Submitted:
19 December 2023
Posted:
20 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
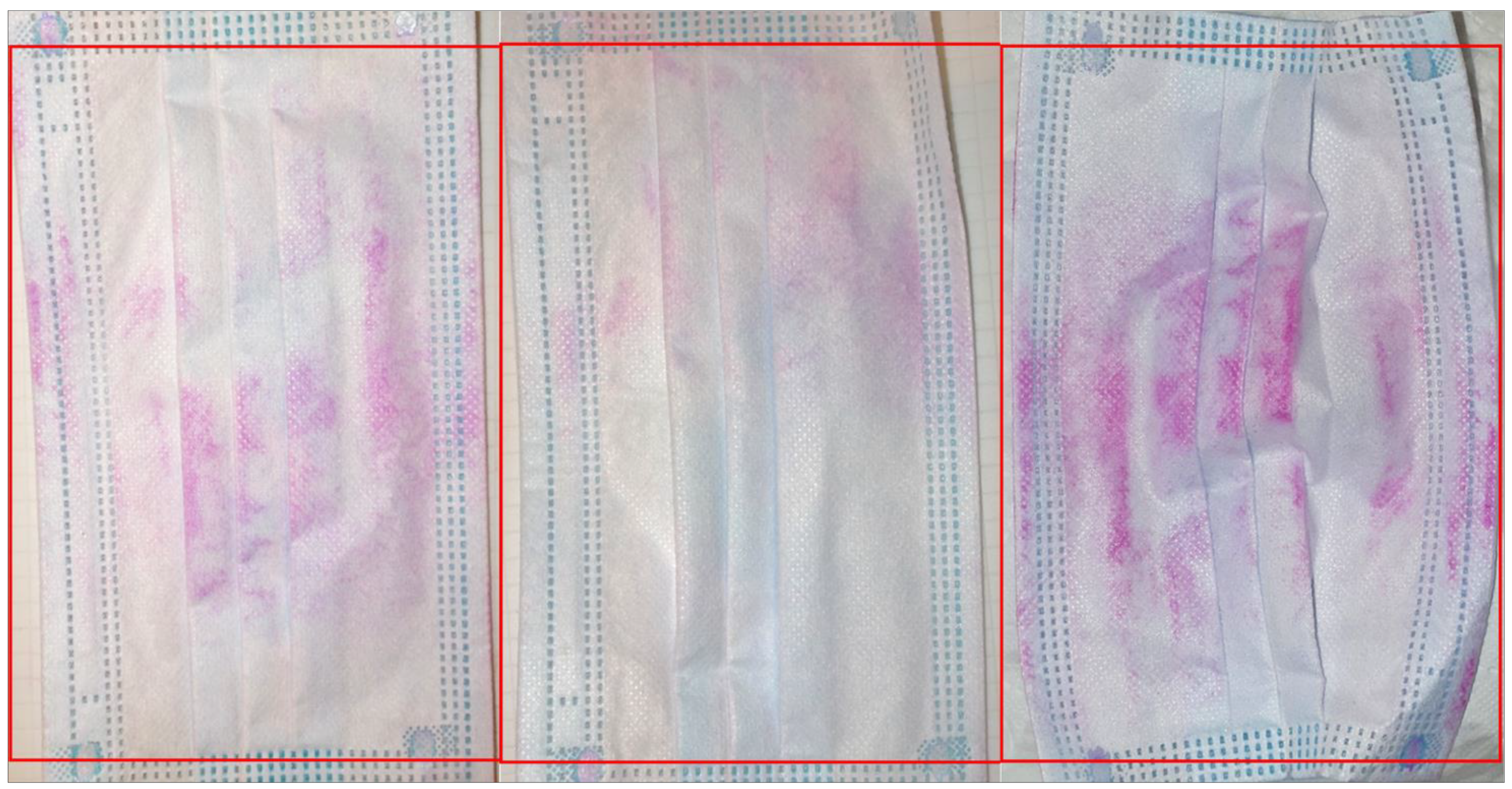
Rose Bengal staining and visualisation of contamination
Microbiological mask study design
Identification of isolates by Sanger Sequencing of the 16S rRNA gene
Biochemical characterisation of isolates
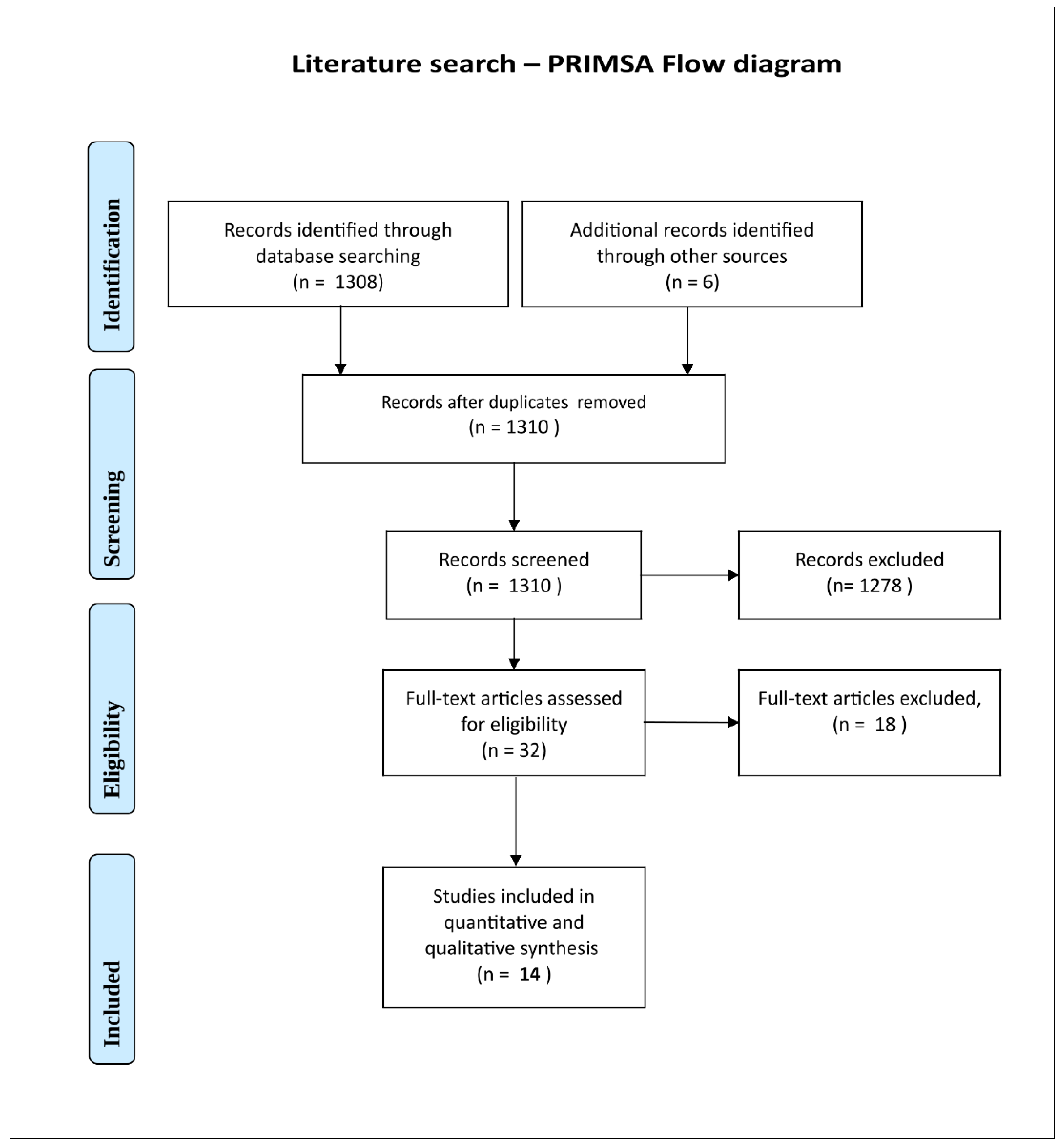
Systematic literature search
3. Results
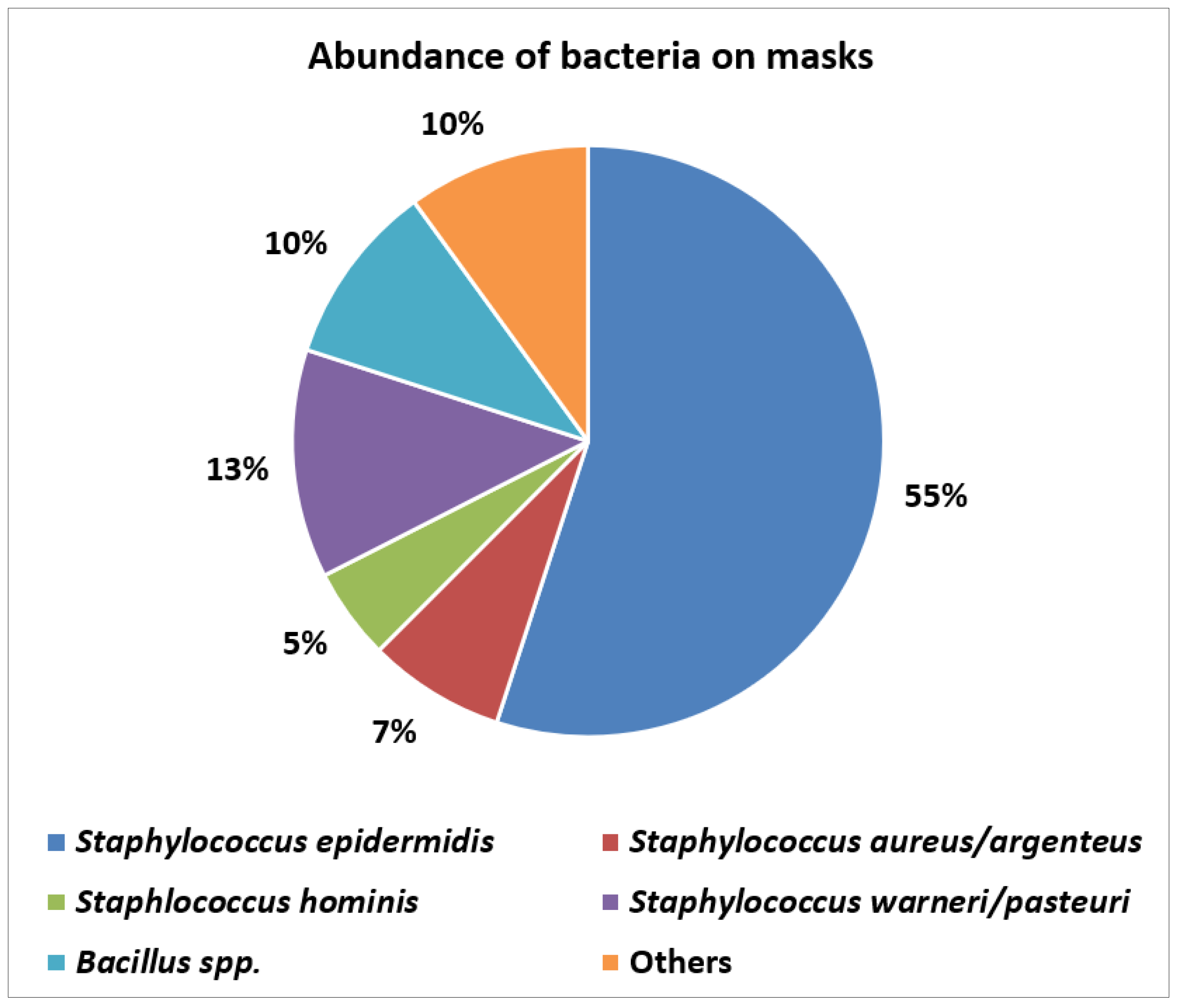
Abundance and types of bacteria on worn masks
Identification of isolates by Sanger Sequencing of 16S rRNA gene
Biochemical identification of isolates
Systematic literature search
4. Discussion
Bacteria detected: potential clinical implications
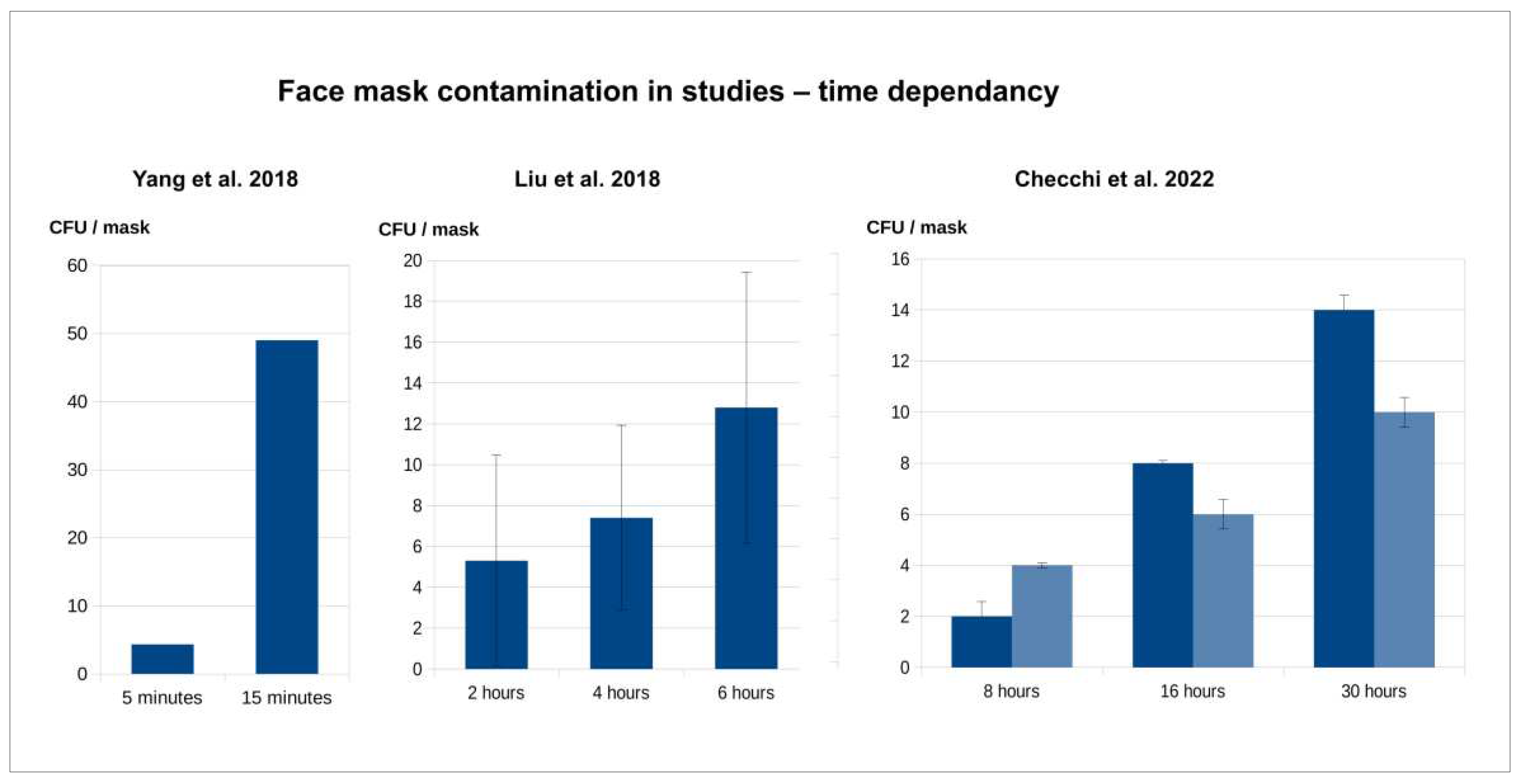
Literature review on mask contamination
Face mask contamination – contributing factors
Face mask contamination – potential clinical implications
Findings in context
5. Limitations and strengths
6. Conclusions
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgements
Research Ethics Statement
Conflict of Interest
References
- Face covering policies during the COVID-19 pandemic, Our World in Data. (2023). Available online: https://ourworldindata.org/grapher/face-covering-policies-covid (accessed on 29 December 2022).
- S.N. Ladhani, Face masking for children - time to reconsider, Journal of Infection. 85 (2022) 623–624. [CrossRef]
- S. Thomson, Mask mandates for children during the COVID-19 pandemic: an international human rights perspective, Scand J Public Health. 50 (2022) 683–685. [CrossRef]
- World Health Organization, U.N.C. Fund (UNICEF), WHO - Advice on the use of masks for children in the community in the context of COVID-19: annex to the advice on the use of masks in the context of COVID-19, 21 August 2020, (2020). Available online: https://apps.who.int/iris/handle/10665/333919 (accessed on 7 November 2020).
- S. Schwarz, E. Jenetzky, H. Krafft, T. Maurer, D. Martin, Corona child studies “Co-Ki”: first results of a Germany-wide register on mouth and nose covering (mask) in children., Monatsschr Kinderheilkd. 169 (2021) 353–365. [CrossRef]
- World Health Organization, WHO - Advice on the use of masks in the context of COVID-19: interim guidance, 5 June 2020, (2020). Available online: https://apps.who.int/iris/handle/10665/332293 (accessed on 7 November 2020).
- K.J. Cummings, J. Cox-Ganser, M.A. Riggs, N. Edwards, K. Kreiss, Respirator Donning in Post-Hurricane New Orleans - Volume 13, Number 5—May 2007 - Emerging Infectious Diseases journal - CDC, (2007). [CrossRef]
- J. Gralton, M.-L. McLaws, Protecting healthcare workers from pandemic influenza: N95 or surgical masks?, Crit Care Med. 38 (2010) 657–667. [CrossRef]
- I. Kappstein, Mund-Nasen-Schutz in der Öffentlichkeit: Keine Hinweise für eine Wirksamkeit, Krankenhaushygiene up2date. 15 (2020) 279–295. [CrossRef]
- R. Roberge, Facemask use by children during infectious disease outbreaks, Biosecur Bioterror. 9 (2011) 225–231. [CrossRef]
- A.P.S. Munro, R.C. Hughes, Face coverings have little utility for young school-aged children, Archives of Disease in Childhood. 108 (2023) 77–78. [CrossRef]
- T. Jefferson, M. Jones, L.A.A. Ansari, G. Bawazeer, E. Beller, J. Clark, J. Conly, C.D. Mar, E. Dooley, E. Ferroni, P. Glasziou, T. Hoffman, S. Thorning, M.V. Driel, Physical interventions to interrupt or reduce the spread of respiratory viruses. Part 1 - Face masks, eye protection and person distancing: systematic review and meta-analysis, medRxiv. (2020) 2020.03.30.20047217. [CrossRef]
- T. Jefferson, L. Dooley, E. Ferroni, L.A. Al-Ansary, M.L. van Driel, G.A. Bawazeer, M.A. Jones, T.C. Hoffmann, J. Clark, E.M. Beller, P.P. Glasziou, J.M. Conly, Physical interventions to interrupt or reduce the spread of respiratory viruses, Cochrane Database of Systematic Reviews. (2023). [CrossRef]
- J. Sandlund, R. Duriseti, S.N. Ladhani, K. Stuart, J. Noble, T.B. Høeg, Child mask mandates for COVID-19: a systematic review, Archives of Disease in Childhood. (2023). [CrossRef]
- K. Kisielinski, S. Wagner, O. Hirsch, B. Klosterhalfen, A. Prescher, Possible toxicity of chronic carbon dioxide exposure associated with face mask use, particularly in pregnant women, children and adolescents – A scoping review, Heliyon. 0 (2023). [CrossRef]
- E. Coma, M. Català, L. Méndez-Boo, S. Alonso, E. Hermosilla, E. Alvarez-Lacalle, D. Pino, M. Medina, L. Asso, A. Gatell, Q. Bassat, A. Mas, A. Soriano-Arandes, F.F. Avilés, C. Prats, Unravelling the role of the mandatory use of face covering masks for the control of SARS-CoV-2 in schools: a quasi-experimental study nested in a population-based cohort in Catalonia (Spain), Archives of Disease in Childhood. (2022). [CrossRef]
- K. Kisielinski, P. Giboni, A. Prescher, B. Klosterhalfen, D. Graessel, S. Funken, O. Kempski, O. Hirsch, Is a Mask That Covers the Mouth and Nose Free from Undesirable Side Effects in Everyday Use and Free of Potential Hazards?, International Journal of Environmental Research and Public Health. 18 (2021) 4344. [CrossRef]
- E.F.E.M. Ahmad, M. Mohammed, A.A. Al Rayes, A. Al Qahtani, A.G. Elzubier, F.A.E. Suliman, The Effect of Wearing the Veil by Saudi Ladies on the Occurrence of Respiratory Diseases, Journal of Asthma. 38 (2001) 423–426. [CrossRef]
- H. Ryu, Y.-H. Kim, Measuring the quantity of harmful volatile organic compounds inhaled through masks, Ecotoxicology and Environmental Safety. 256 (2023) 114915. [CrossRef]
- P. Sukul, J. Bartels, P. Fuchs, P. Trefz, R. Remy, L. Rührmund, S. Kamysek, J.K. Schubert, W. Miekisch, Effects of COVID-19 protective face masks and wearing durations on respiratory haemodynamic physiology and exhaled breath constituents, European Respiratory Journal. 60 (2022). [CrossRef]
- R. G. M. Al-Allaff, S. M. Y. Al-Taee, S. T. D. Baker, Some Immunological Impacts of Face Mask Usage During the COVID-19 Pandemic, Pakistan Journal of Biological Sciences. 24 (2021) 920–927. [CrossRef]
- R.J. Vakharia, I. Jani, S. Yadav, T. Kurian, To Study Acute Changes in Brain Oxygenation on MRI in Healthcare Workers Using N95 Mask and PPE Kits for Six Hours a Day, Indian J Radiol Imaging. 31 (2021) 893–900. [CrossRef]
- C.S.W. Law, P.S. Lan, G.H. Glover, Effect of wearing a face mask on fMRI BOLD contrast, NeuroImage. 229 (2021) 117752. [CrossRef]
- S. Patel, E. Mohapatra, A.K. Suganthy, S. Shah, J. Abraham, R. Nanda, A.K. Behera, A. Gupta, A pilot study to evaluate the changes in venous blood gas parameters and hypoxia biomarkers in health care workers using different kinds of masks, Lung India. 40 (2023) 134–142. [CrossRef]
- D. Prousa, Studie zu psychischen und psychovegetativen Beschwerden mit den aktuellen Mund-Nasenschutz-Verordnungen, (2020). [CrossRef]
- M.A. Pavlova, C.-C. Carbon, Y. Coello, A.A. Sokolov, A.M. Proverbio, Editorial: Impact of face covering on social cognition and interaction, Front Neurosci. 17 (2023) 1150604. [CrossRef]
- C.-C. Carbon, M.J. Held, A. Schütz, Reading Emotions in Faces With and Without Masks Is Relatively Independent of Extended Exposure and Individual Difference Variables, Frontiers in Psychology. 13 (2022). Available online: https://www.frontiersin.org/articles/10.3389/fpsyg.2022.856971 (accessed on 1 February 2023).
- F. Schönweitz, J. Eichinger, J. Kuiper, F. Ongolly, W. Spahl, B. Prainsack, B. Zimmermann, The social meanings of artefacts: Face masks in the COVID-19 pandemic, Frontiers in Public Health. 10 (2022). Available online: https://www.frontiersin.org/articles/10.3389/fpubh.2022.829904 (accessed on 14 October 2023).
- C. Villani, S. D’Ascenzo, E. Scerrati, P. Ricciardelli, R. Nicoletti, L. Lugli, Wearing the face mask affects our social attention over space, Frontiers in Psychology. 13 (2022). Available online: https://www.frontiersin.org/articles/10.3389/fpsyg.2022.923558 (accessed on 14 October 2023).
- A.M. Proverbio, A. Cerri, The Recognition of Facial Expressions Under Surgical Masks: The Primacy of Anger, Frontiers in Neuroscience. 16 (2022). Available online: https://www.frontiersin.org/articles/10.3389/fnins.2022.864490 (accessed on 14 October 2023).
- F. Grundmann, K. Epstude, S. Scheibe, Face masks reduce emotion-recognition accuracy and perceived closeness, PLOS ONE. 16 (2021) e0249792. [CrossRef]
- L. Mathis, The Effects of Face Masks on Emotion Interpretation in Socially Anxious Individuals, Graduate Student Journal of Psychology. 20 (2023). [CrossRef]
- T.L. Truong, S.D. Beck, A. Weber, The impact of face masks on the recall of spoken sentences, J Acoust Soc Am. 149 (2021) 142–144. [CrossRef]
- R. Sönnichsen, G. Llorach Tó, S. Hochmuth, V. Hohmann, A. Radeloff, How Face Masks Interfere With Speech Understanding of Normal-Hearing Individuals: Vision Makes the Difference, Otol Neurotol. 43 (2022) 282–288. [CrossRef]
- V.S. McKenna, C.L. Kendall, T.H. Patel, R.J. Howell, R.L. Gustin, Impact of Face Masks on Speech Acoustics and Vocal Effort in Healthcare Professionals, The Laryngoscope. 132 (2022) 391–397. [CrossRef]
- Education recovery in early years providers: spring 2022, GOV.UK. (2022). Available online: https://www.gov.uk/government/publications/education-recovery-in-early-years-providers-spring-2022/education-recovery-in-early-years-providers-spring-2022 (accessed on 1 February 2023).
- B. Spira, Correlation Between Mask Compliance and COVID-19 Outcomes in Europe, Cureus. 14 (2022) e24268. [CrossRef]
- Z. Fögen, The Foegen effect: A mechanism by which facemasks contribute to the COVID-19 case fatality rate, Medicine (Baltimore). 101 (2022) e28924. [CrossRef]
- L. Delanghe, E. Cauwenberghs, I. Spacova, I. De Boeck, W. Van Beeck, K. Pepermans, I. Claes, D. Vandenheuvel, V. Verhoeven, S. Lebeer, Cotton and Surgical Face Masks in Community Settings: Bacterial Contamination and Face Mask Hygiene, Front Med (Lausanne). 8 (2021) 732047. [CrossRef]
- J. Szostak-Kotowa, Biodeterioration of textiles, International Biodeterioration & Biodegradation. 53 (2004) 165–170. [CrossRef]
- A. Buzzin, G. Domènech-Gil, E. Fraschetti, E. Giovine, D. Puglisi, D. Caputo, Assessing the consequences of prolonged usage of disposable face masks, Sci Rep. 12 (2022) 16796. [CrossRef]
- Q. Yang, H. Li, S. Shen, G. Zhang, R. Huang, Y. Feng, J. Yang, S. Ma, Study of the micro-climate and bacterial distribution in the deadspace of N95 filtering face respirators, Sci Rep. 8 (2018) 17382. [CrossRef]
- P. Luksamijarulkul, N. Aiempradit, P. Vatanasomboon, Microbial Contamination on Used Surgical Masks among Hospital Personnel and Microbial Air Quality in their Working Wards: A Hospital in Bangkok, Oman Med J. 29 (2014) 346–350. [CrossRef]
- A.A. Chughtai, S. Stelzer-Braid, W. Rawlinson, G. Pontivivo, Q. Wang, Y. Pan, D. Zhang, Y. Zhang, L. Li, C.R. MacIntyre, Contamination by respiratory viruses on outer surface of medical masks used by hospital healthcare workers, BMC Infect Dis. 19 (2019) 491. [CrossRef]
- D. Monalisa, Microbial Contamination of the Mouth Masks Used By Post- Graduate Students in a Private Dental Institution: An In-Vitro Study, (2017) 7.
- K. Kisielinski, B. Wojtasik, Suitability of Rose Bengal sodium salt staining for visualisation of face mask contamination by living organisms, AIMSES. 9 (2022) 218–231. [CrossRef]
- Z. Liu, Y. Chang, W. Chu, M. Yan, Y. Mao, Z. Zhu, H. Wu, Z. Jie, K. Dai, H. Li, F. Liu, Z. Zhai, Surgical masks as source of bacterial contamination during operative procedures, Journal of Orthopaedic Translation. 14 (2018) 57–62. [CrossRef]
- P. Monciardini, M. Sosio, L. Cavaletti, C. Chiocchini, S. Donadio, New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes1, FEMS Microbiology Ecology. 42 (2002) 419–429. [CrossRef]
- Calc | LibreOffice - Free Office Suite - Based on OpenOffice - Compatible with Microsoft, (2023). Available online: https://www.libreoffice.org/discover/calc (accessed on 17 March 2023).
- R.P.G. Feenstra, S.C.G. Tseng, What Is Actually Stained by Rose Bengal?, Archives of Ophthalmology. 110 (1992) 984–993. [CrossRef]
- H.J. Conn, Rose Bengal as a General Bacterial Stain, J Bacteriol. 6 (1921) 253–254.
- W.E. Maneval, Staining Bacteria and Yeasts with Acid Dyes, Stain Technology. 16 (1941) 13–19. [CrossRef]
- D.C. Saha, A Rapid Staining Method for Detection of Endophytic Fungi in Turf and Forage Grasses, Phytopathology. 78 (1988) 237. [CrossRef]
- B. Wojtasik, M. Zbawicka, L. Grabarczyk, W. Juzwa, Flow cytometric approach to evaluate the impact of hydro-technical concrete compounds’ release to the freshwater microbiome, Environ Monit Assess. 193 (2021) 698. [CrossRef]
- A.L. Byrd, Y. Belkaid, J.A. Segre, The human skin microbiome, Nat Rev Microbiol. 16 (2018) 143–155. [CrossRef]
- N.A. Logan, Bacillus and relatives in foodborne illness, J Appl Microbiol. 112 (2012) 417–429. [CrossRef]
- W.J. Wolfgang, A. Coorevits, J.A. Cole, P. De Vos, M.C. Dickinson, G.E. Hannett, R. Jose, E.J. Nazarian, P. Schumann, A. Van Landschoot, S.E. Wirth, K.A. Musser, Sporosarcina newyorkensis sp. nov. from clinical specimens and raw cow’s milk, International Journal of Systematic and Evolutionary Microbiology. 62 (2012) 322–329. [CrossRef]
- P. Kämpfer, A. Albrecht, S. Buczolits, H.-J. Busse, Psychrobacter faecalis sp. nov., a new species from a bioaerosol originating from pigeon faeces, Syst Appl Microbiol. 25 (2002) 31–36. [CrossRef]
- P. Deschaght, M. Janssens, M. Vaneechoutte, G. Wauters, Psychrobacter isolates of human origin, other than Psychrobacter phenylpyruvicus, are predominantly Psychrobacter faecalis and Psychrobacter pulmonis, with emended description of P. faecalis, International Journal of Systematic and Evolutionary Microbiology. 62 (2012) 671–674. [CrossRef]
- A.-M. Park, S. Khadka, F. Sato, S. Omura, M. Fujita, K. Hashiwaki, I. Tsunoda, Bacterial and fungal isolation from face masks under the COVID-19 pandemic, Sci Rep. 12 (2022) 11361. [CrossRef]
- R. Sachdev, K. Garg, G. Singh, V. Mehrotra, Is safeguard compromised? Surgical mouth mask harboring hazardous microorganisms in dental practice, J Family Med Prim Care. 9 (2020) 759–763. [CrossRef]
- M.P. Gund, G. Boros, M. Hannig, S. Thieme-Ruffing, B. Gärtner, T.R. Rohrer, A. Simon, S. Rupf, Bacterial contamination of forehead skin and surgical mask in aerosol-producing dental treatment, J Oral Microbiol. 13 (2021) 1978731. [CrossRef]
- M.P. Gund, J. Naim, M. Hannig, A. Halfmann, B. Gärtner, G. Boros, S. Rupf, CHX and a Face Shield Cannot Prevent Contamination of Surgical Masks, Front Med (Lausanne). 9 (2022) 896308. [CrossRef]
- V. Checchi, M. Montevecchi, L. Valeriani, L. Checchi, Bioburden Variation of Filtering Face Piece Respirators over Time: A Preliminary Study, Materials. 15 (2022) 8790. [CrossRef]
- M. Yousefimashouf, R. Yousefimashouf, M.S. Alikhani, H. Hashemi, P. Karami, Z. Rahimi, S.M. Hosseini, Evaluation of the bacterial contamination of face masks worn by personnel in a center of COVID 19 hospitalized patients: A cross-sectional study, New Microbes New Infect. 52 (2023) 101090. [CrossRef]
- M. Nightingale, M. Mody, A.H. Rickard, M. Cassone, Bacterial contamination on used face masks among nursing home healthcare personnel, Antimicrob Steward Healthc Epidemiol. 3 (2023) e54. [CrossRef]
- V.C. Keri, A. Kumar, G. Singh, A. Mandal, H. Ali, P. Ranjan, N. Wig, Pilot study on burden of fungal contamination in face masks: need for better mask hygiene in the COVID-19 era, Infez Med. 29 (2021) 557–561. [CrossRef]
- Y. Merad, Z. Belmokhtar, O. Hadjazi, M. Belkacemi, D. Matmour, Z. Merad, A. Bassaid, O. Megherbi, Fungal contamination of medical masks among forensic healthcare workers in the COVID19 era, New Microbes and New Infections. 53 (2023) 101134. [CrossRef]
- VDI 6022, VDI. (2023). Available online: https://www.vdi.de/richtlinien/unsere-richtlinien-highlights/vdi-6022 (accessed on 14 October 2023).
- S. Rengasamy, A. Miller, B.C. Eimer, R.E. Shaffer, Filtration Performance of FDA-Cleared Surgical Masks, J Int Soc Respir Prot. 26 (2009) 54–70.
- J. Fernández-Arribas, T. Moreno, R. Bartrolí, E. Eljarrat, COVID-19 face masks: A new source of human and environmental exposure to organophosphate esters, Environment International. 154 (2021) 106654. [CrossRef]
- P. Ostrowski, H. Masiuk, P. Kulig, A. Skoryk, A. Wcisłek, J. Jursa-Kulesza, A. Sarna, M. Sławiński, M. Kotowski, K. Tejchman, K. Kotfis, J. Sieńko, Medical Face Masks Do Not Affect Acid–Base Balance Yet Might Facilitate the Transmission of Staphylococcus aureus in Hospital Settings during the COVID-19 Pandemic, International Journal of Environmental Research and Public Health. 20 (2023) 2474. [CrossRef]
- A. Sakr, F. Brégeon, J.-L. Mège, J.-M. Rolain, O. Blin, Staphylococcus aureus Nasal Colonization: An Update on Mechanisms, Epidemiology, Risk Factors, and Subsequent Infections, Frontiers in Microbiology. 9 (2018). Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02419 (accessed on 14 October 2023).
- R.R. Dietert, J.M. Dietert, The Human Superorganism: Using Microbes for Freedom vs. Fear, Applied Microbiology. 3 (2023) 883–905. [CrossRef]
- D. Nowicka, E. Grywalska, Staphylococcus aureus and Host Immunity in Recurrent Furunculosis, Dermatology. 235 (2019) 295–305. [CrossRef]
- L. Akhtar Danesh, Z. Saiedi Nejad, H. Sarmadian, S. Fooladvand, A. van Belkum, E. Ghaznavi-Rad, Elimination of Staphylococcus aureus nasal carriage in intensive care patients lowers infection rates, Eur J Clin Microbiol Infect Dis. 39 (2020) 333–338. [CrossRef]
- C. Han, J. Shi, Y. Chen, Z. Zhang, Increased flare of acne caused by long-time mask wearing during COVID-19 pandemic among general population, Dermatologic Therapy. 33 (2020) e13704. [CrossRef]
- P. Bortoluzzi, V. Boneschi, S. Veraldi, “Mask” Tinea: An Increasing Infection during COVID-19 Pandemic, Mycopathologia. 187 (2022) 141–142. [CrossRef]
- R.Z. Silkiss, M.K. Paap, S. Ugradar, Increased incidence of chalazion associated with face mask wear during the COVID-19 pandemic, Am J Ophthalmol Case Rep. 22 (2021) 101032. [CrossRef]
- W. Akioud, S. Sebbata, Y. Mozarie, A. Oubaaz, Chalazion and Face Mask Wear during COVID-19 Pandemic: Is There A Link?, European Journal of Medical and Health Sciences. 5 (2023) 17–19. [CrossRef]
- M. Molero-Senosiain, S. Tiew, A. Patel, I. Houben, N. Dhillon, Impact of face mask wear on bacterial keratitis, Journal Français d’Ophtalmologie. 46 (2023) e37–e39. [CrossRef]
- T. Sakamoto, H. Terasaki, T. Yamashita, H. Shiihara, R. Funatsu, A. Uemura, Increased incidence of endophthalmitis after vitrectomy relative to face mask wearing during COVID-19 pandemic, British Journal of Ophthalmology. 107 (2023) 1472–1477. [CrossRef]
- M.I. Goncheva, R.M. Gibson, A.C. Shouldice, J.D. Dikeakos, D.E. Heinrichs, The Staphylococcus aureus protein IsdA increases SARS CoV-2 replication by modulating JAK-STAT signaling, iScience. 26 (2023). [CrossRef]
- M. Otto, Staphylococcus epidermidis – the “accidental” pathogen, Nat Rev Microbiol. 7 (2009) 555–567. [CrossRef]
- S.M.K. Schoenfelder, C. Lange, M. Eckart, S. Hennig, S. Kozytska, W. Ziebuhr, Success through diversity – How Staphylococcus epidermidis establishes as a nosocomial pathogen, International Journal of Medical Microbiology. 300 (2010) 380–386. [CrossRef]
- C. Heilmann, W. Ziebuhr, K. Becker, Are coagulase-negative staphylococci virulent?, Clin Microbiol Infect. 25 (2019) 1071–1080. [CrossRef]
- F. Khorvash, F. Abdi, H.H. Kashani, F.F. Naeini, T. Narimani, Staphylococcus aureus in Acne Pathogenesis: A Case-Control Study, N Am J Med Sci. 4 (2012) 573–576. [CrossRef]
- K. Findley, E.A. Grice, The Skin Microbiome: A Focus on Pathogens and Their Association with Skin Disease, PLOS Pathogens. 10 (2014) e1004436. [CrossRef]
- R.D. Bjerre, J. Bandier, L. Skov, L. Engstrand, J.D. Johansen, The role of the skin microbiome in atopic dermatitis: a systematic review, Br J Dermatol. 177 (2017) 1272–1278. [CrossRef]
- C.C.I. Foo, A.T.J. Goon, Y. Leow, C. Goh, Adverse skin reactions to personal protective equipment against severe acute respiratory syndrome – a descriptive study in Singapore, Contact Dermatitis. 55 (2006) 291–294. [CrossRef]
- E. Rosner, Adverse Effects of Prolonged Mask Use among Healthcare Professionals during COVID-19, J Infect Dis Epidemiol. 6:130 (2020). [CrossRef]
- L. Techasatian, S. Lebsing, R. Uppala, W. Thaowandee, J. Chaiyarit, C. Supakunpinyo, S. Panombualert, D. Mairiang, S. Saengnipanthkul, K. Wichajarn, P. Kiatchoosakun, P. Kosalaraksa, The Effects of the Face Mask on the Skin Underneath: A Prospective Survey During the COVID-19 Pandemic, J Prim Care Community Health. 11 (2020) 2150132720966167. [CrossRef]
- M. Abduljabbar, D.E. Kalthoum, M. Bakarman, I. Wahby Salem, Z. Alsulaimani, W. Alharbi, S. Shawish, R. Alsobhi, The Correlation Between Wearing Face Masks and Skin Damage in Adults During the COVID-19 Pandemic: A Cross-Sectional Study in Jeddah, Saudi Arabia, Cureus. 14 (2022) e31521. [CrossRef]
- A. Villani, G. Fabbrocini, M.C. Annunziata, L. Potestio, Maskne prevalence and risk factors during the COVID-19 pandemic, J Eur Acad Dermatol Venereol. (2022) 10.1111/jdv.18248. [CrossRef]
- R. a Bakhsh, S.Y. Saddeeg, K.M. Basaqr, B.M. Alshammrani, B.S. Zimmo, Prevalence and Associated Factors of Mask-Induced Acne (Maskne) in the General Population of Jeddah During the COVID-19 Pandemic, Cureus. 14 (n.d.) e26394. [CrossRef]
- A. Dani, A. Eseonu, K. Bibee, Risk factors for the development of acne in healthcare workers during the COVID-19 pandemic, Arch Dermatol Res. 315 (2023) 1067–1070. [CrossRef]
- O. Falodun, N. Medugu, L. Sabir, I. Jibril, N. Oyakhire, A. Adekeye, An epidemiological study on face masks and acne in a Nigerian population, PLOS ONE. 17 (2022) e0268224. [CrossRef]
- Y.-F. Cheng, H. Zhao, J. Li, K.E. Lipa, H.-F. Xie, B. Wang, Y.-X. Huang, Factors aggravating acne vulgaris during the COVID-19 pandemic in China: a web-based cross-sectional survey, Eur Rev Med Pharmacol Sci. 26 (2022) 7305–7312. [CrossRef]
- Sandra M. Tallent, Ann Knolhoff, E. Jeffery Rhodehamel, Stanley M. Harmon, Reginald W. Bennett, Bacteriological Analytical Manual (BAM); Chapter 14: Bacillus cereus., FDA, 1996. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-14-bacillus-cereus (accessed on 15 October 2023).
- R. Roberge, S. Benson, J.-H. Kim, Thermal burden of N95 filtering facepiece respirators, Ann Occup Hyg. 56 (2012) 808–814. [CrossRef]
- R.J. Roberge, J.-H. Kim, S.M. Benson, Absence of consequential changes in physiological, thermal and subjective responses from wearing a surgical mask, Respiratory Physiology & Neurobiology. 181 (2012) 29–35. [CrossRef]
- J.-H. Kim, S.M. Benson, R.J. Roberge, Pulmonary and heart rate responses to wearing N95 filtering facepiece respirators, Am J Infect Control. 41 (2013) 24–27. [CrossRef]
- A. Scarano, F. Inchingolo, F. Lorusso, Facial Skin Temperature and Discomfort When Wearing Protective Face Masks: Thermal Infrared Imaging Evaluation and Hands Moving the Mask, Int J Environ Res Public Health. 17 (2020). [CrossRef]
- S.-R. Park, J. Han, Y.M. Yeon, N.Y. Kang, E. Kim, Effect of face mask on skin characteristics changes during the COVID-19 pandemic, Skin Res Technol. 27 (2021) 554–559. [CrossRef]
- Y.-H. Lee, H. Kim, D.W. Heo, I.-S. Ahn, H.-K. Park, Oral microbiome of the inner surface of face masks and whole saliva during the COVID-19 pandemic, Frontiers in Oral Health. 4 (2023). Available online: https://www.frontiersin.org/articles/10.3389/froh.2023.1178020 (accessed on 28 October 2023).
- S. Szunerits, H. Dӧrfler, Q. Pagneux, J. Daniel, S. Wadekar, E. Woitrain, D. Ladage, D. Montaigne, R. Boukherroub, Exhaled breath condensate as bioanalyte: from collection considerations to biomarker sensing, Anal Bioanal Chem. 415 (2023) 27–34. [CrossRef]
- G. Xiang, K. Xu, Y. Jian, L. He, Z. Shen, M. Li, Q. Liu, Prolonged mask wearing changed nasal microbial characterization of young adults during the COVID-19 pandemic in Shanghai, China, Frontiers in Immunology. 14 (2023). Available online: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1266941 (accessed on 28 October 2023).
- I.M. Viola, B. Peterson, G. Pisetta, G. Pavar, H. Akhtar, F. Menoloascina, E. Mangano, K.E. Dunn, R. Gabl, A. Nila, E. Molinari, C. Cummins, G. Thompson, T.-Y.M. Lo, F.C. Denison, P. Digard, O. Malik, M.J.G. Dunn, C.M. McDougall, F.V. Mehendale, Face Coverings, Aerosol Dispersion and Mitigation of Virus Transmission Risk, IEEE Open J Eng Med Biol. 2 (2021) 26–35. [CrossRef]
- F. Drewnick, J. Pikmann, F. Fachinger, L. Moormann, F. Sprang, S. Borrmann, Aerosol filtration efficiency of household materials for homemade face masks: Influence of material properties, particle size, particle electrical charge, face velocity, and leaks, Aerosol Science and Technology. 55 (2021) 63–79. [CrossRef]
- Y. Shah, J.W. Kurelek, S.D. Peterson, S. Yarusevych, Experimental investigation of indoor aerosol dispersion and accumulation in the context of COVID-19: Effects of masks and ventilation, Physics of Fluids. 33 (2021) 073315. [CrossRef]
- R. Datta, Use of Surgical Facemasks in the Operation Theatre: Effective or Habit?, Medical Journal Armed Forces India. 66 (2010) 163–165. [CrossRef]
- M.H. Barbosa, K.U. Graziano, Influence of wearing time on efficacy of disposable surgical masks as microbial barrier, Braz. J. Microbiol. 37 (2006) 216–217. [CrossRef]
- U.S. Kelkar, B. Gogate, S. Kurpad, P. Gogate, M. Deshpande, How effective are face masks in operation theatre? A time frame analysis and recommendations., International Journal of Infection Control. 9 (2013).
- A. Tcharkhtchi, N. Abbasnezhad, M. Zarbini Seydani, N. Zirak, S. Farzaneh, M. Shirinbayan, An overview of filtration efficiency through the masks: Mechanisms of the aerosols penetration, Bioactive Materials. 6 (2021) 106–122. [CrossRef]
- N.V. McCullough, L.M. Brosseau, D. Vesley, Collection of three bacterial aerosols by respirator and surgical mask filters under varying conditions of flow and relative humidity, The Annals of Occupational Hygiene. 41 (1997) 677–690. [CrossRef]
- A. Hadayer, A. Zahavi, E. Livny, O. Gal-Or, A. Gershoni, K. Mimouni, R. Ehrlich, PATIENTS WEARING FACE MASKS DURING INTRAVITREAL INJECTIONS MAY BE AT A HIGHER RISK OF ENDOPHTHALMITIS, Retina. 40 (2020) 1651–1656. [CrossRef]
- C. Huber, Masks, false safety and real dangers, Part 4: Proposed mechanisms by which masks increase risk of COVID-19, Primary Doctor Medical Journal. (2020) 1–9.
- Boris Borovoy, Colleen Huber, Maria Crisler, Masks, false safety and real dangers, Part 2: Microbial challenges from masks, PDMJ. (2020) 1–19.
- M. Wyszyńska, A. Czelakowska, P. Rosak, E. Białożyt-Bujak, O. Gruca, J. Rosak-Szyrocka, J. Kasperski, M. Skucha-Nowak, Changes in the Oral Cavity Mucosal Surface under the Influence of Wearing Protective Face Masks—Nitric Oxide Concentration Analysis—Preliminary Report, Coatings. 12 (2022) 1164. [CrossRef]
- ICRP: Human respiratory tract model for radiological protection. A report of a Task Group of the International Commission on Radiological Protection, Ann ICRP. 24 (1994) 1–482.
- M.L. Everard, J.G. Hardy, A.D. Milner, Comparison of nebulised aerosol deposition in the lungs of healthy adults following oral and nasal inhalation, Thorax. 48 (1993) 1045–1046. [CrossRef]
- C. Cengiz, İ.H. Can, The effect of N95 and surgical masks on mucociliary clearance function and sinonasal complaints, Eur Arch Otorhinolaryngol. 279 (2022) 759–764. [CrossRef]
- S. Sangkham, O. Faikhaw, N. Munkong, P. Sakunkoo, C. Arunlertaree, M. Chavali, M. Mousazadeh, A. Tiwari, A review on microplastics and nanoplastics in the environment: Their occurrence, exposure routes, toxic studies, and potential effects on human health, Marine Pollution Bulletin. 181 (2022) 113832. [CrossRef]
- K. Kisielinski, S. Hockertz, O. Hirsch, S. Korupp, B. Klosterhalfen, A. Schnepf, G. Dyker, Wearing Face Masks as a Potential Source for Inhalation and Oral Uptake of Inanimate Toxins: a Scoping Review, (2023). [CrossRef]
- A. Khan, Z. Jia, Recent insights into uptake, toxicity, and molecular targets of microplastics and nanoplastics relevant to human health impacts, iScience. 26 (2023). [CrossRef]
- H. Liang, Y. Ji, W. Ge, J. Wu, N. Song, Z. Yin, C. Chai, Release kinetics of microplastics from disposable face masks into the aqueous environment, Science of The Total Environment. 816 (2022) 151650. [CrossRef]
- J. Ma, F. Chen, H. Xu, H. Jiang, J. Liu, P. Li, C.C. Chen, K. Pan, Face masks as a source of nanoplastics and microplastics in the environment: Quantification, characterization, and potential for bioaccumulation, Environmental Pollution. 288 (2021) 117748. [CrossRef]
- S. Wieland, A. Balmes, J. Bender, J. Kitzinger, F. Meyer, A.F. Ramsperger, F. Roeder, C. Tengelmann, B.H. Wimmer, C. Laforsch, H. Kress, From properties to toxicity: Comparing microplastics to other airborne microparticles, Journal of Hazardous Materials. 428 (2022) 128151. [CrossRef]
- W.-L. Teo, The “Maskne” microbiome – pathophysiology and therapeutics, International Journal of Dermatology. 60 (2021) 799–809. [CrossRef]
- Y. Sawada, Occupational Skin Dermatitis among Healthcare Workers Associated with the COVID-19 Pandemic: A Review of the Literature, International Journal of Molecular Sciences. 24 (2023) 2989. [CrossRef]
- 131. A. Tunçer Vural, The development of acne vulgaris due to face masks during the pandemic, risk awareness and attitudes of a group of university students, Journal of Cosmetic Dermatology. 21 (2022) 5306–5313. [CrossRef]
- Y. Liu, H. Zhao, H. Chen, X. Li, C. Ran, H. Sun, L. Wang, Does mask wearing affect skin health? An untargeted skin metabolomics study, Environ Int. 178 (2023) 108073. [CrossRef]
- J.K. Brooks, A.S. Sultan, M.A. Jabra-Rizk, Prolonged facial mask wear is a concern for the development of dysbiotic microbiome, Respiratory Medicine and Research. 81 (2022) 100877. [CrossRef]
- V.A. Koshevarova, Z.K. Westenhaver, M. Schmitz-Brown, B.J. McKinnon, K.H. Merkley, P.K. Gupta, Blepharoconjunctivitis and Otolaryngological Disease Trends in the Context of Mask Wearing during the COVID-19 Pandemic, Clinics and Practice. 12 (2022) 619–627. [CrossRef]
- W.G. Schultheis, J.E. Sharpe, Q. Zhang, S.N. Patel, A.E. Kuriyan, A. Chiang, S.J. Garg, J. Hsu, Effect of Taping Face Masks on Quantitative Particle Counts Near the Eye: Implications for Intravitreal Injections in the COVID-19 Era, American Journal of Ophthalmology. 225 (2021) 166–171. [CrossRef]
- B. Burgos-Blasco, P. Arriola-Villalobos, J.I. Fernandez-Vigo, C. Oribio-Quinto, M. Ariño-Gutierrez, D. Diaz-Valle, J.M. Benitez-del-Castillo, Face mask use and effects on the ocular surface health: A comprehensive review, The Ocular Surface. 27 (2023) 56–66. [CrossRef]
- M. Moshirfar, W.B. West, D.P. Marx, Face Mask-Associated Ocular Irritation and Dryness, Ophthalmol Ther. 9 (2020) 397–400. [CrossRef]
- L. Boccardo, Self-reported symptoms of mask-associated dry eye: A survey study of 3,605 people, Contact Lens and Anterior Eye. 45 (2022). [CrossRef]
- S.R. Islam, D. Prusty, S. Maiti, R. Dutta, P. Chattopadhyay, S.K.K. Manna, Effect of short-term use of FFP2 (N95) mask on salivary metabolome of young healthy volunteers: A pilot study., Mol. Omics. (2023). [CrossRef]
- U. Arora, M. Priyadarshi, V. Katiyar, M. Soneja, P. Garg, I. Gupta, V. Bharadiya, P. Berry, T. Ghosh, L. Patel, R. Sarda, S. Garg, S. Agarwal, V. Arora, A. Ramprasad, A. Kumar, R.K. Garg, P. Kodan, N. Nischal, G. Singh, P. Jorwal, A. Kumar, U. Baitha, V.P. Meena, A. Ray, P. Sethi, I. Xess, N. Vikram, S. Sinha, A. Biswas, A. Thakar, S. Bhatnagar, A. Trikha, N. Wig, Risk factors for Coronavirus disease-associated mucormycosis, J Infect. 84 (2022) 383–390. [CrossRef]
- N.L. Belkin, The evolution of the surgical mask: filtering efficiency versus effectiveness, Infect Control Hosp Epidemiol. 18 (1997) 49–57. [CrossRef]
- C. Matuschek, F. Moll, H. Fangerau, J.C. Fischer, K. Zänker, M. van Griensven, M. Schneider, D. Kindgen-Milles, W.T. Knoefel, A. Lichtenberg, B. Tamaskovics, F.J. Djiepmo-Njanang, W. Budach, S. Corradini, D. Häussinger, T. Feldt, B. Jensen, R. Pelka, K. Orth, M. Peiper, O. Grebe, K. Maas, E. Bölke, J. Haussmann, The history and value of face masks, European Journal of Medical Research. 25 (2020) 23. [CrossRef]
- S.-A. Lee, S.A. Grinshpun, T. Reponen, Respiratory Performance Offered by N95 Respirators and Surgical Masks: Human Subject Evaluation with NaCl Aerosol Representing Bacterial and Viral Particle Size Range, Ann Occup Hyg. 52 (2008) 177–185. [CrossRef]
- M.G.L. Ntlailane, J. Wichmann, Effectiveness of N95 respirators for nanoparticle exposure control (2000–2016): a systematic review and meta-analysis, J Nanopart Res. 21 (2019) 170. [CrossRef]
- L.P. Samaranayake, K.S. Fakhruddin, H.C. Ngo, J.W.W. Chang, C. Panduwawala, The effectiveness and efficacy of respiratory protective equipment (RPE) in dentistry and other health care settings: a systematic review, Acta Odontologica Scandinavica. 78 (2020) 626–639. [CrossRef]
- K. Willeke, Y. Qian, J. Donnelly, S. Grinshpun, V. Ulevicius, Penetration of Airborne Microorganisms Through a Surgical Mask and a Dust/Mist Respirator, American Industrial Hygiene Association Journal. 57 (1996) 348–355. [CrossRef]
- T.K. Hodous, C.C. Coffey, The role of respiratory protective devices in the control of tuberculosis, Occup Med. 9 (1994) 631–657.
- Y. Qian, K. Willeke, S.A. Grinshpun, J. Donnelly, C.C. Coffey, Performance of N95 Respirators: Filtration Efficiency for Airborne Microbial and Inert Particles, American Industrial Hygiene Association Journal. 59 (1998) 128–132. [CrossRef]
- M. Loeb, N. Dafoe, J. Mahony, M. John, A. Sarabia, V. Glavin, R. Webby, M. Smieja, D.J.D. Earn, S. Chong, A. Webb, S.D. Walter, Surgical Mask vs N95 Respirator for Preventing Influenza Among Health Care Workers: A Randomized Trial, JAMA. 302 (2009) 1865. [CrossRef]
- J.D. Smith, C.C. MacDougall, J. Johnstone, R.A. Copes, B. Schwartz, G.E. Garber, Effectiveness of N95 respirators versus surgical masks in protecting health care workers from acute respiratory infection: a systematic review and meta-analysis, CMAJ. 188 (2016) 567–574. [CrossRef]
- I. Liu, V. Prasad, J. Darrow, Evidence for Community Face Masking to Limit the Spread of SARS-CoV-2: A Critical Review, Health Matrix: The Journal of Law-Medicine. 33 (2023) 1.
- M. Vincent, P. Edwards, Disposable surgical face masks for preventing surgical wound infection in clean surgery, Cochrane Database Syst Rev. 4 (2016) CD002929. [CrossRef]
- H.N. Burdick, H. Maibach, Clinical relevance of masks in the operating room? A systematic review, Clinical Infection in Practice. 12 (2021) 100087. [CrossRef]
- C.-C. Carbon, Wearing Face Masks Strongly Confuses Counterparts in Reading Emotions, Frontiers in Psychology. 11 (2020). Available online: https://www.frontiersin.org/articles/10.3389/fpsyg.2020.566886 (accessed on 14 October 2023).
- World Medical Association, WMA - The World Medical Association-Declaration of Helsinki, (2013). Available online: https://www.wma.net/what-we-do/medical-ethics/declaration-of-helsinki/ (accessed on 8 November 2021).
- WHO, World Medical Association (WMA): Declaration of Helsinki. Ethical principles for medical research involving human subjects., Bulletin of the World Health Organization. 79 (2001) 373–374.
- I.H. Elgersma, A. Fretheim, P. Elstrøm, P. Aavitsland, Association between face mask use and risk of SARS-CoV-2 infection: Cross-sectional study, Epidemiol Infect. 151 (2023) e194. [CrossRef]
- A. Boretti, Efficacy of Generalized Face Masking Mandates, Health Services Research and Managerial Epidemiology. 8 (2021) 23333928211058023. [CrossRef]
- P. Galanis, I. Vraka, D. Fragkou, A. Bilali, D. Kaitelidou, Impact of personal protective equipment use on health care workers’ physical health during the COVID-19 pandemic: A systematic review and meta-analysis, American Journal of Infection Control. 49 (2021) 1305–1315. [CrossRef]
- T. Unoki, H. Sakuramoto, R. Sato, A. Ouchi, T. Kuribara, T. Furumaya, J. Tatsuno, Y. Wakabayashi, A. Tado, N. Hashimoto, N. Inagaki, Y. Sasaki, Adverse Effects of Personal Protective Equipment Among Intensive Care Unit Healthcare Professionals During the COVID-19 Pandemic: A Scoping Review, SAGE Open Nursing. 7 (2021) 23779608211026164. [CrossRef]
- H. Dirol, E. Alkan, M. Sindel, T. Ozdemir, D. Erbas, The physiological and disturbing effects of surgical face masks in the COVID-19 era, BLL. 122 (2021) 821–825. [CrossRef]
- R.P. Gaikwad, A.B. Banodkar, V.P. Nandgaonkar, Respiratory Consequences of N95 Mask during Covid-19 Pandemic- An Observational Study, International Journal of Health Sciences and Research. 11 (2021) 55.
- H. Walach, H. Traindl, J. Prentice, R. Weikl, A. Diemer, A. Kappes, S. Hockertz, Carbon dioxide rises beyond acceptable safety levels in children under nose and mouth covering: Results of an experimental measurement study in healthy children, Environmental Research. 212 (2022) 113564. [CrossRef]
- C. Acuti Martellucci, M.E. Flacco, M. Martellucci, F.S. Violante, L. Manzoli, Inhaled CO2 Concentration While Wearing Face Masks: A Pilot Study Using Capnography, Environ Health Insights. 16 (2022) 11786302221123573. [CrossRef]
- M.D.F. Ahmad, S. Wahab, F. Ali Ahmad, M. Intakhab Alam, H. Ather, A. Siddiqua, S. Amir Ashraf, M. Abu Shaphe, M. Idreesh Khan, R. Ali Beg, A novel perspective approach to explore pros and cons of face mask in prevention the spread of SARS-CoV-2 and other pathogens, Saudi Pharmaceutical Journal. 29 (2021) 121–133. [CrossRef]
- N. Shobako, Lessons from the health policies for children during the pandemic in Japan, Frontiers in Public Health. 10 (2022). Available online: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1015955 (accessed on 25 September 2023).
- J.D. Beauchamp, C.A. Mayhew, Revisiting the rationale of mandatory masking, J. Breath Res. 17 (2023) 042001. [CrossRef]
- L. Mastropasqua, M. Lanzini, L. Brescia, R. D’Aloisio, M. Nubile, M. Ciancaglini, C. D’Amario, L. Agnifili, R. Mastropasqua, Face Mask-Related Ocular Surface Modifications During COVID-19 Pandemic: A Clinical, In Vivo Confocal Microscopy, and Immune-Cytology Study, Translational Vision Science & Technology. 10 (2021) 22. [CrossRef]
- S. D’Souza, T. Vaidya, A.P. Nair, R. Shetty, N.R. Kumar, A. Bisht, T. Panigrahi, T.S. J, P. Khamar, M.M. Dickman, R. Agrawal, S. Mahajan, S. Sengupta, R.M.M.A. Nuijts, S. Sethu, A. Ghosh, Altered Ocular Surface Health Status and Tear Film Immune Profile Due to Prolonged Daily Mask Wear in Health Care Workers, Biomedicines. 10 (2022) 1160. [CrossRef]
- S. Jin, D. Wetzel, M. Schirmer, Deciphering mechanisms and implications of bacterial translocation in human health and disease, Current Opinion in Microbiology. 67 (2022) 102147. [CrossRef]
- S. Asadi, C.D. Cappa, S. Barreda, A.S. Wexler, N.M. Bouvier, W.D. Ristenpart, Efficacy of masks and face coverings in controlling outward aerosol particle emission from expiratory activities, Scientific Reports. 10 (2020) 15665. [CrossRef]
- S. Bagchi, S. Basu, S. Chaudhuri, A. Saha, Penetration and secondary atomization of droplets impacted on wet facemasks, Phys. Rev. Fluids. 6 (2021) 110510. [CrossRef]
- T. Rebmann, R. Carrico, J. Wang, Physiologic and other effects and compliance with long-term respirator use among medical intensive care unit nurses, Am J Infect Control. 41 (2013) 1218–1223. [CrossRef]
- Ł. Matusiak, M. Szepietowska, P. Krajewski, R. Białynicki-Birula, J.C. Szepietowski, Inconveniences due to the use of face masks during the COVID-19 pandemic: A survey study of 876 young people, Dermatologic Therapy. 33 (2020) e13567. [CrossRef]
- Naylor, L.A. Burke, J.A. Holman, Covid-19 Lockdown Affects Hearing Disability and Handicap in Diverse Ways: A Rapid Online Survey Study, Ear Hear. 41 (2020) 1442–1449. [CrossRef]
- F. Thomas, C. Allen, W. Butts, C. Rhoades, C. Brandon, D.L. Handrahan, Does wearing a surgical facemask or N95-respirator impair radio communication?, Air Med J. 30 (2011) 97–102. [CrossRef]
- C.A. Heider, M.L. Álvarez, E. Fuentes-López, C.A. González, N.I. León, D.C. Verástegui, P.I. Badía, C.A. Napolitano, Prevalence of Voice Disorders in Healthcare Workers in the Universal Masking COVID-19 Era, The Laryngoscope. n/a (2020). [CrossRef]
- H. Sezer, D. Çınar, N. Kılıç Akça, The effect of prolonged use of surgical masks during face-to-face teaching on cognitive and physiological parameters of nursing students: A cross-sectional and descriptive study, Nurse Education in Practice. 72 (2023) 103779. [CrossRef]
- C. Elbl, J.X. Brunner, D. Schier, A. Junge, H. Junge, Protective face masks add significant dead space, European Respiratory Journal. 58 (2021). [CrossRef]
- A. Chandra, T.B. Høeg, Lack of correlation between school mask mandates and paediatric COVID-19 cases in a large cohort, Journal of Infection. 85 (2022) 671–675. [CrossRef]
- J. Xiao, E.Y.C. Shiu, H. Gao, J.Y. Wong, M.W. Fong, S. Ryu, B.J. Cowling, Nonpharmaceutical Measures for Pandemic Influenza in Nonhealthcare Settings—Personal Protective and Environmental Measures - Volume 26, Number 5—May 2020 - Emerging Infectious Diseases journal - CDC, (2020). [CrossRef]
- M.X. Wang, S.X.W. Gwee, P.E.Y. Chua, J. Pang, Effectiveness of Surgical Face Masks in Reducing Acute Respiratory Infections in Non-Healthcare Settings: A Systematic Review and Meta-Analysis, Frontiers in Medicine. 7 (2020). Available online: https://www.frontiersin.org/articles/10.3389/fmed.2020.564280 (accessed on 26 November 2022).
- M.S. Fønhus, T.K. Dalsbø, K.G. Brurberg, Facemasks to prevent transmission of respiratory illness, such as COVID-19, Norwegian Institute of Public Health, 2021. Available online: https://fhi.brage.unit.no/fhi-xmlui/handle/11250/2756758 (accessed on 7 November 2021).
- A.E. Aiello, V. Perez, R.M. Coulborn, B.M. Davis, M. Uddin, A.S. Monto, Facemasks, hand hygiene, and influenza among young adults: a randomized intervention trial, PLoS One. 7 (2012) e29744. [CrossRef]
- E. Zain Alabdeen, A. Choudhry, D. Al-Naji, Effect of use of Face mask on Hajj related Acute Respiratory Infection among Hajjis from Riyadh -A Health Promotion Intervention study, FETP Saudi Epidemiology Bulletin. 12 (2005) 27–28.
- M. Alfelali, E.A. Haworth, O. Barasheed, A.-M. Badahdah, H. Bokhary, M. Tashani, M.I. Azeem, J. Kok, J. Taylor, E.H. Barnes, H. El Bashir, G. Khandaker, E.C. Holmes, D.E. Dwyer, L.G. Heron, G.J. Wilson, R. Booy, H. Rashid, Hajj Research Team, Facemask against viral respiratory infections among Hajj pilgrims: A challenging cluster-randomized trial, PLoS One. 15 (2020) e0240287. [CrossRef]
- L. Canini, L. Andréoletti, P. Ferrari, R. D’Angelo, T. Blanchon, M. Lemaitre, L. Filleul, J.-P. Ferry, M. Desmaizieres, S. Smadja, A.-J. Valleron, F. Carrat, Surgical mask to prevent influenza transmission in households: a cluster randomized trial, PLoS One. 5 (2010) e13998. [CrossRef]
- C.R. MacIntyre, S. Cauchemez, D.E. Dwyer, H. Seale, P. Cheung, G. Browne, M. Fasher, J. Wood, Z. Gao, R. Booy, N. Ferguson, Face mask use and control of respiratory virus transmission in households, Emerg Infect Dis. 15 (2009) 233–241. [CrossRef]
- C.R. MacIntyre, H. Seale, T.C. Dung, N.T. Hien, P.T. Nga, A.A. Chughtai, B. Rahman, D.E. Dwyer, Q. Wang, A cluster randomised trial of cloth masks compared with medical masks in healthcare workers, BMJ Open. 5 (2015) e006577. [CrossRef]
- J.M. Simmerman, P. Suntarattiwong, J. Levy, R.G. Jarman, S. Kaewchana, R.V. Gibbons, B.J. Cowling, W. Sanasuttipun, S.A. Maloney, T.M. Uyeki, L. Kamimoto, T. Chotipitayasunondh, Findings from a household randomized controlled trial of hand washing and face masks to reduce influenza transmission in Bangkok, Thailand, Influenza Other Respir Viruses. 5 (2011) 256–267. [CrossRef]
- B.J. Cowling, R.O.P. Fung, C.K.Y. Cheng, V.J. Fang, K.H. Chan, W.H. Seto, R. Yung, B. Chiu, P. Lee, T.M. Uyeki, P.M. Houck, J.S.M. Peiris, G.M. Leung, Preliminary Findings of a Randomized Trial of Non-Pharmaceutical Interventions to Prevent Influenza Transmission in Households, PLoS One. 3 (2008). [CrossRef]
- B.J. Cowling, K.-H. Chan, V.J. Fang, C.K.Y. Cheng, R.O.P. Fung, W. Wai, J. Sin, W.H. Seto, R. Yung, D.W.S. Chu, B.C.F. Chiu, P.W.Y. Lee, M.C. Chiu, H.C. Lee, T.M. Uyeki, P.M. Houck, J.S.M. Peiris, G.M. Leung, Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial, Ann Intern Med. 151 (2009) 437–446. [CrossRef]
- T. Suess, C. Remschmidt, S.B. Schink, B. Schweiger, A. Nitsche, K. Schroeder, J. Doellinger, J. Milde, W. Haas, I. Koehler, G. Krause, U. Buchholz, The role of facemasks and hand hygiene in the prevention of influenza transmission in households: results from a cluster randomised trial; Berlin, Germany, 2009-2011, BMC Infect Dis. 12 (2012) 26. [CrossRef]
- E.L. Larson, Y. Ferng, J. Wong-McLoughlin, S. Wang, M. Haber, S.S. Morse, Impact of Non-Pharmaceutical Interventions on URIs and Influenza in Crowded, Urban Households, Public Health Rep. 125 (2010) 178–191.
- J.L. Jacobs, S. Ohde, O. Takahashi, Y. Tokuda, F. Omata, T. Fukui, Use of surgical face masks to reduce the incidence of the common cold among health care workers in Japan: a randomized controlled trial, Am J Infect Control. 37 (2009) 417–419. [CrossRef]
- H. Bundgaard, J.S. Bundgaard, D.E.T. Raaschou-Pedersen, C. von Buchwald, T. Todsen, J.B. Norsk, M.M. Pries-Heje, C.R. Vissing, P.B. Nielsen, U.C. Winsløw, K. Fogh, R. Hasselbalch, J.H. Kristensen, A. Ringgaard, M. Porsborg Andersen, N.B. Goecke, R. Trebbien, K. Skovgaard, T. Benfield, H. Ullum, C. Torp-Pedersen, K. Iversen, Effectiveness of Adding a Mask Recommendation to Other Public Health Measures to Prevent SARS-CoV-2 Infection in Danish Mask Wearers, Ann Intern Med. (2020). [CrossRef]
- A. Juutinen, E. Sarvikivi, P. Laukkanen-Nevala, O. Helve, Face mask recommendations in schools did not impact COVID-19 incidence among 10–12-year-olds in Finland – joinpoint regression analysis, BMC Public Health. 23 (2023) 730. [CrossRef]
- S.A. Gómez-Ochoa, T. Muka, Meta-analysis on facemask use in community settings to prevent respiratory infection transmission shows no effect, International Journal of Infectious Diseases. 103 (2021) 257–259. [CrossRef]

| Mask number | Average CFU* recovered/mask and standard deviation** |
|---|---|
| 3 clean masks | 0.1 ± 0.09 × 103 |
| 1 | 4.8 ± 0.3 × 103 |
| 2 | 9.29 ± 0.17 × 104 |
| 3 | 1.35 ± 0.15 × 103 |
| 4 | 9.3 ± 0.3 × 103 |
| 5 | 1.03 ± 0.03 × 105 |
| 6 | 2.85 ± 0.05 × 105 |
| 7 | 1.79 ± 0.14 × 104 |
| 8 | 9.15 ± 0.15 × 103 |
| 9 | 0.45 ± 0 × 103 |
| 10 | 5.55 ± 0.45 × 103 |
| 11 | 3.47 ± 0.29 × 104 |
| 12 | 1.76 ± 0.05 × 104 |
| 13 | 3.45 ± 1.05 × 103 |
| 14 | 1.55 ± 0.05 × 104 |
| 15 | 3.53 ± 0.08 × 104 |
| Author and year | Mask types (population) |
Duration of wear | n | Contamination | Method | Maximum level of contamination* | Principal microorganisms detected |
|---|---|---|---|---|---|---|---|
| Present study 2023 | surgical, disposable (general population) |
5 min- 12 h |
15 | bacterial (nearly whole mask) |
PBS, agar plates 16S rRNA |
2.85 ×105 / mask |
Staphylococcus aureus, Staphylococcus warneri, Staphylococcus epidermidis, B. cereus, B. thuringiensis, B. altitudinis, B. megaterium |
| Checchi 2022 [64] | N95 (HCW, dental practice) |
30 h | 6 | bacterial (outer surface samples) |
agar plates, eye sighting and counting |
101-141 / outer mask area sample | not specified |
| Delanghe 2021 [39] | cotton, surgical (general population) |
4 h | 21 | bacterial (half mask) |
PBS, agar plates 16S rRNA |
1.53×105 ± 1.96×105 / cotton mask 1.79×104 ± 1.63×104 / surgical mask |
Staphylococcus spp., Bacillus spp., Acinetobacter spp. |
| Gund 2021 [62] | surgical (HCW, dental practice) |
45-60 min |
32 | bacterial, (external surface samples) |
agar plates, MALDI-TOF MS, colony counter | <102 / contact sample external surface |
S. epidermidis, S. capitis, S. saprophyticus, B. cereus |
|
Gund 2022 [63] |
surgical over N95 (HCW, dental practice) |
60-90 min | 102 | bacterial, (external surface samples) |
agar plates, MALDI-TOF MS, colony counter |
80 ± 130 / imprint external surface |
Streptococcus, Staphylococcus spp., Micrococcus spp., Bacillus spp. |
| Keri 2021 [67] | cloth, surgical, N95 (general population) |
4-72 h | 50 | fungi, (inner and outer surface samples) |
agar plates, microscopy, lactophenol cotton blue |
fungi: 64% outside (32 in 50 masks) 67% inside (14 in 21 masks) |
Aspergillus niger, Rhizopus arrhizus, Syncephalastrum spp., Mucor spp. |
| Liu 2018 [47] | Surgical (HCW, orthopaedic surgery) |
2-6 h | 40 | bacterial, (first outer layer, second outer layer samples) |
agar plates, eye sighting and counting |
bacteria: 5.3 (0-2h) / layer samples 7.4 (2-4h) /layer samples 12.8 (4-6h) / layer samples |
not specified |
| Lukasmijarkul 2014 [43] | surgical (HCW) |
not given (working day) |
203 | bacterial, fungal, (outer side and inner side) |
agar plates, Gram´s stain, lactophenol cotton blue. microscopic morphology |
bacteria: 47 ± 56 / inside area sample 166 ± 199 / outside area sample fungi: 15 ± 9 / inside area sample 34 ± 18 / outside area sample |
Staphylococcus spp., Pseudomonas spp. Aspergillus spp., Penicillium spp. |
| Merad 2023 [68] | surgical, N95 (HCW) |
1-7 h | 52 | fungal, inner side samples | agar plates, macroscopic and microscopic features of growing colonies |
fungi: 88% surgical 8% KN95 |
Alternaria spp. (32%), Penicillium spp. (20%), Aspergillus spp. (16%) |
| Monalisa 2017 [45] | surgical (HCW, dental practice) |
not given (working day) |
36 | bacterial, fungal, (outer side and face-side samples) |
agar plates, colony counter, biochemical tests | bacteria: 31.7×102 / outer sample 22.8 ×105 / internal sample |
E. coli (54%), S. aureus (25%), Klebsiella spp. (5%), Enterococcus spp. (4%), Pseudomonas spp.(3%), Enterobacter spp. (2%), Candida (6%), Aspergillus spp., Cladosporium spp., Alternaria spp. |
| Nightingale 2023 [66] | surgical (HCW) |
4-8 h | 69 | bacterial (whole mask) |
selective plates, catalase and coagulase tests |
bacterial: 44.9% |
Enterococcus spp. (44.9%) S. aureus (15.9%), Klebsiella pneumoniae (14.5%) |
| Park 2022 [60] | non-woven, gauze, polyurethane (general population) |
1 day (3-6 h) up to 3 days |
109 | bacterial, fungal, (outer side and face-side samples) |
agar plates, 16S rRNA sampling, Gram staining |
bacteria: 168.6 ± 24.7 / face-side 36.0 ± 7.0 / outer side fungi: 4.6 ± 1.9 / face-side 6.1 ± 1.9 / outer side |
S. epidermidis, S. aureus, B. cereus, S. saprophyticus Aspergillus, Microsporum, Cladosporium |
| Sachdev 2020 [61] | surgical (HCW, dental practice) |
30 min | 240 | bacterial, fungal, (outside and inside samples) |
agar plates, Gram´s stain, lactophenol cotton blue, microscopic morphology |
bacteria: 48±26 / inside mask 180±110 / outside mask fungi: 14±6 / inside mask 32±13 / outside mask |
Staphylococcus spp. (26.35%), Pseudomonas spp. (17.82%), Streptococcus spp. (15.50%), Aspergillus (6.97%) |
| Yang 2018 [42] | N95 (general population) |
5+15 min | 2 | bacterial, (inner surface samples) |
agar plates, eye sighting and counting |
bacteria: 4.33 (5 min sample) 49 (15 min sample) |
not specified |
| Yousefimashouf 2023 [65] | surgical, N95 (HCW) |
≤2-8 h | 175 | bacterial, (inner and outer surface samples) |
immersion physiological serum, agar plates, analytical profile index kit |
Bacteria distribution on 471 positive isolates: 52.2 % (N95) 47.8 % (surgical) counts N95, inner vs outer: 128:118 counts surgical, inner vs. outer: 106:119 |
Coagulase-negative Staphylococcus (28%) Acinetobacter baumannii (20.8%), Pseudomonas aeruginosa (13.8%), E. coli (10.8%), S. aureus (10.1%), ß-Haemolytic Streptococcus (5.9%), Enterobacter (5.4%), Klebsiella (3.8%), Enterococcus (1.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





