1. Introduction
Neurological disorders are quite common, with congenital/inborn disorders very difficult to diagnose or detect[
1]. Intellectual disability (ID), autism, and other associated disorders are known to affect 1%-3% of the world population, with an incidence of 1 in 12000 live births[
2]. Although the NCBI's dbSNP (
https://www.ncbi.nlm.nih.gov/snp/ last accessed on July 1, 2024) is validated, and ClinVar (
https://www.ncbi.nlm.nih.gov/clinvar/ last accessed on July 1, 2024) has a compendium of
bona fide records, not all data is reviewed thoroughly. For example, several disease-causing genes that are known, with specific genetic diagnosis remain elusive in many cases[
3], and as these are all heterogeneous groups of disorders, various mutations associated with developmental defects and dysfunction are nor properly reviewed and annotated. The last decade has led to a better understanding of disease diagnosis, owing to chromosomal breakpoint mapping, multi-omics integration, systems genomics approaches, whole-genome array-based copy-number analysis and nanostring panels besides the emphasis on understanding the pedigree structure for molecular diagnosis[
4]. In this work, we attempt to identify pathogenic mutations in three neurological diseases,
viz. arthrogryposis, bilateral congenital cataracts with global developmental delay (GDD) and autism spectrum disorders (ASD). The autism spectrum is a neurodevelopmental condition that is visible at the beginning of early childhood and lasts throughout a person's life[
5]. As it affects the nervous system, the affected person lacks cognitive, emotional, social and physical health. The severity and duration of symptoms might vary substantially with severe communication and social interaction issues with repetitive behavior patterns. Knowing the genetic origin of ASD could be one of the most critical aspects of future diagnosis and therapy. Likewise, arthrogryposis is a condition that causes a variety of joint contractures. It is a complicated aetiological illness that affects one in 3000 live babies, although the prenatal frequency is higher, signifying a high intrauterine mortality rate[
6]. The disease's genetic diversity has been demonstrated by linking it to 400 distinct genes. Intrinsic/primary/fetal etiology is caused by abnormalities in different body sections, including the brain, nerve cells, muscles, bones, tendons, joints,
etc. Amongst the 400 genes implicated, nine newly found genes including
CNTNAP1, MAGEL2, ADGRG6,
ASXL3 and
STAC3 harbor pathogenic variants[
7].
2. Case Presentation
Chief complaints
Case 1: Arthrogryposis congenita with delayed development.
Case 2: Small head and autism.
Case 3: Bilateral cataract, failure to thrive (FTT), and some developmental delay.
History of past illness
Case 1: In antenatal ultrasonography, he was found to have a short femur and humerus, and the mother had hyperemesis during the whole pregnancy. He was born full term by the vaginal route and had meconium-stained liquor along with crossed leg and contracture of the elbow and bilateral right hamstring and iliotibial band. During six days of hospitalization, no hypoglycemia and convulsion were noted. For contracture,he was operated on at the age of 3 years.
Case 2: Parents noticed developmental delay at 18 months of age and the development still lagged behind.
Case 3: The mother had a fever without a rash during the first two months of pregnancy for 2-3 days of admission. While antenatal ultrasound showed signs of early onset intrauterine growth retardation, he was born preterm with low birth weight (1.7 kg), requiring neonatal intensive care unit admission for eight days.
Personal and family history
Case 1: Our further counseling in lieu of familial history revealed that his sister was similarly but only mildly affected than him.
Case 2: He was born from a non consanguineous marriage without significant family history.
Case 3: Born of a non-consanguineous marriage without a significant family history.
Physical examination
Case 1: He had GDD and microcephaly (head circumference of 50 cm) and other examination findings indicate broad nose, long toes, laxity of fingers, with hyperextensibility of knees and elbow.
Case 2: The clinical examination showed microcephaly, hypertonia, failure to recognize things, with significant speech delay.
Case 3: The clinical examination showed microcephaly, FTT and small anterior fontanelle with a prominent metopic suture. He had normal motor development but had significant speech delay (only cooning).
Laboratory examinations
Case 1: On referral to the previous investigation from a neurologist, an investigation showed normal karyotype, electromyography and nerve conduction study.
Case 2: Thyroid and routine blood investigations were normal.
Case 3: The ophthalmic examination showed bilateral cataracts (right > left) with intraocular calcification. Other investigations showed normal to complete hemogram, toxoplasma and other, rubella, cytomegalovirus, and herpes simplex negative, normal renal and liver function test except increased alkaline phosphatase level (567 U/L).
Imaging examinations
Case 1: Magnetic resonance imaging (MRI) of the brain showed paucity of white matter.
Case 2: MRI brain was normal.
Case 3: The computed tomography of the brain showed generalized cerebral shrinkage with prominence of cortical sulci and cisternal spaces with bilateral periventricular volume loss and ex vacuo prominence of frontal horns.
FINAL DIAGNOSIS
Case 1
Patient was thought to be suffering from arthrogryposis congenita most probably due to neurological causes.
Case 2
Case of complex autism phenotype with microcephaly probably due to genetic cause.
Case 3
Case of GDD with bilateral cataract to rule out genetic or metabolic etiology.
TREATMENT
All of them were advised on physiotherapy and developmental and stimulation therapy but none of them on regular treatment.
OUTCOME AND FOLLOW-UP
Case 1
Able to walk with difficulty. No further deterioration or improvement from previous level.
Case 2
Had some improvement in speech and social communication but still significant lag present.
Case 3
Had more neurological deterioration. Not able to sit, stand or communicate.
3. DISCUSSION
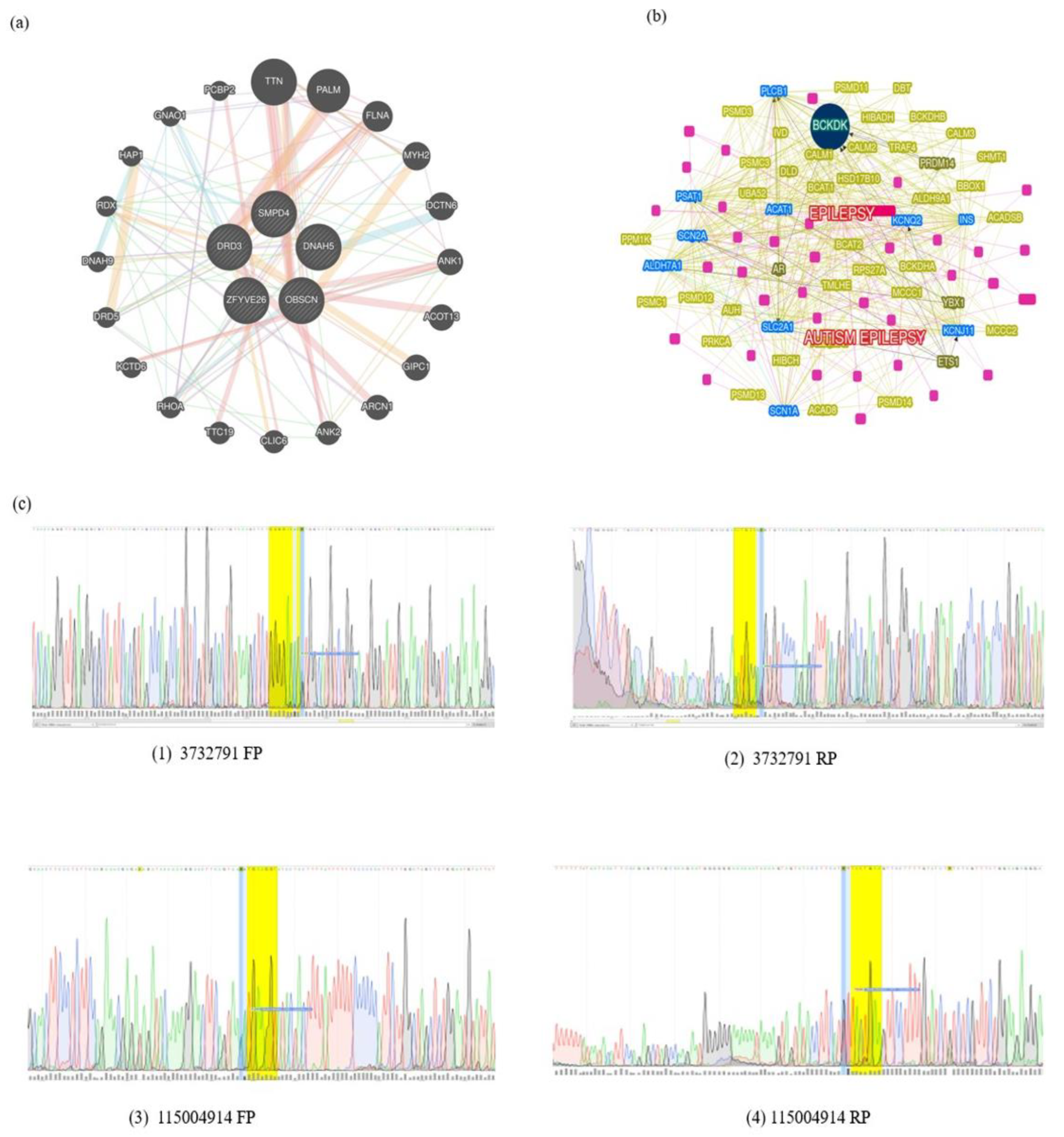
Three variants in SMPD4 are known to be associated with Arthrogryposis
SMPD4 is the only known gene implicated in Arthrogryposis from our exome sequencing analyses and further, we found three variants derived from the same gene,
SMPD4 (
Table 1)[
8]. These are found on chromosome 2 (rs766318490, rs780446128, and rs1391542283) with minor allele frequency (MAF) for the first two variants attributing to ≤ 0.0001%, showing that they are extremely rare variants. While the MAF of rs766318490 and rs780446128 in the GnomAD attribute minor allele to T = 0.000016/4 and A = 0.000004/1, respectively, the Allele Frequency Aggregator (ALFA) database showed T = 0.000051/1 and A = 0./0. rs1391542283, GeneMANIA[
9] yielded distinct interactions for
SMPD4, and many pathways,
viz. transcription regulation, factors, cell adhesion, chromatin binding, and neurodegeneration are associated (
Figure 1).
SMPD4 is associated with ceramide and is produced by sphingomyelinases as a secondary messenger in intracellular signaling pathways involved in the cell cycle, differentiation, or death.
SMPD4 mediates tumour necrosis factor-stimulated oxidant generation in skeletal muscle. Genomic research showed bi-allelic loss-of-function mutations in
SMPD4, which codes for the neutral sphingomyelinase-3/
SMPD4. However, we could not find any
SMPD4 variants attributing to pathogenesis from our CONVEX pipeline. Proteomics research on human Myc-tagged
SMPD4 overexpression demonstrated localization to both the outer nuclear envelope and the endoplasmic reticulum (ER) and interactions with multiple nuclear pore complex proteins. Fibroblasts from afflicted people had aberrant ER cisternae, suggesting enhanced autophagy and were more vulnerable to apoptosis under stress circumstances, whereas
SMPD4 therapy slowed cell cycle progression. It has been demonstrated that
SMPD4 connects membrane sphingolipid homeostasis to cell fate by regulating the cross-talk between the ER and the outer nuclear envelope and that its absence indicates a pathogenic mechanism in microcephaly[
10].
Pathogenic variants associated with microcephaly and autism
The pathogenic variants were screened for case 2, and a final list of 15 variants was filtered across subpopulation databases and specific phenotype matches based on their MAF, clinical significance, and phenotypes. The four pathogenic variants,
viz. NM_001104.4(ACTN3):c.1729C>T(p.Arg577Ter); NM_015346.4(ZFYVE26):c.-70A>T; NM_000796.6(DRD3):c.1077C>T(p.His359=); NM_001369.3(DNAH5):c.2253C>A (p.Asn751Lys). All the variants mentioned above were found on chromosome positions 11, 14, 3, 5, respectively, and were linked with neurodevelopmental disorders like Schizophrenia and structural brain anomalies. A few extremely rare variants with MAF ≤ 0.001 were identified for the filtered set matching the index case. These variants were further searched in the Indian genome variant database, Indigen (
https://clingen.igib.res.in/indigen last accessed on July 1, 2024) in addition to ALFA and GnomAD_exomes reporting these variants (
Table 1). However, from the latest ClinVar mapping, we found them to be benign. Further shortlisting to three pathogenic variants was done and were probed to check whether they are associated with inherent pathways, The genes harboring mutations are alpha-actinin-3 (
ACTN3), dopamine receptor D3 (
DRD3), dynein axonemal heavy chain 5 (
DNAH5) and zinc finger five types containing 26 (
ZFYVE26) were the candidates inherent to congenital bilateral cataract even as we obtained D3 subtype receptor proteins inhibiting adenylyl cyclase pathways (
Figure 1). The receptor is localized to the limbic areas of the brain, associated with cognitive, emotional, and endocrine function. Several literature studies show that this gene has some association with ASD[
11,
12]. A single nucleotide polymorphism of the
DRD3 gene (rs167771) was recently associated with ASD with different polymorphisms corresponding to varying degrees of behavior[
13]. In contrast, the other two genes are the normal genes unrelated to neurological disease. We further sought to check whether or not Phenolyzer pathways revealed the diagnosis of PWS and how four genes,
viz.
DRD3, DNAH5, ZFYVE26, and
ACTN3 are associated with the phenotypes represented by human phenotype ontology terms[
14]. The branched-chain ketoacid dehydrogenase kinase
(BCKDK) is a seed gene, on chromosome 16 associated with mitochondrial protein kinases family and regulates the catabolic pathways for valine, leucine, and isoleucine. By searching two related types of syndromic ASD-one caused by mutations in
BCKDK and the other by mutations in
BCKDK. Lower BCAA levels may also be detrimental to brain development, as evidenced by the discovery of BCKDH mutations in families with ASD, ID, and seizures. Other genes
ACAT1,
SLC2AL,
ALDH7A1,
SCN2A,
PSAT1,
SCNIA, and
KCNJ11 are all connected and may likely cause some neurological disorders. ASD is a complex neurodevelopmental condition characterized by social communication deficits and repetitive behaviors[
10,
11]. Recent research has highlighted the involvement of molecular factors in ASD, with particular attention to the glycoprotein Reelin. Encoded by the
RELN gene, Reelin plays a crucial role in neuronal migration and synaptic plasticity during brain development, notably in the cerebral cortex and cerebellum, influencing neural circuit formation. Altered Reelin expression in individuals with autism suggests its potential contribution to atypical neural connectivity. The downstream effects of Reelin on molecular pathways, including the modulation of GABAergic interneurons, further enhance its role in autism pathogenesis. Recent findings underscore the importance of considering Reelin's role in the biological etiology of autism, emphasizing the intricate interplay of genetic and environmental factors[
15]. These insights not only advance our understanding of autism's biological underpinnings but also offer promising avenues for targeted therapeutic interventions,shaping the future of diagnostic and therapeutic strategies for individuals with autism
4. Conclusions
Genetic variation attributing to pathogenesis is a significant bottleneck. In this work, we attempted to understand rare neurological disorders in an Indian pediatric cohort using exome studies. The CONVEX pipeline did not yield pathogenic variants for consensus variant calling tools, nevertheless,
EIF2B2 is inherently pathogenic. While these are the diseased correlations, we find that neuroleptic malignant syndrome may cause brain damage, which is a matching phenotype. Our study has certain limitations,
viz. (1) Parental genotyping or family exome analyses was not done, which could bring candidate germline mutations associated with these disorders. Despite the lack of parental data and the potential for misdiagnosis, this study identifies the inherent pathogenicity of
EIF2B2 and its association with neuroleptic malignant syndrome and brain damage. However, we acknowledge that our findings are specific to a particular region in India and may not be attributed to the entire population; and (2) the potential presence of intronic variants and the limited scope of exome sequencing necessitate further investigation with whole-genome sequencing and functional studies. Identifying
EIF2B2 as a disease-causing gene offers a promising avenue for future research and development of targeted therapies for these rare neurological disorders. Small sample sizes may not be typical for the general population, especially in rare diseases. We argue that if the research population is not diverse, the results may not be applied to other ethnic or demographic groups. The clinical exome panel may not cover all crucial genes or areas, and variation categorization might be complex and subjective. Further exome sequencing may overlook regulatory regions and non-coding variations that may play a role in illness, and variant identification methods may yield false positives or false negatives. Using ClinVar alone to evaluate variants has limits, and prediction methods may not always adequately represent
in vivo biological importance. Finally, as monogenic disorders are rare and poorly understood, it hints at a general restriction in our understanding of rare diseases. Because the CONVEX pipeline is considered a consensus variant pipeline, it is critical to confirm its performance by comparing it to known standards and datasets[
16]. As genomics is a dynamic field, new genes and variant classifications might develop, affecting study findings' relevance and accuracy over time. In conclusion, this research lays the groundwork for further explorations into the genetic landscape of rare neurological disorders in the Indian population, paving the path for improved diagnosis, treatment, and, ultimately, a brighter future for those affected by these debilitating conditions.
Author Contributions
Tayade N and Krishna A A contributed equally to this work as co-first authors; Tayade N and Suravajhala P conceptualized the work and proofread the manuscript; Suravajhala P, Krishna A A, Manoj G and Kewat A performed the analyses; Devulapalli R, Kumar S, Polipalli SK, Nair BG and Bandapalli OR contributed to write-up figures and writing the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank the parents for their vivid support of samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Warmerdam HAG, Termeulen-Ferreira EA, Tseng LA, Lee JY, van Eeghen AM, Ferreira CR, van Karnebeek CDM. A Scoping Review of Inborn Errors of Metabolism Causing Progressive Intellectual and Neurologic Deterioration (PIND). Front Neurol 2019; 10: 1369 [PMID: 32132962 DOI: 10.3389/fneur.2019.01369]. [CrossRef]
- Charman T, Pickles A, Simonoff E, Chandler S, Loucas T, Baird G. IQ in children with autism spectrum disorders: data from the Special Needs and Autism Project (SNAP). Psychol Med 2011; 41: 619-627 [PMID: 21272389 DOI: 10.1017/S0033291710000991]. [CrossRef]
- Jackson M, Marks L, May GHW, Wilson JB. The genetic basis of disease. Essays Biochem 2018; 62: 643-723 [PMID: 30509934 DOI: 10.1042/EBC20170053]. [CrossRef]
- Lunke S, Bouffler SE, Patel CV, Sandaradura SA, Wilson M, Pinner J, Hunter MF, Barnett CP, Wallis M, Kamien B, Tan TY, Freckmann ML, Chong B, Phelan D, Francis D, Kassahn KS, Ha T, Gao S, Arts P, Jackson MR, Scott HS, Eggers S, Rowley S, Boggs K, Rakonjac A, Brett GR, de Silva MG, Springer A, Ward M, Stallard K, Simons C, Conway T, Halman A, Van Bergen NJ, Sikora T, Semcesen LN, Stroud DA, Compton AG, Thorburn DR, Bell KM, Sadedin S, North KN, Christodoulou J, Stark Z. Integrated multi-omics for rapid rare disease diagnosis on a national scale. Nat Med 2023; 29: 1681-1691 [PMID: 37291213 DOI: 10.1038/s41591-023-02401-9]. [CrossRef]
- Faras H, Al Ateeqi N, Tidmarsh L. Autism spectrum disorders. Ann Saudi Med 2010; 30: 295-300 [PMID: 20622347 DOI: 10.4103/0256-4947.65261]. [CrossRef]
- Hall JG, Kiefer J. Arthrogryposis as a Syndrome: Gene Ontology Analysis. Mol Syndromol 2016; 7: 101-109 [PMID: 27587986 DOI: 10.1159/000446617]. [CrossRef]
- Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet 2014; 15: 195-213 [PMID: 25184530 DOI: 10.1146/annurev-genom-090413-025600]. [CrossRef]
- Magini P, Smits DJ, Vandervore L, Schot R, Columbaro M, Kasteleijn E, van der Ent M, Palombo F, Lequin MH, Dremmen M, de Wit MCY, Severino M, Divizia MT, Striano P, Ordonez-Herrera N, Alhashem A, Al Fares A, Al Ghamdi M, Rolfs A, Bauer P, Demmers J, Verheijen FW, Wilke M, van Slegtenhorst M, van der Spek PJ, Seri M, Jansen AC, Stottmann RW, Hufnagel RB, Hopkin RJ, Aljeaid D, Wiszniewski W, Gawlinski P, Laure-Kamionowska M, Alkuraya FS, Akleh H, Stanley V, Musaev D, Gleeson JG, Zaki MS, Brunetti-Pierri N, Cappuccio G, Davidov B, Basel-Salmon L, Bazak L, Shahar NR, Bertoli-Avella A, Mirzaa GM, Dobyns WB, Pippucci T, Fornerod M, Mancini GMS. Loss of SMPD4 Causes a Developmental Disorder Characterized by Microcephaly and Congenital Arthrogryposis. Am J Hum Genet 2019; 105: 689-705 [PMID: 31495489 DOI: 10.1016/j.ajhg.2019.08.006]. [CrossRef]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38: W214-W220 [PMID: 20576703 DOI: 10.1093/nar/gkq537]. [CrossRef]
- Smits DJ, Schot R, Krusy N, Wiegmann K, Utermöhlen O, Mulder MT, den Hoedt S, Yoon G, Deshwar AR, Kresge C, Pletcher B, van Mook M, Ferreira MS, Poot RA, Slotman JA, Kremers GJ, Ahmad A, Albash B, Bastaki L, Marafi D, Dekker J, van Ham TJ, Nguyen L, Mancini GMS. SMPD4 regulates mitotic nuclear envelope dynamics and its loss causes microcephaly and diabetes. Brain 2023; 146: 3528-3541 [PMID: 36732302 DOI: 10.1093/brain/awad033]. [CrossRef]
- Liu J, Fu H, Kong J, Yu H, Zhang Z. Association between autism spectrum disorder and polymorphisms in genes encoding serotine and dopamine receptors. Metab Brain Dis 2021; 36: 865-870 [PMID: 33644845 DOI: 10.1007/s11011-021-00699-3]. [CrossRef]
- Jiang CC, Lin LS, Long S, Ke XY, Fukunaga K, Lu YM, Han F. Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications. Signal Transduct Target Ther 2022; 7: 229 [PMID: 35817793 DOI: 10.1038/s41392-022-01081-0]. [CrossRef]
- Staal WG, Langen M, van Dijk S, Mensen VT, Durston S. DRD3 gene and striatum in autism spectrum disorder. Br J Psychiatry 2015; 206: 431-432 [PMID: 25792691 DOI: 10.1192/bjp.bp.114.148973]. [CrossRef]
- Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods 2015; 12: 841-843 [PMID: 26192085 DOI: 10.1038/nmeth.3484]. [CrossRef]
- Scala M, Grasso EA, Di Cara G, Riva A, Striano P, Verrotti A. The Pathophysiological Link Between Reelin and Autism: Overview and New Insights. Front Genet 2022; 13: 869002 [PMID: 35422848 DOI: 10.3389/fgene.2022.869002]. [CrossRef]
- 1Raju RM, Singh UP, Suravajhala P. Benchmarking whole exome sequencing pipeline for predicting pathogenic variants of significance. bioRxiv 2023 [DOI: 10.1101/2023.10.07.561328]. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).