycin |
Lincocin |
No |
1.259 |
2.593 |
3.405 |
yes |
No |
0.285 |
3.463 |
4. Conclusion
This work focused on the investigation of 60 different antibiotics evaluating their main toxicity parameters through pkCSM database, including Max. Tolerated Dose (Human) (log mg/kg/day), Oral Rat Acute Toxicity (LD50) (mol/kg), Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day), and Minnow Toxicity (LC50) (log mM). From these results four potential antibiotics are showed less toxicity effects which are cefotaxime, ceftriaxone, imipenem, and meropenem respectively. They have demonstrated lower toxicity compared to other compounds. These findings suggest that these antibiotics may have a relatively favorable safety profile when considering these specific toxicity parameters. However, it's crucial to interpret these results in the context of the specific clinical application and to consider additional factors for a comprehensive safety assessment.
It's important to note that the assessment of toxicity is complex, and the determination of what is considered "less toxic" can depend on specific criteria and the context of use. Additionally, individual responses to antibiotics can vary, and the selection of an antibiotic should be based on various factors, including the specific infection being treated, the patient's health, and potential side effects.
References
- Hutchings, M. I., Truman, A. W., & Wilkinson, B. Antibiotics: past, present and future. Current opinion in microbiology, 2019, 51, 72-80. [CrossRef]
- Mohr, K. I. (2016). History of antibiotics research. How to Overcome the Antibiotic Crisis: Facts, Challenges, Technologies and Future Perspectives, 237-272. [CrossRef]
- Larsson, D. J. (2014). Antibiotics in the environment. Upsala journal of medical sciences, 119(2), 108-112. [CrossRef]
- Hamad, B. (2010). The antibiotics market. Nature reviews Drug discovery, 9(9), 675. [CrossRef]
- Masouda, M. S., Alib, A. E., & Nasrc, N. M. (2014). Chemistry, classification, pharmacokinetics, clinical uses and analysis of beta lactam antibiotics: a review. Journal of Chemical and Pharmaceutical Research, 6(11), 28-58.
- Kümmerer, K. (2003). Significance of antibiotics in the environment. Journal of Antimicrobial Chemotherapy, 52(1), 5-7. [CrossRef]
- Etebu, E., & Arikekpar, I. (2016). Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res, 4(2016), 90-101.
- 8. Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of medicinal chemistry, 58(9), 4066-4072. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).