1. Introduction
The paddy field ecosystem is an artificial wetland created inland where water is supplied during a specific season (from spring to summer), and it is a unique form of wetland ecosystem connected to the rivers and reservoirs, where various aquatic species spawn and thrive [
1,
2]. The paddy field ecosystem, accounting for approximately 18% of all wetlands on Earth, is a crucial component of a comprehensive use plan that utilizes wetlands to address environmental pollution and preserve biodiversity worldwide, and because the Ramsar Convention adopted "Enhancing biodiversity in rice paddies as a wetland system," there has been a growing interest in promoting biodiversity using paddy fields [
3,
4].
Most of the biological surveys conducted on paddy field ecosystems aim to understand the status of agricultural pests. In the early days, research surveys were mainly conducted to determine the incidence of mosquitoes in paddy fields, and a full-fledged research survey on aquatic organisms in paddy fields has been conducted from 1994 [
5]. Moreover, since the Ramsar Convention in 2008, NGOs have also formed networks and conducted biological surveys of paddy fields [
5,
6,
7]. Even today, surveys are being conducted on the actual condition of the paddy field ecosystem to investigate the emergence of amphibians, reptiles, spiders, and aquatic insects living in the paddy field ecosystem, and such a process identifies the date and time of appearance of different types of biota due to climate change and regional changes [
8,
9].
In general, the state survey of organisms in these paddy field ecosystems utilizes a traditional method that involves directly collecting and observing individuals. Amphibians, like frogs, are identified by the investigator through direct filming of the individuals on the spot or by listening to their croak sounds [
10]. However, these traditional collection methods are highly labor-intensive, and they heavily rely on the timing of the investigation as well as on the investigator's competence. Moreover, conducting a survey by searching for individuals in a wide range can be time consuming, and if the population is small, identifying individuals in the paddy field ecosystem becomes highly challenging.
The organism screening method using environmental genes was proposed as a solution to the problems and difficulties associated with traditional investigation methods [
11,
12,
13]. Environmental DNA (eDNA) refers to the genetic material that exists in large quantities in various environments, such as water, air, and soil, and this genetic material includes traces of organisms living in the environment [
14,
15,
16]. Thus, it can be used as a clue to determine the existence of organisms in the environment without the need for collecting them directly. Currently, organism screening using eDNA is most actively used in fish taxa, and it is also actively utilized to search for endangered, foreign, and harmful species [
16,
17,
18]. Identification of biodiversity using eDNA was primarily conducted in river and lake environments, with only a few instances of exploring the biodiversity using eDNA in paddy field ecosystems [
19,
20,
21,
22]. Amphibians, such as frogs, in particular, live, spawn, and reproduce around paddy fields or nearby ecosystems, which means that their genetic traces may exist within the paddy field ecosystem. Frogs, as amphibians, require water for breeding and refuge from predators. The paddy field ecosystem, classified as a freshwater habitat, serves not only as a breeding ground for frogs but also plays a crucial role as a hiding place. In the paddy field ecosystem, frogs occupy a vital ecological niche as predators of pests such as rice planthoppers.
Therefore, this study utilizes eDNA to identify and verify the frog community residing in the aquatic ecosystem of paddy fields in order to assess the applicability of eDNA for screening the biodiversity of the aquatic ecosystem of paddy fields.
2. Materials and Methods
2.1. eDNA sampling and extraction
The survey sites for investigation were selected as paddy fields (site U; Upper paddy, site L; Lower paddy) located in areas where paddy fields are surrounded by forests and had utilized stream water flowing from mountains as irrigation water (agricultural water) (
Figure 1 and
Figure 2). Considering the activity characteristics of amphibians, eDNA was collected from the water at sunset and immediately filtered and collected on-site using Dual eDNA Filter
TM (Sphylum, Netherlands, 0.45 ㎛ pore size) and a 50 ㎖ syringe. A single filter was used for each area of paddy fields, and at least three points of eDNA were collected. In addition, eDNA was collected from a nearby river, which was utilized as a source of agricultural water. Immediately after collection, the outlet of the filter was sealed with a Luer lock, and then it was maintained at a low temperature (4°C) state during immediate transportation to the laboratory. Extraction of eDNA was performed within 24 hours. We conducted eDNA sampling once during a rain-free period lasting one week in May. The reason for dividing the rice paddies into two locations in this study is to compare ponds of different sizes. The Upper paddy is more than four times smaller in scale than the Lower paddy, which may not be suitable for frog habitat. Therefore, by exploring larger ponds downstream, we aimed to ensure a comprehensive detection of frog eDNA.
2.2. PCR amplification and metabarcoding
To identify the genes of amphibians present in the eDNA of the aquatic ecosystem in paddy fields, a primary PCR of the mitochondrial 16S rDNA (mt16S rDNA) gene was conducted using the primer presented by Sakata
, et al. [
23]. The DNA polymerase used for this purpose was the 2X GainBlue
TM Hot Start Max Master Mix PCR Master Mix (Gainbio, Korea), and Forward and Reverse primers were added at a concentration of 10 pmol/㎕. Further, 3 ㎕ of template eDNA was added and PCR was performed using the PCR Thermal Cycler (LongGen, Taiwan). The product of primary PCR amplification was analyzed using the 2% agarose gel of E-gel
TM Power Snap Electrophoresis system (Thermo Fisher Scientific, USA) to perform electrophoresis, and the amplification product was then identified using the E-gel
TM Power Snap Camera (Thermo Fisher Scientific, USA).
2.3. eDNA meta-barcoding analysis
For meta-barcoding analysis, electrophoresis was performed on the primary PCR amplification product using a 2% size selection gel of the E-gel
TM Power Snap Electrophoresis system (Thermo Fisher Scientific, USA), followed by gel extraction of the target amplification product. The primary PCR amplification product was separated through gel extraction, and then a barcode index (Illumina Nextrea kit V2) was attached through a secondary PCR process. Samples with the barcode index were analyzed using metabarcoding based on the Illumina MiSeq System by a company specializing in analysis (Metagenom bio life Co. ltd, Waterloo, ON, Canada), and species were identified based on sequencing of the mitochondrial 16S rDNA using IdTaxa classifier [
24] of the DECIPHER package [
25].
2.4. Field survey of frogs
Individuals found based on eDNA were verified by directly observing them in the area where eDNA was collected or by identifying their cry. The survey was conducted in accordance with the amphibian survey method provided in the "Standards and Manuals for Ecological Environment Survey and Evaluation in the Agricultural Sector" to identify amphibians living in the paddy field environment [
10]. During the daytime, adult frogs were visually examined in paddy fields, along the ridges between paddies, and waterways within the investigated area, and species were identified by taking pictures of the individuals [
26].
3. Results
3.1. Amphibian diversity based on eDNA metabarcoding
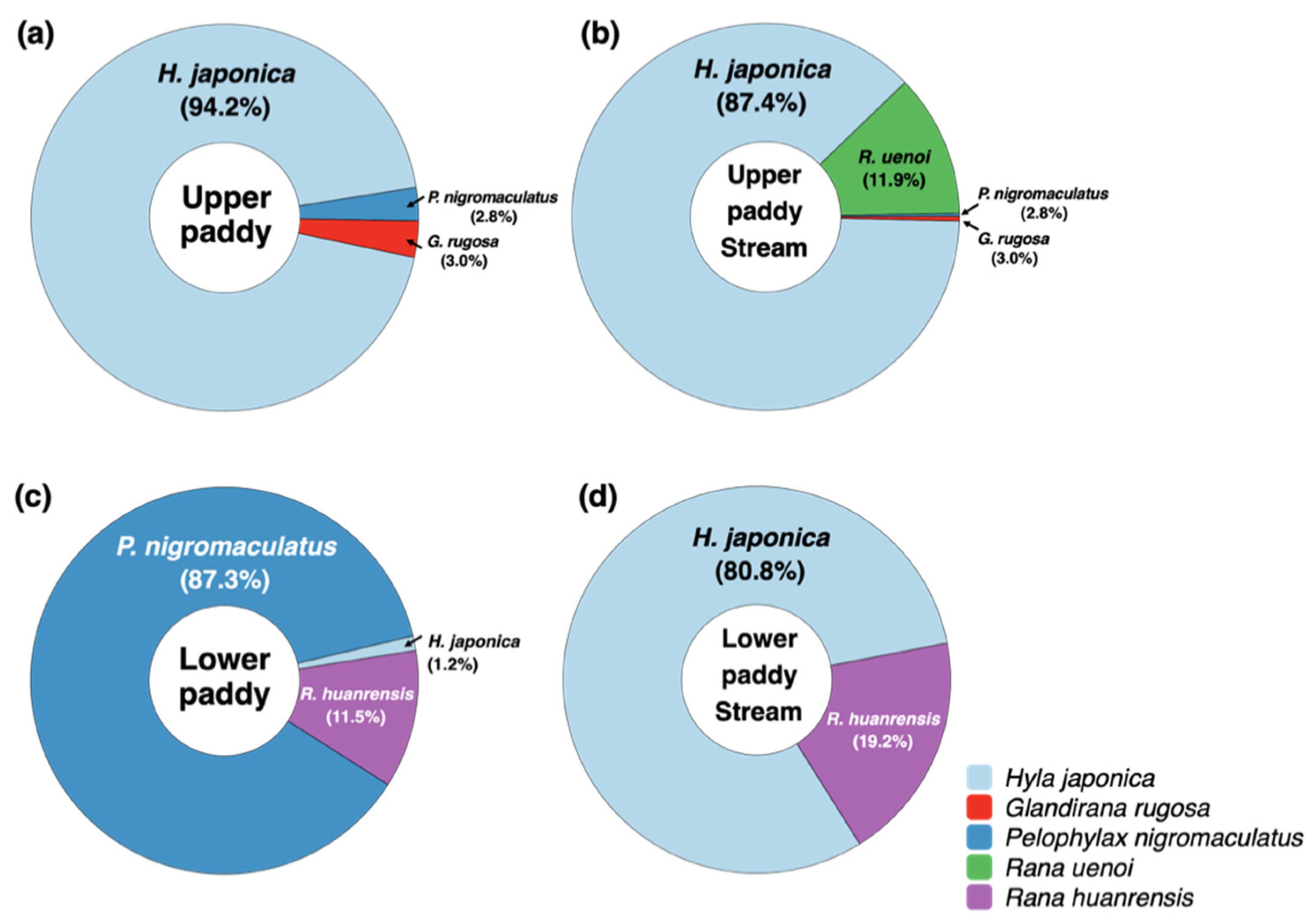
As a result of analyzing the genes of frogs included in Anura's mitochondrial 16S rDNA (mt 16S rDNA) in the eDNA collected from the paddy field environment, we detected the DNAs of 5 species of frogs, including black-spotted pond frogs (
Pelophylax nigromaculatus), tree frogs (
Hyla japonica), rough-skinned frogs (
Glandirana rugosa), Korean large brown frogs (
Rana uenoi), and Huanren brown frog (
Rana huanrensis) (
Figure 2). In site U (Upper paddy), we detected four frog species in the paddy and stream, and tree frog DNA accounted for the largest proportion of frog mt 16S rDNA, with a percentage of 94.2% (
Figure 2a) in the paddy. A relatively small proportion of black-spotted pond frogs and rough-skinned frogs were found at 2.8% and 3.0%, respectively. In the waterways leading to Site U, the DNA of tree frogs accounted for the highest proportion at 87.4%, similar as in the paddy, while the proportions of DNA of black-spotted pond frogs and rough-skinned frogs were significantly lower at 0.3% and 0.4%, respectively (
Figure 2b). In addition, DNAs of Korean large brown frogs, which were not found within the water at site U, were discovered in the waterways of site U. On the other hand, at site L (Lower paddy), we detected three frog species in the paddy and stream, and the DNA of black-spotted pond frogs accounted for the largest proportion at 87.3%, while tree frogs accounted for a lowest proportion at 1.2% (
Figure 2c). In addition, the DNAs of Huanren brown frogs, which were not found at site U, accounted for 11.5% at site L, whereas the DNAs of Korean large brown frogs and rough-skinned frogs were not detected. In the waterways near site L, similar to site U, the proportion of DNA of tree frogs was the highest at 80.8%, while the proportion of DNA of Huanren brown frogs accounted for 19.2% (
Figure 2d). However, DNAs of black-spotted pond frogs were not found in the waterway.
3.2. Verification of eDNA detection by field survey
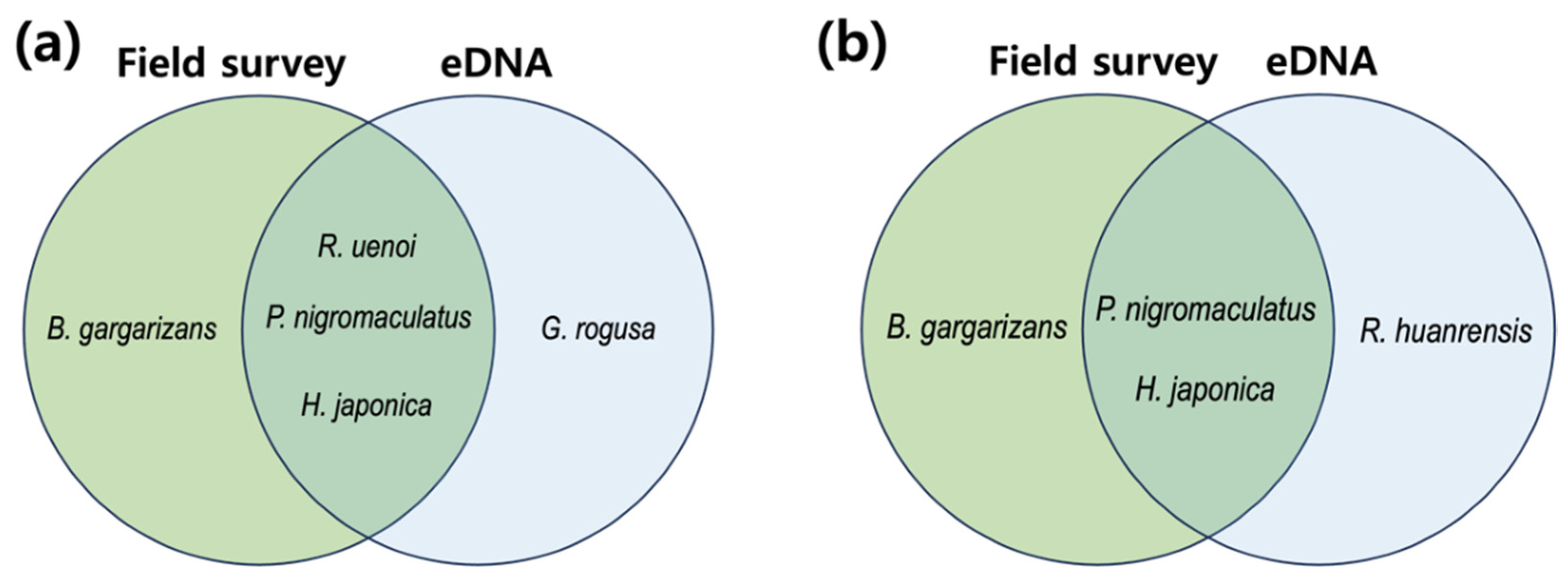
The results of eDNA investigation were verified by comparing the colony of frogs found using eDNA with the colony identified through direct observation and calls (FigS1,
Figure S2). When adult frogs and tadpoles were found in two areas of paddy fields through direct observation of individuals and by identifying frog species living in the investigation site through their calls, the list of species directly observed did not completely match with the list of species in the eDNA-based frog community (
Figure 3).
At site U, three adult tree frogs (Dryophytes japonica) and four adult black-spotted pond frogs were spotted, and one adult Korean large brown frog was found in a puddle upstream of the upper paddy. Rough-skinned frogs were not found inside the upper paddy, and croak sounds were not identified either. Thirty adult tree frogs were found at site L, but only eight black-spotted pond frogs were found - a number lesser than that of the adult tree frogs. The Huanren brown frog, whose DNA was found in the lower paddy, was not found in the paddies or waterways, and croak sounds were not heard. At the investigation site, tree frogs were primarily discovered on the ridges surrounding the paddy and around the paddy field, with adult tree frogs primarily being located within the paddy. Korean large brown frogs were found around the puddle upstream rather than around the paddy. Adult frogs were not found in the waterways at the investigation site, and 12 larvae (tadpoles) of Korean large brown frogs were discovered. In the process of searching for frog adults, one toad was directly identified in the area between site U and site L, and toad adults or larvae were not found inside the paddies or waterways.
4. Discussion
Agricultural water is supplied through irrigation systems from agricultural reservoirs or nearby streams. Therefore, in the agricultural water of paddy fields, DNAs of organisms living in the upper stream of the reservoirs and organisms living in paddy fields coexist [
27,
28,
29,
30,
31]. In this study, neither rough-skinned frogs nor Huanren brown frogs were identified through direct observation or croak sounds, but a very low proportion of their DNAs was identified. Therefore, it is believed that the DNAs of rough-skinned frogs and Huanren brown frogs, which were found not inside the paddy fields, were identified because there were individuals living in or near paddy fields, but the DNAs of frogs who lived in forest areas outside the fields flowed into the paddy fields through nearby streams. In addition, rough-skinned frogs were found only in the upper paddy and stream water, while the DNA of Huanren brown frogs was found exclusively in the lower paddy and stream water. This suggests that the DNA of organisms, such as frogs and fish, which roam within their boundaries, can also be found in areas near their habitat, and even the DNA collected from paddy fields in the same area may vary depending on the specific location from where the eDNA was collected [
32,
33,
34]. Therefore, the DNA of rough-skinned frogs found only in the upper paddy suggests that rough-skinned frogs live exclusively in the upper part of the stream, and Huanren brown frogs are believed to inhabit the upper stream of the valley, with their DNAs flowing into the stream near the paddy fields below.
The number of amphibians is not proportional to the number of contigs in eDNA, and the number of contigs in eDNA differs depending on the ecological characteristics of each taxon [
35,
36,
37]. In this study, although there were more than twice as many tree frogs in the lower paddy, a higher amount of DNA from black-spotted pond frogs was collected in the eDNA of water samples. After undergoing metamorphosis from tadpoles to adults, tree frogs are primarily active in areas with leaves, grass, and shrubs, with their presence in the paddy fields limited to the breeding season [
38,
39,
40,
41]. On the contrary, the habitat of black-spotted pond frogs is closely related to the freshwater environment. Black-spotted Pond frogs have a protective color similar to that of paddy water, and they hide in aquatic plants, such as duckweeds, which are present in the paddy fields [
42]. Thus, although the population of tree frogs was higher around the paddy fields, it is believed that the DNA of black-spotted pond frogs dwelling underwater accounted for a larger proportion than that of tree frogs. However, in the upper paddy, tree frogs accounted for a larger proportion of genetic contigs, despite the similar population sizes of tree frogs and black-spotted pond frogs. This is believed to be because the DNA of tree frogs, which constitutes a significant proportion of the stream water, flowed into the paddy fields, thereby affecting the proportion of tree frog DNA in the fields. In addition, Korean large brown frogs mainly inhabit the mountain slopes, streams, and arable land in the valley, and they spawn in still water rather than in flowing water [
42,
43]. In this research, a few individuals were also found near water pools located upstream of the fields. However, given that Korean large brown frogs constitute the second largest proportion of DNA found in the upstream water, it is estimated that there will be a greater abundance of Korean large brown frogs in the upper valley at the survey site. Moreover, although DNA was not found in the eDNA of the survey site, toads (
Bufo gargarizans) were directly observed and identified at the site, which mainly live crawling on land as adults and breed in groups only during the spawning season [
44]. Toads spawn in still water between March and April, but during the research period, there was no water in the paddy fields during this season at the survey site. Therefore, toads did not spawn in paddy fields at the survey site of this study, and it is estimated that they spawned in puddles located outside the paddy fields.
5. Conclusions
The aquatic ecosystems in paddy fields are used as spawning grounds and habitats for organisms, such as frogs. Use of eDNA can help search for traces of organisms at the DNA level, allowing detection of organisms in the paddy field without direct observation of individuals. In this study, it was possible to determine the presence of creatures living adjacent to paddy fields by detecting the DNA of Huanren brown frogs and rough-skinned frogs that were not observed directly. In addition, research has led to the possibility that more individuals of the Korean large brown frog could inhabit the upstream of site U. This use of eDNA-based biological exploration of paddy fields provides the advantage of high analytical resolution, making it a valuable method for studying biodiversity of the agricultural ecosystem. However, due to the nature of the paddy field environment, various organisms pass through and DNAs from humans (Homo sapiens) are introduced during rice planting and management; and thus, DNA introduced from external sources can impact the analysis of biological communities at the survey site. In addition, similar to the case of toads, it can be very difficult to identify the DNA of organisms that live mostly on land, except during the larval period, in eDNA research. Therefore, eDNA-based surveys on biological communities and on-site censuses should be treated as complementary research methods that can interact with each other, rather than as separate research methods. In other words, to supplement the search results at the DNA level through eDNA, it is necessary to extract the list of species observed at the survey site and proceed with direct observation and investigation. Hence, future utilization of both eDNA and traditional direct observation methods is expected to yield crucial data for confirming biodiversity in the paddy field ecosystem.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Figure S1: Field survey results of the study area; Figure S2: The photographs of frogs that were taken in the study area.
Author Contributions
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Funding
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ015071042023)" Rural Development Administration, Republic of Korea.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable.
Data Availability Statement
Data can be made available upon reasonable request to the corresponding author.
Acknowledgments
The author is very grateful to Sera Kwon who help with the identification of frogs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kim, S.K.; Park, H.S.; Park, S.R. Distribution of fish and amphibian in rice fields near the Yedang Reservoir in Korea. Korean Journal of Environment and Ecology 2016, 30, 48–57. [Google Scholar] [CrossRef]
- Saito, K. Movement and spawning of several freshwater fishes in temporary waters around paddy fields. Jpn. J. Ecol. 1988, 38, 35–47. [Google Scholar]
- COP10, R. Enhancing biodiversity in rice paddies as wetland systems. In Proceedings of the 10th Meeting of the Conference of the Parties to the Convention on Wetlands (Ramsar, Iran, 1971), 2008.
- Bang, H.S.; Han, M.S.; Na, Y.E.; Kang, K.K. Biodiversity of fauna and flora in Korean paddy field. In Proceedings of the Marco symposium ₂₀₀₉'Challenges for agro-environmental research in monsoon asia; 2010. [Google Scholar]
- Kang, K.K.; Han, M.S.; Na, Y.E.; Kim, M.H.; Kim, M.R. Development of technologies for the management and restoration of paddy ecosystem to improve biodiversity in agro-ecosystem; National Institute of Agricultural Sciences: NIAS, 2013; pp. 1–134. [Google Scholar]
- Sagong, J.H.; Jung, O.S.; Yeo, H.B. Ecosystem service value evaluation study of paddy wetlands in Chungcheongnam-do; Chungnam Development Institute: 2014.
- Jang, K.S.; Kim, J.O.; Lee, S.H.; Ji, K.J.; Seo, J.B.; Shin, H.S.; Yu, J.H. A study on the development of ecological infrastructure to improve bio-diversity in paddy wetlands; Korea Rural Community Corporation: Rural research institute, 2010.12.01 2010.
- Eea, J.W.; Kim, M.H.; Song, Y.J.; Kim, S.T.; Lee, J.H.; Jang, I.K.; Kweon, S.R.; Kang, H.K.; Kim, Y.i. Survey on the current status of biodiversity and phenology for impact assessment to climate change; Rural Development Administration: National Institute of Agricultural Sciences, 2019. [Google Scholar]
- Kim, M.H. Climate change impact assessment through long-term monitoring of biological seasons of indicator organisms; TRKO202200009107; Rural Development Administration: National Institute of Agricultural Sciences, 2021. [Google Scholar]
- Kang, H.S.; Kim, C.; Kim, J.S.; Kim, J.O.; Kim, M.H. Agricultural ecological environment survey/evaluation criteria and manual; National Institute of Agricularal Science: NIAS, 2020. [Google Scholar]
- Yoccoz, N.G. The future of environmental DNA in ecology. Molecular ecology 2012, 21, 2031–2038. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For biodiversity research and monitoring; Oxford University Press: 2018.
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biology letters 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Douglas, W.Y.; De Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends in ecology & evolution 2014, 29, 358–367. [Google Scholar]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conservation letters 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Doi, H. Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PloS one 2013, 8, e56584. [Google Scholar] [CrossRef]
- Kim, J.-H.; Jo, H.; Chang, M.-H.; Woo, S.-H.; Cho, Y.; Yoon, J.-D. Application of environmental DNA for monitoring of freshwater fish in Korea. Korean Journal of Ecology and Environment 2020, 53, 63–72. [Google Scholar] [CrossRef]
- Edmunds, R.C.; Cooper, M.; Huerlimann, R.; Robson, H.; Burrows, D. Environmental DNA survey of Eureka Creek, Upper Mitchell, and Walsh River for Two Invasive Tilapia Species. 2019.
- Marshall, K.M. Stream-Associated Amphibian Detection and Fine-Scale Distribution: Inferences from Environmental DNA; Washington State University: 2021.
- Uchida, N.; Kubota, K.; Aita, S.; Kazama, S. Aquatic insect community structure revealed by eDNA metabarcoding derives indices for environmental assessment. PeerJ 2020, 8, e9176. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, R.; Nakao, R.; Ushimaru, A.; Minamoto, T. Development of environmental DNA detection assays for snakes in paddy fields in Japan. Landscape and Ecological Engineering 2023, 19, 3–10. [Google Scholar] [CrossRef]
- Cai, W.; Ma, Z.; Yang, C.; Wang, L.; Wang, W.; Zhao, G.; Geng, Y.; Yu, D.W. Using eDNA to detect the distribution and density of invasive crayfish in the Honghe-Hani rice terrace World Heritage site. PloS one 2017, 12, e0177724. [Google Scholar] [CrossRef] [PubMed]
- Sakata, M.K.; Kawata, M.U.; Kurabayashi, A.; Kurita, T.; Nakamura, M.; Shirako, T.; Kakehashi, R.; Nishikawa, K.; Hossman, M.Y.; Nishijima, T. Development and evaluation of PCR primers for environmental DNA (eDNA) metabarcoding of Amphibia. bioRxiv 2021. [Google Scholar] [CrossRef]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R Journal 2016, 8. [Google Scholar] [CrossRef]
- 26. RDA. An Illustrated guide to paddy ecology fauna - Fish, amphibians and reptiles, RDA (Rural Development Administration), 2019.
- Altieri, M.A. The ecological role of biodiversity in agroecosystems. In Invertebrate biodiversity as bioindicators of sustainable landscapes; Elsevier: 1999; pp. 19-31.
- Moonen, A.C.; Bàrberi, P. Functional biodiversity: An agroecosystem approach. Agriculture, ecosystems & environment 2008, 127, 7–21. [Google Scholar]
- Randall, N.; Smith, B. The biology of agroecosystems; Oxford University Press, USA: 2019.
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.; Olds, B.; Mahon, A.R.; Jerde, C.L.; Bolster, D. Controls on eDNA movement in streams: Transport, retention, and resuspension. Scientific reports 2017, 7, 5065. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.A.; Olds, B.; Jerde, C.; Bolster, D. Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. Journal of the Royal Society Interface 2016, 13, 20160290. [Google Scholar] [CrossRef] [PubMed]
- Biggs, J.; Ewald, N.; Valentini, A.; Gaboriaud, C.; Griffiths, R.; Foster, J.; Wilkinson, J.; Arnett, A.; Williams, P.; Dunn, F. Analytical and methodological development for improved surveillance of the Great Crested Newt. Defra Project WC1067 2014. [Google Scholar]
- Herder, J.; Kranenbarg, J.; De Bruin, A.; Valentini, A. Op jacht naar DNA–Effectief zoeken naar grote modderkruipers. RAVON, Nijmegen, 2013. [Google Scholar]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic PCR to detect B urmese python in F lorida waters and an assessment of persistence of environmental DNA. Molecular ecology resources 2014, 14, 374–380. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PloS one 2011, 6, e22746. [Google Scholar] [CrossRef] [PubMed]
- Laramie, M.B.; Pilliod, D.S.; Goldberg, C.S. Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biological Conservation 2015, 183, 29–37. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Molecular ecology 2016, 25, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Matsui, M. Feeding habits of the pond frog, Rana nigromaculata, inhabiting rice fields in Kyoto, Japan. Copeia 1999, 940–947. [Google Scholar] [CrossRef]
- Naito, R.; Yamasaki, M.; Natuhara, Y.; Morimoto, Y. Effects of water management, connectivity, and surrounding land use on habitat use by frogs in rice paddies in Japan. Zoological Science 2012, 29, 577–584. [Google Scholar] [CrossRef]
- Ra, N. Habitat and behavioral characteristics, captive breeding and recovery strategy of the endangered Gold-spotted pond frog (Rana plancyi chosenica). PhD thesis (Kangwon Natl Univ., 2010), 2010.
- Sawahata, T. Seasonal changes in number of Rana porosa brevipoda killed on the road. Bull Herpetol Soc Jpn 2002, 2, 72–74. [Google Scholar]
- Lee, J.H.; Park, D. The Encyclopedia of Korean Amphibians; Cho, Yeong Kwon: Nature and Ecology, 2016; p. 248. [Google Scholar]
- Ko, Y. Study of life-cycle of Rana dyvowskii in JEJU island. Dissertation, Graduate School Jeju National University, 2011.
- Günther, R. Die amphibien und reptilien Deutschlands; Springer: 1996.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).