Submitted:
13 December 2023
Posted:
14 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant material
2.2. DNA and RNA extraction and cDNA synthesis
2.3. Primer design and gene cloning and sequencing
2.4. Phylogeny analysis
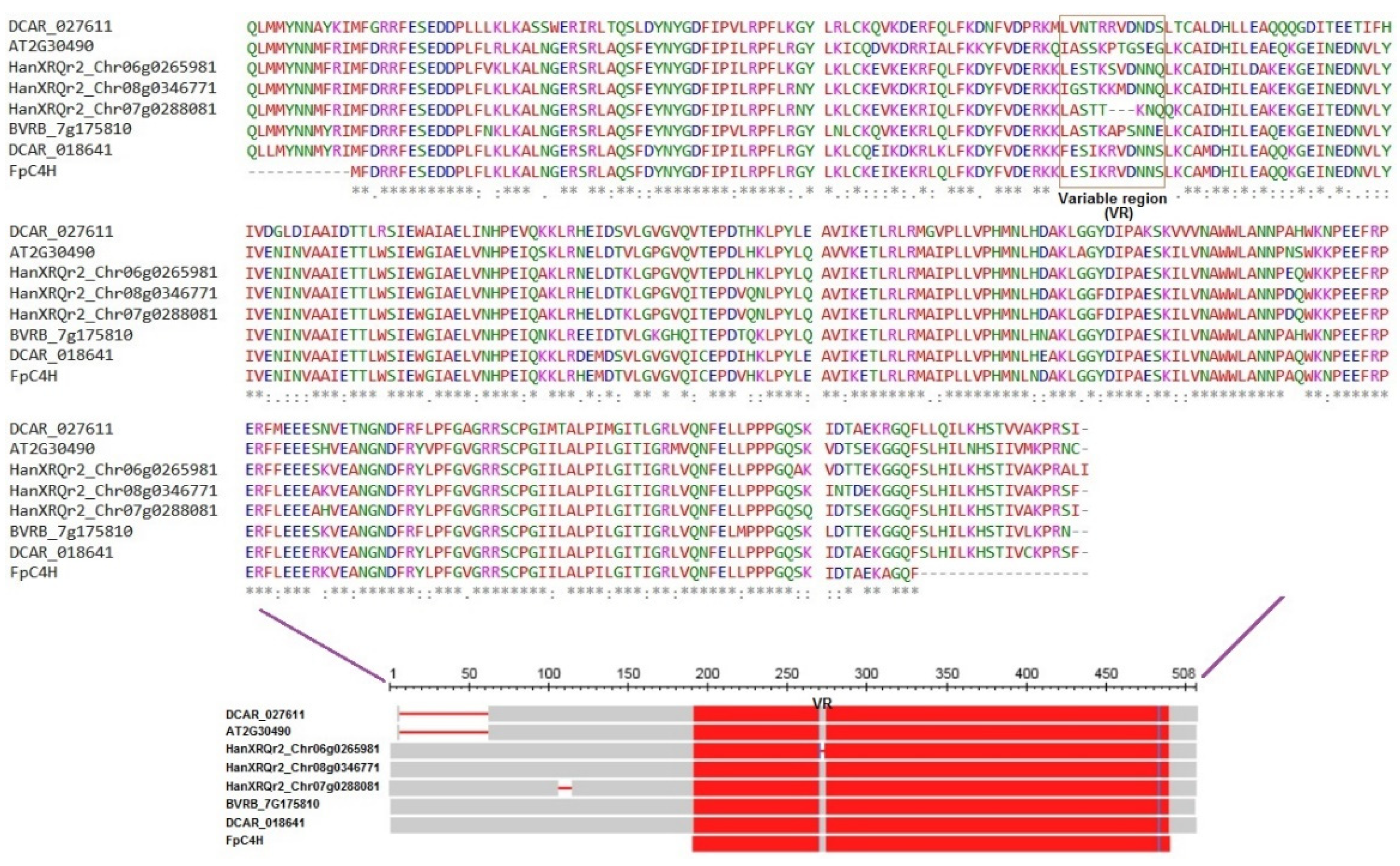
2.5. Sequence structure analysis
2.6. Real-time PCR analysis
3. Results
3.1. Screening and confirmation of clones
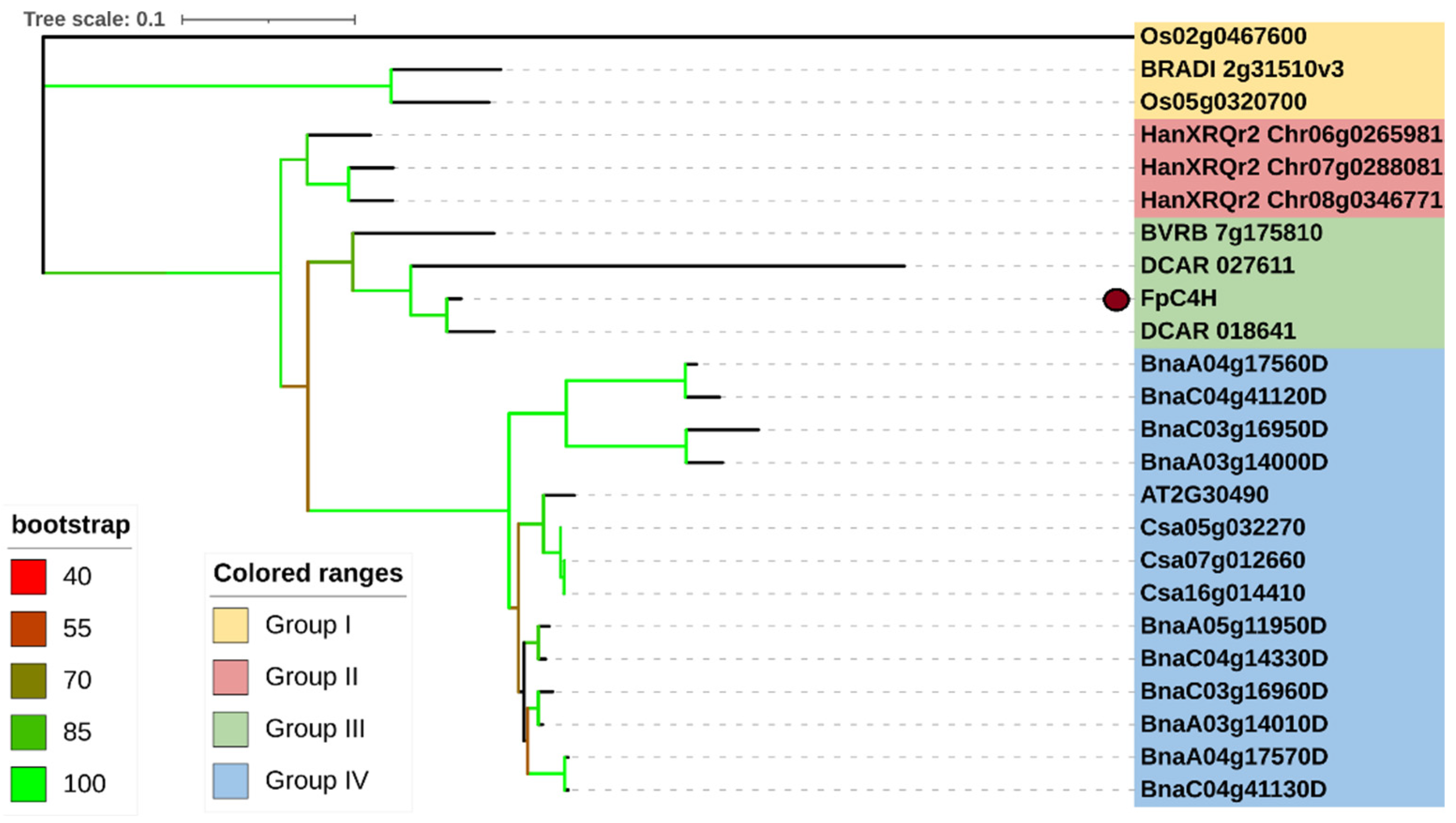
3.2. Phylogenetic analysis
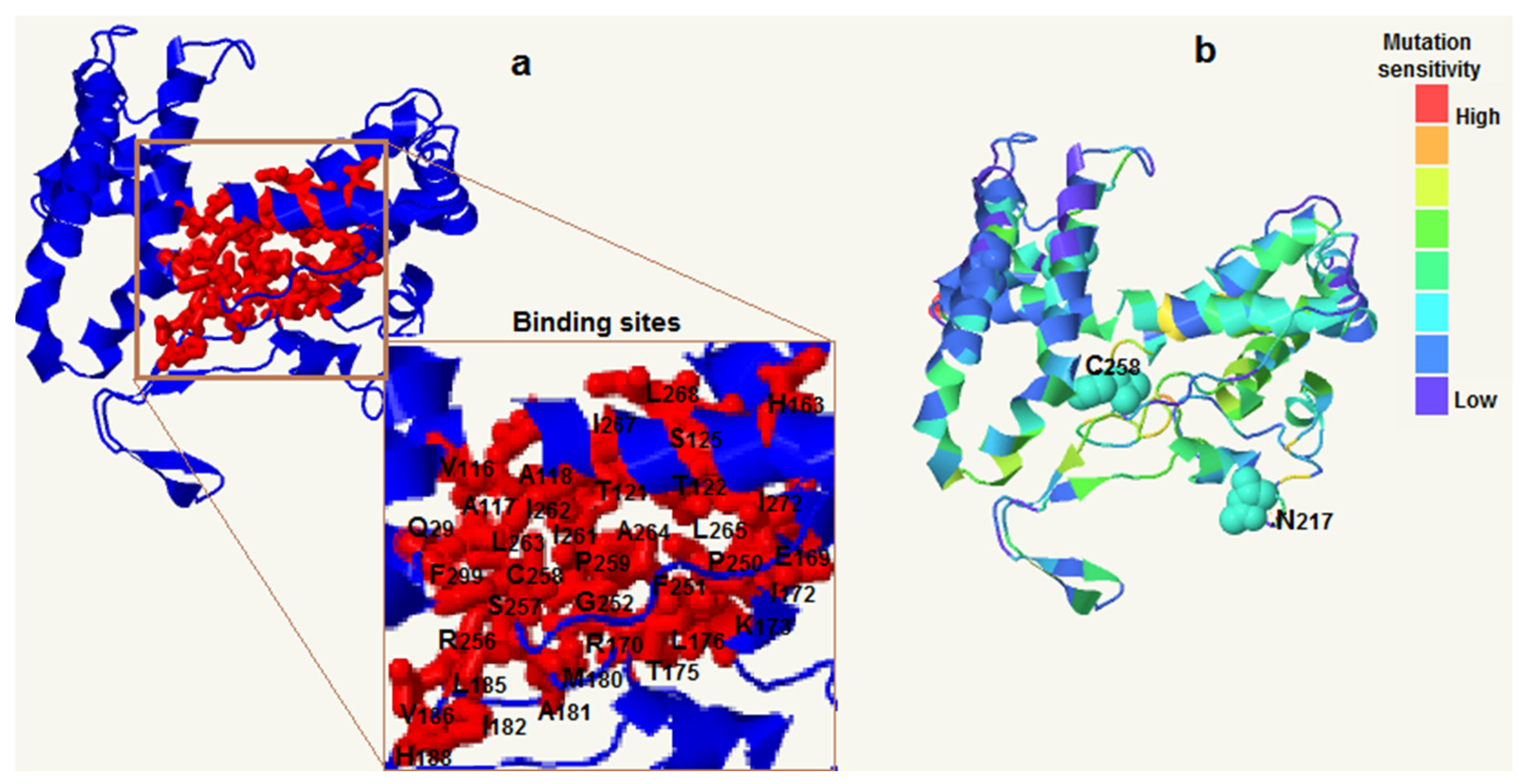
3.3. Three-dimensional structure and binding sites of FpC4H
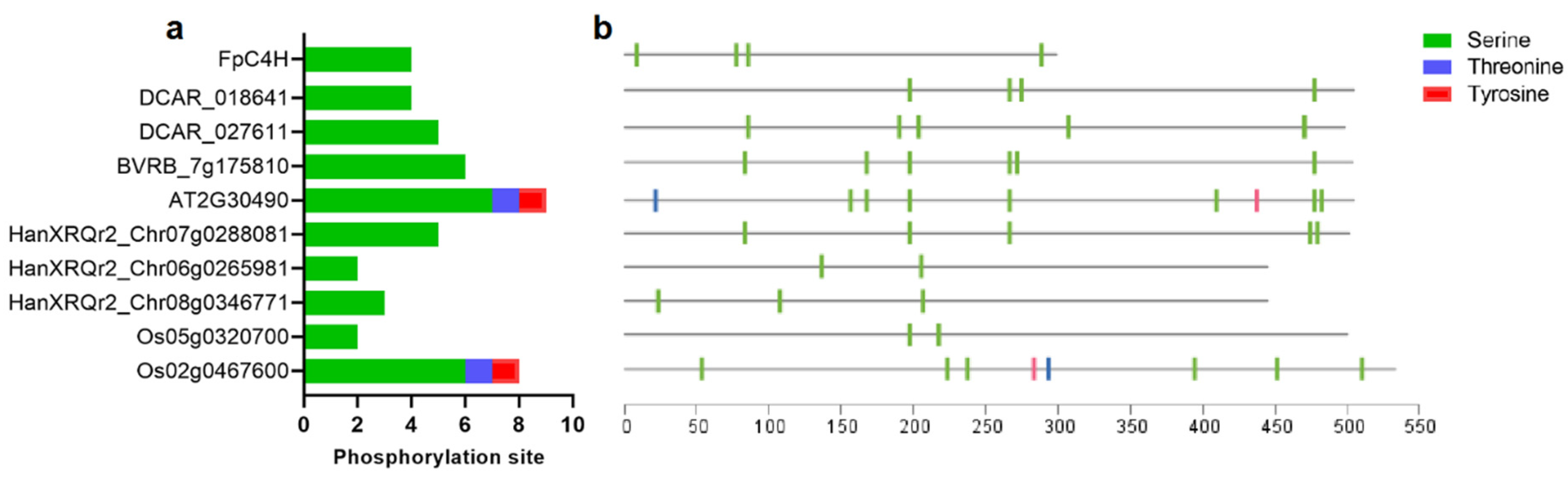
3.4. FpC4H protein phosphorylation events
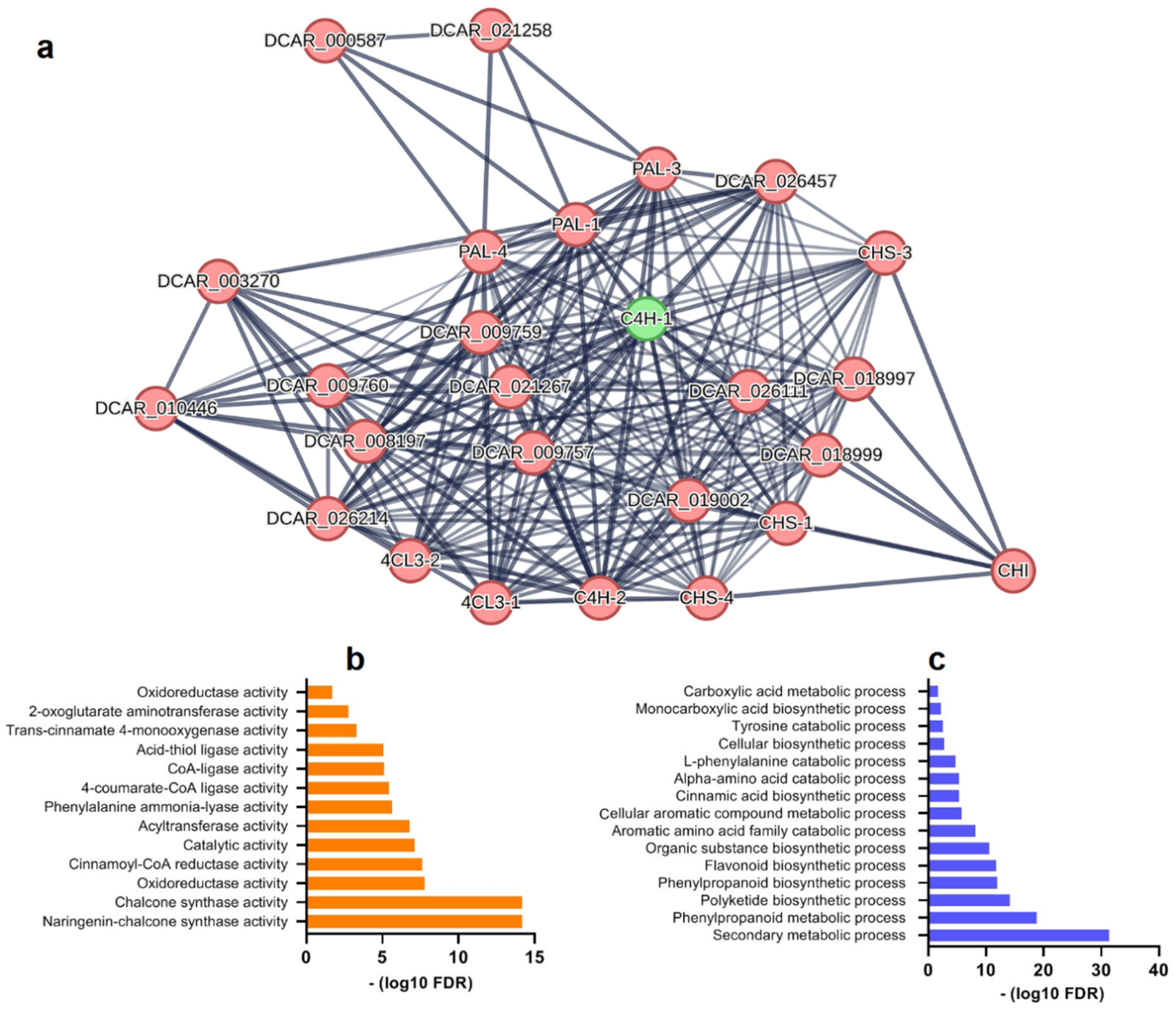
3.5. Interaction network of FpC4H
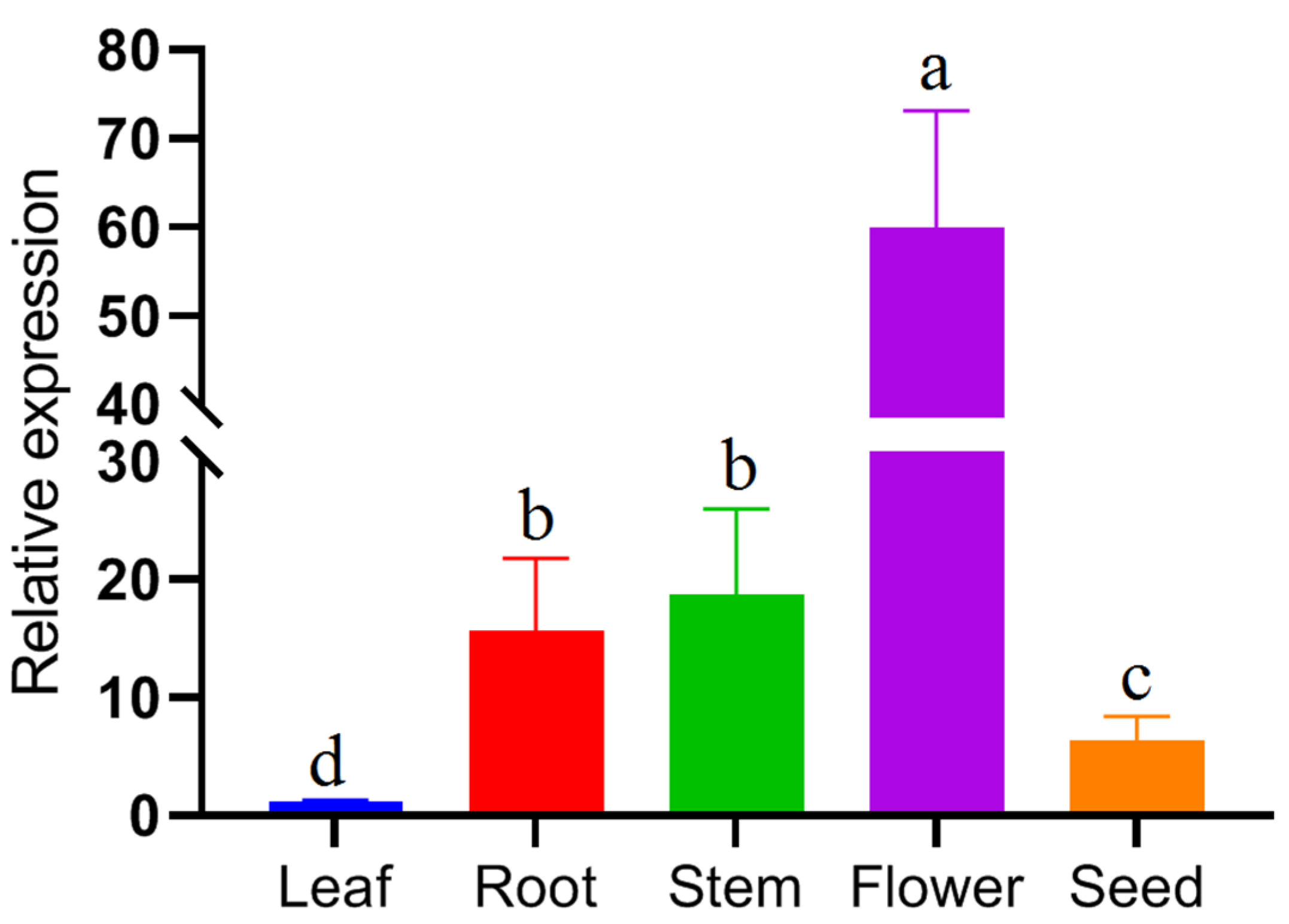
3.6. Analysis of FpC4H gene expression
4. Discussion
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buso, P.; Manfredini, S.; Reza Ahmadi-Ashtiani, H.; Sciabica, S.; Buzzi, R.; Vertuani, S.; Baldisserotto, A. Iranian medicinal plants: From ethnomedicine to actual studies. Medicina 2020, 56, 97. [Google Scholar] [CrossRef]
- Amiri, M.S.; Yazdi, M.E.T.; Rahnama, M. Medicinal plants and phytotherapy in Iran: Glorious history, current status and future prospects. Plant Sci. Today 2021, 8, 95–111. [Google Scholar] [CrossRef]
- Hassanpouraghdam, M.B.; Ghorbani, H.; Esmaeilpour, M.; Alford, M.H.; Strzemski, M.; Dresler, S. Diversity and distribution patterns of endemic medicinal and aromatic plants of Iran: Implications for conservation and habitat management. Int. J. Environ. Res. Public Health 2022, 19, 1552. [Google Scholar] [CrossRef] [PubMed]
- Amiri, M.S.; Joharchi, M.R. Ethnobotanical knowledge of Apiaceae family in Iran: A review. Avicenna J. Phytomedicine 2016, 6, 621. [Google Scholar]
- Sayed-Ahmad, B.; Talou, T.; Saad, Z.; Hijazi, A.; Merah, O. The Apiaceae: Ethnomedicinal family as source for industrial uses. Ind. Crops Prod. 2017, 109, 661–671. [Google Scholar] [CrossRef]
- Wang, X.-J.; Luo, Q.; Li, T.; Meng, P.-H.; Pu, Y.-T.; Liu, J.-X.; Zhang, J.; Liu, H.; Tan, G.-F.; Xiong, A.-S. Origin, evolution, breeding, and omics of Apiaceae: A family of vegetables and medicinal plants. Hortic. Res. 2022, 9, uhac076. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, F.; Vannozzi, A.; Barcaccia, G. Impact of genomic and transcriptomic resources on apiaceae crop breeding strategies. Int. J. Mol. Sci. 2021, 22, 9713. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.J.; Chou, S.-C. The structural diversity of phthalides from the Apiaceae. J. Nat. Prod. 2007, 70, 891–900. [Google Scholar] [CrossRef]
- Salehi, M.; Naghavi, M.R.; Bahmankar, M. A review of Ferula species: Biochemical characteristics, pharmaceutical and industrial applications, and suggestions for biotechnologists. Ind. Crops Prod. 2019, 139, 111511. [Google Scholar] [CrossRef]
- Mohammadhosseini, M.; Venditti, A.; Sarker, S.D.; Nahar, L.; Akbarzadeh, A. The genus Ferula: Ethnobotany, phytochemistry and bioactivities—A review. Ind. Crops Prod. 2019, 129, 350–394. [Google Scholar] [CrossRef]
- Dastan, D.; Hamah-Ameen, B.A.; Salehi, P.; Ghaderi, H.; Miran, M. Chemical composition and bioactivities of essential oils from different plant parts of Ferula pseudalliacea Rech. f. as an endemic plant from Iran. Nat. Prod. Res. 2022, 36, 1311–1316. [Google Scholar] [CrossRef]
- Dastan, D.; Salehi, P.; Gohari, A.R.; Zimmermann, S.; Kaiser, M.; Hamburger, M.; Khavasi, H.R.; Ebrahimi, S.N. Disesquiterpene and sesquiterpene coumarins from Ferula pseudalliacea, and determination of their absolute configurations. Phytochemistry 2012, 78, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Dastan, D.; Salehi, P.; Gohari, A.R.; Ebrahimi, S.N.; Aliahmadi, A.; Hamburger, M. Bioactive sesquiterpene coumarins from Ferula pseudalliacea. Planta Med. 2014, 80, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Mahaki, H.; Tanzadehpanah, H.; Abou-Zied, O.K.; Moghadam, N.H.; Bahmani, A.; Salehzadeh, S.; Dastan, D.; Saidijam, M. Cytotoxicity and antioxidant activity of Kamolonol acetate from Ferula pseudalliacea, and studying its interactions with calf thymus DNA (ct-DNA) and human serum albumin (HSA) by spectroscopic and molecular docking techniques. Process Biochem. 2019, 79, 203–213. [Google Scholar] [CrossRef]
- JafariA, S.H.; SepehryB, A.; SoltanlooC, H.; KarimianD, A.A. Effect of topography and soil properties on distribution of Ferula pseudalliacea (Bitter Asafetida) in Yazd Province, Iran. J. Rangel. Sci. 2019, 9, 184. [Google Scholar]
- Ehlting, J.; Hamberger, B.; Million-Rousseau, R.; Werck-Reichhart, D. Cytochromes P450 in phenolic metabolism. Phytochem. Rev. 2006, 5, 239–270. [Google Scholar] [CrossRef]
- Shuab, R.; Lone, R.; Koul, K.K. Cinnamate and cinnamate derivatives in plants. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Kumar, K.; Debnath, P.; Singh, S.; Kumar, N. An overview of plant phenolics and their involvement in abiotic stress tolerance. Stresses 2023, 3, 570–585. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- Khatri, P.; Chen, L.; Rajcan, I.; Dhaubhadel, S. Functional characterization of Cinnamate 4-hydroxylase gene family in soybean (Glycine max). PLoS ONE 2023, 18, e0285698. [Google Scholar] [CrossRef]
- Huang, B.; Duan, Y.; Yi, B.; Sun, L.; Lu, B.; Yu, X.; Sun, H.; Zhang, H.; Chen, W. Characterization and expression profiling of cinnamate 4-hydroxylase gene from Salvia miltiorrhiza in rosmarinic acid biosynthesis pathway. Russ. J. Plant Physiol. 2008, 55, 390–399. [Google Scholar] [CrossRef]
- Rostami, Z.; Fazeli, A.; Hojati, Z. The isolation and expression analysis of cinnamate 4-hydroxylase and chalcone synthase genes of Scrophularia striata under different abiotic elicitors. Sci. Rep. 2022, 12, 8128. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.B.; Huang, S.H.; Zou, J.X.; Liu, H.S.; Quan, W.J.; Yang, H. Cloning and analysis of cinnamate acid 4-hydroxylase gene and its promoter from Euphorbia maculata L. Mol Plant Breed 2023, 21, 819–825. [Google Scholar]
- Hou, Y.; Wang, Y.; Liu, X.; Ahmad, N.; Wang, N.; Jin, L.; Yao, N.; Liu, X. A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 5393. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from plant tissues. In Plant Molecular Biology Manual; Springer: Dordrecht, The Netherlands, 1989; pp. 73–83. [Google Scholar]
- Mazzara, M.; James, D.J. The influence of photoperiodic growth condition on isolation of RNA from strawberry (Fragaria× ananassa Duch.) tissue. Mol. Biotechnol. 2000, 15, 237–241. [Google Scholar]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinforma. 2014, 48, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115–e115. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mizutani, M.; Ohta, D.; Sato, R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997, 113, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.K.; Lee, S.Y.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.-H.; Chung, B.-Y.; Kim, J.-H.; Kim, J.-S.; Lee, S.-S.; An, B.-C.; Lee, I.-J.; Kim, T.-H. cDNA cloning and expression pattern of cinnamate-4-hydroxylase in the Korean black raspberry. BMB Rep. 2008, 41, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, L.; Jiang, L.; Zhang, G.; Luo, Y. Molecular cloning and functional characterization of a cinnamate 4-hydroxylase-encoding gene from Camptotheca acuminata. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Kumar, S.; Omer, S.; Patel, K.; Khan, B.M. Cinnamate 4-Hydroxylase (C4H) genes from Leucaena leucocephala: A pulp yielding leguminous tree. Mol. Biol. Rep. 2013, 40, 1265–1274. [Google Scholar] [CrossRef]
- Chen, A.-H.; Chai, Y.-R.; Li, J.-N.; Chen, L. Molecular cloning of two genes encoding cinnamate 4-hydroxylase (C4H) from oilseed rape (Brassica napus). J. Biochem. Mol. Biol. 2007, 40, 247–260. [Google Scholar] [CrossRef]
- Lu, S.; Zhou, Y.; Li, L.; Chiang, V.L. Distinct roles of cinnamate 4-hydroxylase genes in Populus. Plant Cell Physiol. 2006, 47, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Yaghobi, M.; Heidari, P. Genome-Wide Analysis of Aquaporin Gene Family in Triticum turgidum and Its Expression Profile in Response to Salt Stress. Genes 2023, 14, 202. [Google Scholar] [CrossRef] [PubMed]
- Puresmaeli, F.; Heidari, P.; Lawson, S. Insights into the Sulfate Transporter Gene Family and Its Expression Patterns in Durum Wheat Seedlings under Salinity. Genes 2023, 14, 333. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Puresmaeli, F.; Vafaee, Y.; Ahmadizadeh, M.; Ensani, M.; Ahmadinia, H. Comparative Analysis of Phospholipase D (PLD) Gene Family in Camelina sativa and Brassica napus and Its Responses in Camelina Seedlings under Salt Stress. Agronomy 2023, 13, 2616. [Google Scholar] [CrossRef]
- Poulos, T.L. Heme enzyme structure and function. Chem. Rev. 2014, 114, 3919–3962. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, S.; Ahmadizadeh, M.; Heidari, P. Genome-wide characterization, expression profiling, and post- transcriptional study of GASA gene family. Gene Rep. 2020, 20, 100795. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Levy, H.L.; Sarkissian, C.N.; Scriver, C.R. Phenylalanine ammonia lyase (PAL): From discovery to enzyme substitution therapy for phenylketonuria. Mol. Genet. Metab. 2018, 124, 223–229. [Google Scholar] [CrossRef]
- Zhao, L.; Mali, G.; Yang, Z.A.; Feng, W.S.; Zheng, X.K. expression analysis of C4H gene from Lepidium apetalum. Acta Pharmacol. Sin 2017, 5, 821–831. [Google Scholar]
- Xia, J.; Liu, Y.; Yao, S.; Li, M.; Zhu, M.; Huang, K.; Gao, L.; Xia, T. Characterization and expression profiling of Camellia sinensis cinnamate 4-hydroxylase genes in phenylpropanoid pathways. Genes 2017, 8, 193. [Google Scholar] [CrossRef]
- Karlson, C.K.S.; Mohd Noor, S.N.; Khalid, N.; Tan, B.C. CRISPRi-mediated down-regulation of the cinnamate-4-hydroxylase (C4H) gene enhances the flavonoid biosynthesis in Nicotiana tabacum. Biology 2022, 11, 1127. [Google Scholar] [CrossRef] [PubMed]

| Primer name | Sequence (5’-3’) | tm °C | Product size |
|---|---|---|---|
| FpC4H-F | TCATGTTTGATAGAAGGTTTGAGAG | 61 | 890 |
| FpC4H-R | GAACTGTCCTSCTTTCTCTGCTG | ||
| Oligo(dT) primer | GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV | ||
| PCR anchor primer | GACCACGCGTATCGATGTCGAC | ||
| F-FpC4H.RT | ACAGCTTGAAGTGTGCCATG | 56 | 185 |
| R-FpC4H.RT | GCACAGTGTCCATCTCATGC | ||
| F-FpACTIN.RT | GCCATCTATGATTGGGAATGG | 56 | 190 |
| R-FpACTIN.RT | GCCACCACCTTGATCTTCATG |
| Gene ID | Plant | Length (aa) | MW (kDa) | Instability index | GRAVY | pI |
|---|---|---|---|---|---|---|
| FpC4H (MH987776) | F. pseudaliacea | 299 | 34.54 | Unstable | -0.342 | 5.88 |
| DCAR_018641 | Daucus carota | 505 | 58.10 | Unstable | -0.215 | 9.00 |
| DCAR_027611 | Daucus carota | 498 | 57.00 | Unstable | -0.216 | 9.21 |
| BVRB_7g175810 | Beta vulgaris | 504 | 58.09 | Unstable | -0.298 | 9.14 |
| HanXRQr2_Chr08g0346771 | Helianthus annuus | 505 | 57.93 | Unstable | -0.216 | 9.16 |
| HanXRQr2_Chr07g0288081 | Helianthus annuus | 502 | 57.55 | Unstable | -0.217 | 8.92 |
| HanXRQr2_Chr06g0265981 | Helianthus annuus | 505 | 57.93 | Unstable | -0.204 | 9.07 |
| AT2G30490 | A. thaliana | 505 | 57.79 | Unstable | -0.185 | 8.89 |
| Os02g0467600 | O. sativa | 533 | 59.48 | Unstable | 0.007 | 8.77 |
| Os05g0320700 | O. sativa | 500 | 56.88 | Unstable | -0.102 | 9.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





