1. Introduction
Microglia-mediated neuroinflammation is an important component of Alzheimer's disease (AD) pathogenesis and has been implicated in neurodegeneration [
1,
2,
3]. Interleukin-1β (IL-1β) is a potent proinflammatory cytokine involved in many important cellular functions. The release of IL-1β is a critical step in inflammation through the induction of other proinflammatory cytokines and chemokines [
4,
5]. IL-1β is chronically upregulated in AD and believed to play a role in the vicious inflammatory cycle that drives AD pathology [
6]. A two-step process is generally necessary for IL-1β production. First step is the synthesis of pro-IL-1β, and second step is the processing of synthesized pro-IL-1β. The TLR-dependent signals first activate nuclear factor-κB (NF-κB) to lead pro-IL-1β synthesis. The Nod-like receptor (NLR) family pyrin domain containing 3 (NLRP3) inflammasome then undergoes post-translational modifications that license its activation. The activation signals activate the NLRP3 inflammasome with subsequent activation of pro-caspase-1, which in turn catalyzes the cleavage of pro-IL-1β.
Activation signals are provided by a variety of stimuli and multiple molecular or cellular events, including K
+ efflux, mitochondrial dysfunction with reactive oxygen species (ROS) generation, and lysosomal damage with cathepsin B (CatB; EC 3.4.22.1) leakage. Several studies have suggested a CatB/NLRP3/caspase-1-dependent pathway for pro-IL-1β processing in BV-2 microglia [
7] and THP-1 cells [
8], and the roles of multiple cathepsins in NLRP3 activation have also been implicated in macrophages/microglia [
9,
10]. However, we previously reported a potential NLRP3-independent role of autolysosomal CatB in pro-caspase-1 activation and subsequent IL-1β secretion by microglia following stimulation with chromogranin A [
11,
12]. Therefore, there are at least three different pathways for pro-IL-1β processing, with a special focus on the role of CatB, depending on the stimulating reagents and cell types: (1) a CatB/NLRP3/ caspase-1-dependent pathway, (2) a CatB/caspase-1-dependent, but NLRP3-independent pathway, and (3) a CatB and other cysteine cathepsins/NLRP3/ caspase-1-dependent pathway. In addition to pro-IL-1β processing, there is some evidence demonstrating that CatB is also involved in pro-IL-1β synthesis through the degradation of IκBα, an endogenous inhibitor of NF-κB in macrophages/microglia at the late stage of inflammation [
9,
13,
14,
15,
16,
17].
The emerging role of microbes and innate immune pathways in AD pathology suggests that antimicrobial peptides may be effective as early therapeutic intervention in future clinical trials [
18,
19]. The salivary proteome contains a complex mixture of over 45 antimicrobial proteins and peptides, including human defensins, histatins and cathelicidin hCAP18/LL-37 [
20]. Human β-defensins (hBDs) are small, cationic antimicrobial peptides produced by the oral mucosa and salivary glands. We previously reported that hBD3 strongly suppresses the delayed type of inflammatory responses by microglia following treatment with lipopolysaccharide (LPS) derived from
Porphyromonas gingivalis (
Pg), a major pathogen of chronic periodontitis. hBD3 suppresses
Pg LPS-induced NF-κB activation through inhibition of CatB and cathepsin L (CatL; EC 3.4.22.15) [
21]. Furthermore, we first reported that chronic systemic exposure to
Pg LPS induces AD-like pathologies, including microglia-mediated neuroinflammation and cognitive decline in middle-aged mice, but not in CatB-deficient mice [
22].
However, whether or not hBD3 and CatB inhibitors can suppress
Pg virulence factor-induced IL-1β production by microglia remains unclear. In addition to
Pg LPS, chronic oral gavage with outer membrane vesicles (OMVs) secreted from
Pg also induced AD-like pathologies in middle-aged mice [
23]. In the present study, we have thus attempted to clarify effects of hBD3 and CatB inhibitors on the IL-1β production by microglia following stimulation with
Pg LPS and OMVs.
2. Materials and Methods
2.1. Reagents
hBD3, CA-074Me, KYT-1 and KYT-36 were purchased from the Peptide Institute Inc (Osaka, Japan), and Z-Arg-Leu-Arg-α-aza-glycil-Ile-Val-OMe (ZRLR) was synthesized by Peptide Institute Inc. Standard Pg LPS was purchased from InvivoGen (San Diego, CA, USA). TAK-242 and C29 were purchased from MedChem Express (Monmouth Junction, NJ, USA). MCC950 was purchased from Adipogen Life Sciences, Inc (San Diego, CA, USA). Wedelolactone was purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan).
2.2. Cell culture
The BV-2 cells, a murine microglial cell line [
24], and a well-accepted alternative to primary microglia [
25,
26], were used in this study. BV-2 microglia were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS), penicillin, and streptomycin. To establish NanoLuc (Nluc) probe-expressing cells (Nluc reporter BV-2 microglia), we infected the cells with a lentiviral vector carrying the Nluc probe, as described previously [
27], during routine culturing. After two passages, the cells were detached using Accutase (Nacalai Tesque, Kyoto, Japan), and EGFP-positive cells were sorted using a BD FACSAria III cell sorter (BD Life Sciences, San Jose, CA, USA). In this assay system, the Nluc luciferase gene was designed to be induced by the promoter activity of the
Il1b gene, and luciferase activity was only induced when proteolytic processing of pro-IL-1β occurred because the Nluc luciferase was fused with the sequence of IL-1β (
Il1b 17-216), which has been subjected to proteolytic processing of various proteases including inflammasome-mediated caspase-1 [
28]. Furthermore, the
C-terminus of
Il1b 17-216 was fused with two protein destabilization sequences (hCL1 and hPEST), allowing for rapid proteasome-mediated degradation. Thus, the Nluc luciferase protein is retained only when the proteolytic processing of IL-1β is successful, as it can be escape proteasome degradation.
2.3. The measurement of the luciferase activity (RLU)
Nluc reporter BV-2 microglia were plated in 96-well culture plates at a density of 3×104 cells per well. After overnight culture, drug treatments were performed, and luciferase activity following treatment with Pg LPS (10 µg/mL) or OMVs (150 µg/mL) for 1 h was measured using a luminometer (GloMax; Promega Corp., Madison, WS, USA) with a Nano-Glo® luciferase assay system (N1110; Promega Corp.) according to the manufacturer's protocol. The luciferase activity (RLU) in BV-2 microglia induced by Pg LPS or OMVs was then measured. Each treatment was repeated in triplicate on the same plate, and at least three independent experiments were performed.
2.4. Bacterial culture and isolation of OMVs
Pg ATCC33277 (wild-type; WT), Kgp-deficient mutant strain (KDP129), and a Kgp- and Rgp (both RgpA and RgpB)-deficient mutant strain (KDP136) were maintained on blood agar plates containing 40 mg/mL trypto-soya agar (Nissui Pharmaceutical, Tokyo, Japan), 5 mg/mL brain heart infusion (BHI) medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), 1 mg/mL cysteine (Wako Pure Chemical Industries, Osaka, Japan), 5 μg/mL hemin (Sigma-Aldrich, St. Louis, MO, USA), 0.5 µg/mL menadione (Sigma-Aldrich), and 5% defibrinated sheep blood (Nippon Bio-Test Laboratories, Tokyo, Japan) in a Bactron anaerobic chamber (Shel Lab, Cornelius, OR, USA) at 5% CO2, 5% H2, and 90% N2.
For immunostaining experiments, OMVs were isolated as described previously [
29]. In brief, the bacteria were grown in enriched BHI broth containing 37 mg/mL BHI medium, 2.5 mg/mL yeast extract, 1 mg/mL cysteine, 5 µg/mL hemin, and 0.5 µg/mL menadione. The culture medium was centrifuged at 2800×
g for 15 min at 4 °C to remove bacterial cells. The supernatant was filtered through a Millipore filter (pore size, 0.22 µm). The filtered supernatant (8 mL) was concentrated to 1 mL using an ultrafiltration column at 10 K (Apro Science, Tokushima, Japan). The concentrate was mixed with the Total Exosome Isolation Reagent (Thermo Fisher Scientific Inc.) and incubated overnight at 4 °C. The samples were then centrifuged at 10,000×
g for 60 min at 4 °C. The OMVs fraction was dissolved in 100 µL of phosphate-buffered saline (PBS) and used for the experiments.
In other experiments, OMVs were prepared by ultracentrifugation as described previously [
30]. In brief,
Pg was grown in enriched BHI broth and then removed from the culture by centrifugation at 10,000×
g for 30 min at 4 °C. The supernatant was filtered through a Millipore filter (pore size, 0.22 µm). OMVs were collected as pellets by ultracentrifugation of the culture supernatant at 100,000×
g for 3 h at 4 °C and then dissolved in PBS.
2.5. Fluorescence imaging of cellular localization of OMVs
OMVs from the WT and KDP136 strains (75 µg of total protein) prepared with the Total Exosome Isolation Reagent were incubated with 5 μM Cy5 Mono NHS Ester (Lumiprobe Corporation, Cockeysville, MD, USA) for 90 min at 37 °C. The labelled OMVs were collected by centrifugation (10,000×g for 30 min at 4 °C), dissolved in PBS, and then centrifuged at 10,000×g for 30 min at 4 °C to remove the unincorporated dye. The precipitated labelled OMVs were dissolved in FBS-free DMEM (0.5 mL). To observe the cellular localization of Cy5-labelled OMVs, Nluc reporter BV-2 microglia (2×104 cells/well) were cultured on collagen-coated cover glass in 24-well plates at 37 °C for 1 day and then incubated with FBS-free DMEM containing Cy5-labelled OMVs (150 µg of protein/mL) for 1 h. After incubation, the cells were fixed with 4% (w/v) paraformaldehyde for 15 min and then permeabilized with PBS containing 0.2% (v/v) Triton X-100. After washing with PBS, the cells were incubated with Acti-stain 488 phalloidin (a F-actin detecting dye, Cytoskeleton, Inc., Denver, CO, USA) and Hoechst 33342 (ThermoFisher Scientific) to visualize the cells. Fluorescence images were obtained using a confocal laser-scanning microscope (CLSM; FV1000, Olympus, Tokyo, Japan).
2.6. Trasfection of CatB shRNA
Nluc reporter BV-2 microglia were transfected to CatB shRNA or control shRNA lentiviral particle (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) according to the manufacture’s transfection protocol. In brief, Nluc reporter BV-2 microglia were cultured with complete optimal medium in a 12-well plate (3×105 cells/well) 24 h prior to lentiviral infection. Media were removed from the plate wells and replaced with 1 mL of polyberene/media mixture (5 µg/mL) per well. CatB or control shRNA lentiviral particles were then added and incubated overnight. The cells were cultured for an additional 48 h in complete medium without polybrene. Stable clones expressing CatB shRNA (Nluc reporter CatB-knockdown BV-2 microglia) or control shRNA were selected using puromycin dihydrochloride (6 µg/mL).
2.7. Fluorescence imaging of enzymatic activities of CatB and CatL
BV-2 microglia were stained with the cell-permeable fluorescently labeled CatB substrate z-Arg-Arg-cresyl violet or CatL substrate z-Phe-Arg-cresyl violet according to the manufacturer’s instructions (cv-CatB detection kit and cv-CatL detection kit; Enzo Life Sciences, Inc., Farmingdale, NY, USA). Chamber slides containing stained live cells were then mounted in PBS. Fluorescence images were obtained using CLSM (FV1000, Olympus).
2.8. Nuclear NF-κB p65 translocation
BV-2 microglia were treated with Pg LPS (10 μg/mL) or OMVs (150 µg/mL) for 1 h in the absence or presence of hBD3 (1 µM) or CA-074Me (30 µM), and were fixed with 4% paraformaldehyde. hBD3 and CA-074Me were pre-incubated with BV-2 microglia for 2 and 4 h, respectively. They were then incubated with a rabbit anti-NF-κB p65 IgG antibody (Abcam, Cambridge, UK). After washing with PBS, the cells were incubated with donkey anti-rabbit Alexa 555 (ThermoFisher Scientific), Hoechst 33342, and mounted in Vectashield anti-fading medium (Vector Laboratories, Newark, CA, USA). Fluorescence images were captured using CLSM (FV1000, Olympus). The line plot profile was analyzed using the Image J software program.
2.9. Immunoblotting analyses
For detecting IκBα and β-actin, BV-2 microglia were used. WT and CatB-KD BV-2 microglia were used to detect CatB and GAPDH. Cells were seeded in a 6 cm petri dish at a density of 2.5-3.3×106 cells/dish for 1 day. After treatment with Pg LPS or OMVs in the absence or presence of hBD3 (1 μM) or CA-074Me (30 μM), the cells were lysed with RIPA buffer, consisting of 10 mM Tris-HCl (pH 7.5), 1% (v/v) NP-40, 0.1% (w/v) sodium deoxycholate, 0.1% (w/v) sodium dodecyl sulfate (SDS), 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan), and then cell lysates were subjected to 10% (w/v) SDS-polyacrylamide gel electrophoresis. The proteins on the SDS–polyacrylamide gels were then transferred to nitrocellulose membranes. After blocking with Blocking One (Nacalai Tesque), the membranes were incubated with rabbit anti-IκBα (Abcam), rabbit anti-β-actin (GENETEX, Irvine, CA), rabbit anti-CatB antibodies (Abcam), and rabbit anti-GAPDH (Proteintech, Tokyo, Japan) antibodies at 4°C overnight. After washing, the membranes were incubated with horseradish peroxidase (HRP)-labeled anti-rabbit IgG antibodies (GE Healthcare, Tokyo, Japan) for 1 h at room temperature. Membrane-bound HRP-labeled antibodies were detected using the Amasham ECL Western blotting detection reagent and analysis system (GE Healthcare) with an imaging analyzer (LAS-4000, Fujifilm, Tokyo, Japan). Signal intensities were determined using the Image Lab 6.0.1 software program (Bio-Rad, Hercules, CA, USA).
2.10. AlphaFold predictions
The AlphaFold models were predicted using the AlphaFold v2.0 algorithm on the Colab server (
https://colab.research.google.com/github/ sokrypton/ColabFold/blob/ main/AlphaFold2.ipynb), accessed on November 20, 2023 [
31]. Predictions were performed with default multiple sequence alignment generation using the MMSeqs2 server, with 48 recycles, and templates (homologous structures). The best of the five predicted models (rank 1) computed by AlphaFold was considered in the present work.
2.11. Statistical analyses
Data are presented as the mean ± standard error (SE). Statistical analyses of the results were performed using a one-way analysis of variance (ANOVA) with a post-hoc Tukey’s test and Student’s t-test using the GraphPad Prism8 (GraphPad Software, Inc., CA, USA) software package. P<0.05 was considered to indicate statistical significance.
4. Discussion
hBD3 significantly suppressed the IL-1β production induced by
Pg LPS but not by OMVs, while CA-074Me significantly inhibited the IL-1β production induced by both
Pg LPS and OMVs. Furthermore, both hBD3 and CA-074Me significantly inhibited
Pg LPS-induced nuclear NF-κB p65 translocation and IκBα degradation. In contrast, neither hBD3 nor CA-074Me blocked OMV-induced nuclear NF-κB p65 translocation and IκBα degradation. CA-074 could have non-cathepsin off-target effects [
9,
34], which may be responsible for the inhibitory effect of CA-074Me on OMV-induced IL-1β production. On the other hand, ZRLR, a specific CatB inhibitor, had no effect on
Pg virulence factor-induced IL-1β production. This was consistent with the results obtained using shRNA-mediated knockdown of CatB expression. These results suggest that
Pg LPS induces the synthesis and processing of pro-IL-1β through CatB/CatL-dependent mechanisms, without activation of the NLRP3 inflammasome. In contrast, OMVs may promote the synthesis and processing of pro-IL-1β through CatB/CatL-independent phagocytic mechanisms.
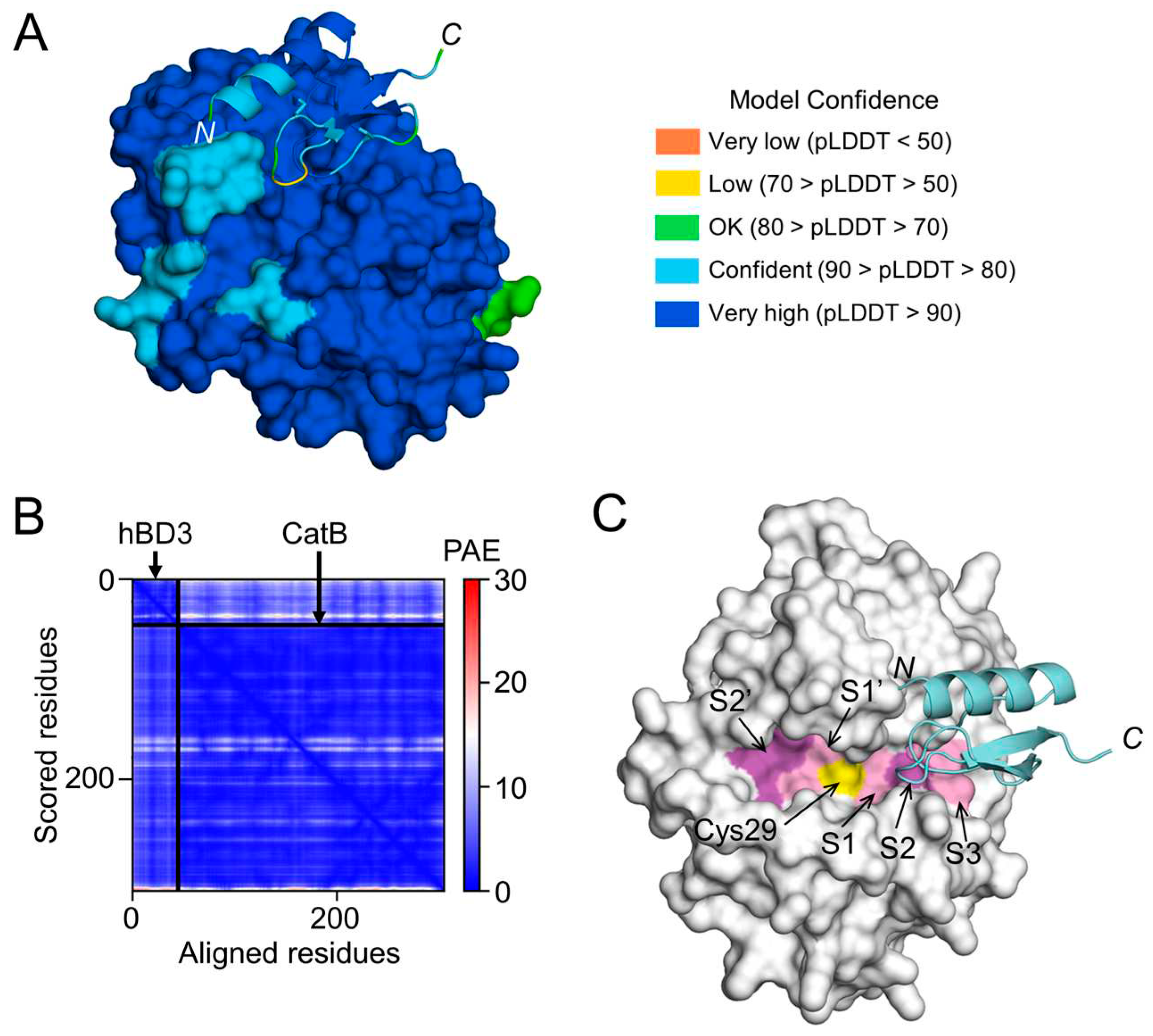
The structural models generated by AlphaFold in this study indicated that hBD3 can bind more strongly to the substrate-binding pocket of CatB than to that of CatL, suggesting that hBD3 can more potently inhibit the enzymatic activity of CatB than that of CatL. This is consistent with our previous observations that hBD3 (1 µM) inhibits the enzymatic activities of human recombinant CatB and CatL by approximately 60% and 10%, respectively [
21]. It was reported that hBD3 can be a substrate for cysteine cathepsins such as CatB and CatL [
35]. Therefore, the present findings extend our previous implication that hBD3 suppresses
Pg LPS-induced oxidative and inflammatory responses in microglia through the suicide substrate-based inhibition of CatB and CatL.
The methyl ester of CA-074Me is considered to be hydrolyzed by intracellular esterases releasing the active inhibitor, CA-074. However, the amount of CA-074 released into BV-2 microglia after treatment with CA-074Me remains unclear. Activity-based cysteine protease labeling using DCG-04 showed that CA-074Me is not a CatB-specific inhibitor in murine fibroblasts [
36]. Therefore, CA-074Me may inhibit CatB and other cysteine cathepsins, especially CatL, at the concentrations used in previous studies (e.g. 10-30 µM). In contrast, ZRLR is a highly potent, irreversible, membrane-permeable, CatB-specific inhibitor. ZRLR is an azapeptide, which are peptide analogs in which the α-CH group of one amino acid resides in the peptide and is replaced by a nitrogen atom. The intrinsic structural prevalence of the
N-terminal part of ZRLR is to remain bent, which allows a covalent attachment of ZRLR onto the active site Cys29 in CatB [
37]. ZRLR exclusively blocks CatB in living primary antigen-presenting cells, in contrast to CA-074Me, by use of DCG-4, which detects active cysteine cathepsins in an activity- based reaction [
33].
Currently, OMVs are considered to be potent vehicles for transmitting virulence factors into the host cells [
38]. OMVs contain gingipains, which covalently bind to anionic LPS on their surface [
32]. Furthermore, gingipains can activate pro-caspase-1 [
39]. However, it is unlikely that the enzymatic activities of gingipains are involved in the IL-1β production, as pharmacological inhibition of both Kgp and Rgp did not block OMV-induced IL-1β production by BV-2 microglia. Our observations here showed that BV-2 microglia did not phagocytose OMVs prepared from KDP136, which is devoid of both Kgp and Rgp. It was reported that Rgp is necessary for the fimbriae formation [
40,
41]. Therefore, BV-2 microglia could not phagocytose OMVs prepared from KDP136 because of a lack of fimbriae, which is necessary for a lipid raft-mediated endocytic pathway [
30,
42]. Of particular note, OMVs prepared from KDP136 did not induce the IL-1β production by BV-2 microglia.
OMVs secreted by Gram-negative bacteria function as a vehicle that delivers LPS into the cytosol and induces caspase-11-dependent inflammatory responses [
43,
44]. It has been suggested that a functional interaction between caspase-11 and the NLRP3 inflammasome, and perhaps involving additional partners, could promote noncanonical activation of the pro-IL-β processing [
45]. It has been reported that phagocytic machinery can activate NF-κB in macrophages [
46] and monocytes [
47]. We previously reported that OMVs can deliver gingipains into the cytosol of hCMEC/D3 cells [
48]. However, whether or not
Pg LPS released into the cytosol is involved in the IL-1β production by BV-2 microglia after phagocytosis of OMVs remains unclear. The precise CatB/CatL -independent mechanisms involved in OMV-induced IL-1β production by BV-2 microglia should be elucidated in future studies.
Author Contributions
Conceptualization, H.N.; investigation, E.I., S.M., S.S, S.M., M.J., K.O., and S.N.; analysis, E.I., S.M., K.O., S.N., and H.N.; writing—original draft preparation, K.O., S.N., and H.N.; writing—review and editing, H.N.; and funding acquisition, H.N. All authors have read and agreed to the published version of the manuscript.
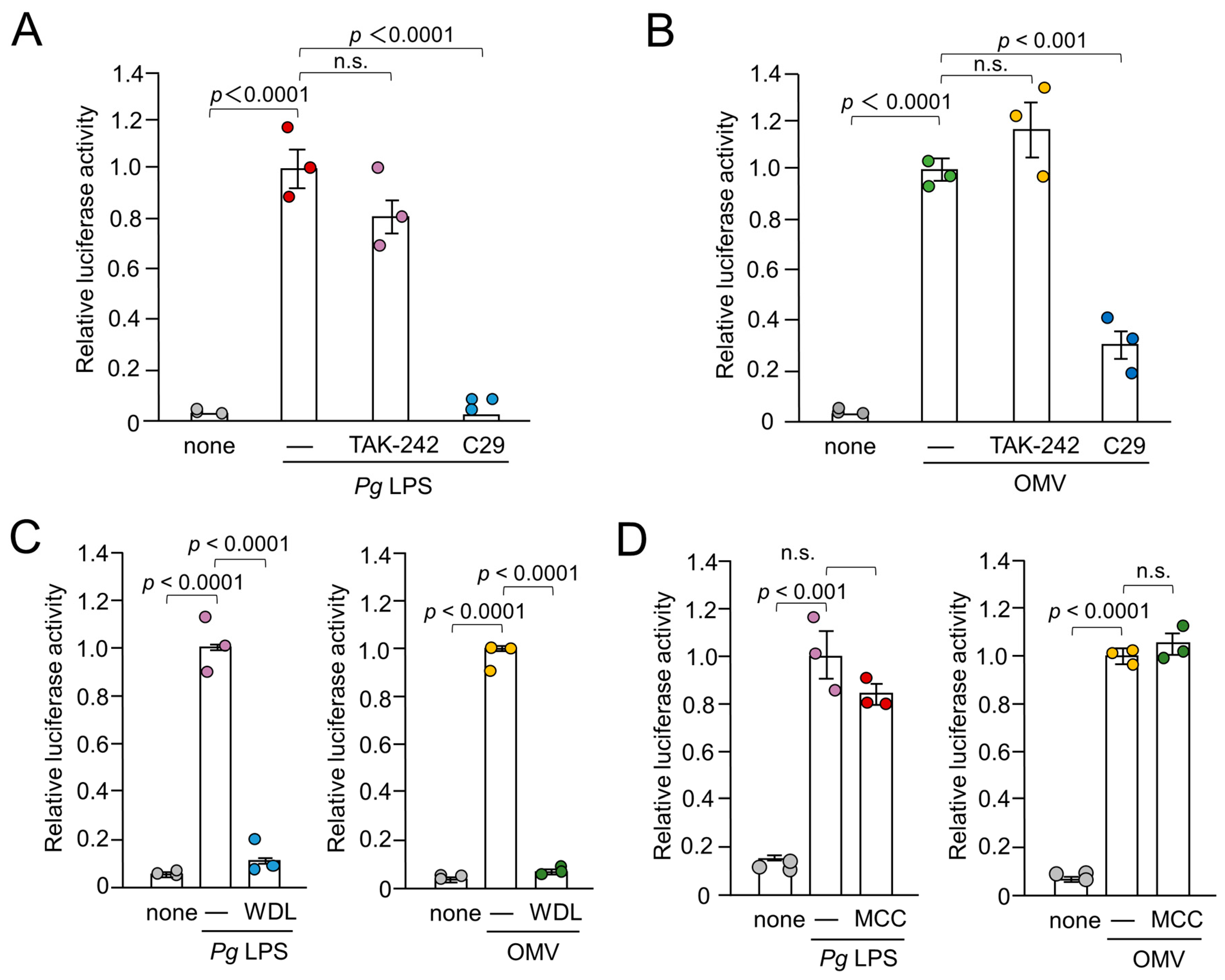
Figure 1.
Effects of inhibitors for TLR4, TLR2, IκB kinase and NRP3 inflammasome on the luciferase activity of the IL-1β probe in BV-2 microglia following stimulation with Pg LPS and OMVs for 1 h. (A), (B) The mean relative luciferase activity of the IL-1β probe induced by PgLPS (A) and OMVs (B) in the absence or presence of the TLR4 inhibitor TAK-242 or the TLR2 inhibitor C29. (C) The mean relative luciferase activity of the IL-1β probe induced by Pg LPS and OMVs in the absence or presence of the IκB kinase inhibitor wedelolactone (WDL). (D). The mean relative luciferase activity of the IL-1β probe induced by Pg LPS and OMVs in the absence or presence of the NLRP3 inflammasome inhibitor MCC950 (MCC). The data relative to the values in Pg LPS or OMV-treated cells are presented as the mean ± SE of three independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p < 0.05 was considered to indicate statistical significance.
Figure 1.
Effects of inhibitors for TLR4, TLR2, IκB kinase and NRP3 inflammasome on the luciferase activity of the IL-1β probe in BV-2 microglia following stimulation with Pg LPS and OMVs for 1 h. (A), (B) The mean relative luciferase activity of the IL-1β probe induced by PgLPS (A) and OMVs (B) in the absence or presence of the TLR4 inhibitor TAK-242 or the TLR2 inhibitor C29. (C) The mean relative luciferase activity of the IL-1β probe induced by Pg LPS and OMVs in the absence or presence of the IκB kinase inhibitor wedelolactone (WDL). (D). The mean relative luciferase activity of the IL-1β probe induced by Pg LPS and OMVs in the absence or presence of the NLRP3 inflammasome inhibitor MCC950 (MCC). The data relative to the values in Pg LPS or OMV-treated cells are presented as the mean ± SE of three independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p < 0.05 was considered to indicate statistical significance.
Figure 2.
Phagocytosis of OMVs by BV-2 microglia and possible involvement of gingipains in OMV-induced IL-1β production. (A) CLSM images of BV-2 microglia after treatment with Cy5-labelled OMVs for 1 h prepared from wild-type strain (WT OMV). F-actin and nuclei were visualized with Acti-stain 488 phalloidin (green) and Hoechst 33342 (blue), respectively. Bottom and right rectangular panels represent z-stack images. Scale bar = 20 μm. (B) The mean relative luciferase activity of the IL-1β probe induced by OMVs for 1 h after pharmacological and genetic inhibition of gingipains. KYT-1: Rgp inhibitor; KYT-136: Kgp inhibitor; KDP129: OMVs prepared from Kgp mutant strain; KDP136: OMVs prepared from Rgp and Kgp mutant strain. The data relative to the values in WT OMV-treated cells are presented as the mean ± SE of three-six independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance. (C) CLSM images of BV-2 microglia after treatment with Cy5-labelled OMVs (red) prepared from wild-type (WT OMV) and gingipain-null mutant KDP136 strains for 1 h. F-actin and nuclei were visualized with Acti-stain 488 phalloidin (green) and Hoechst 33342 (blue), respectively. Scale bar = 20 μm.
Figure 2.
Phagocytosis of OMVs by BV-2 microglia and possible involvement of gingipains in OMV-induced IL-1β production. (A) CLSM images of BV-2 microglia after treatment with Cy5-labelled OMVs for 1 h prepared from wild-type strain (WT OMV). F-actin and nuclei were visualized with Acti-stain 488 phalloidin (green) and Hoechst 33342 (blue), respectively. Bottom and right rectangular panels represent z-stack images. Scale bar = 20 μm. (B) The mean relative luciferase activity of the IL-1β probe induced by OMVs for 1 h after pharmacological and genetic inhibition of gingipains. KYT-1: Rgp inhibitor; KYT-136: Kgp inhibitor; KDP129: OMVs prepared from Kgp mutant strain; KDP136: OMVs prepared from Rgp and Kgp mutant strain. The data relative to the values in WT OMV-treated cells are presented as the mean ± SE of three-six independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance. (C) CLSM images of BV-2 microglia after treatment with Cy5-labelled OMVs (red) prepared from wild-type (WT OMV) and gingipain-null mutant KDP136 strains for 1 h. F-actin and nuclei were visualized with Acti-stain 488 phalloidin (green) and Hoechst 33342 (blue), respectively. Scale bar = 20 μm.
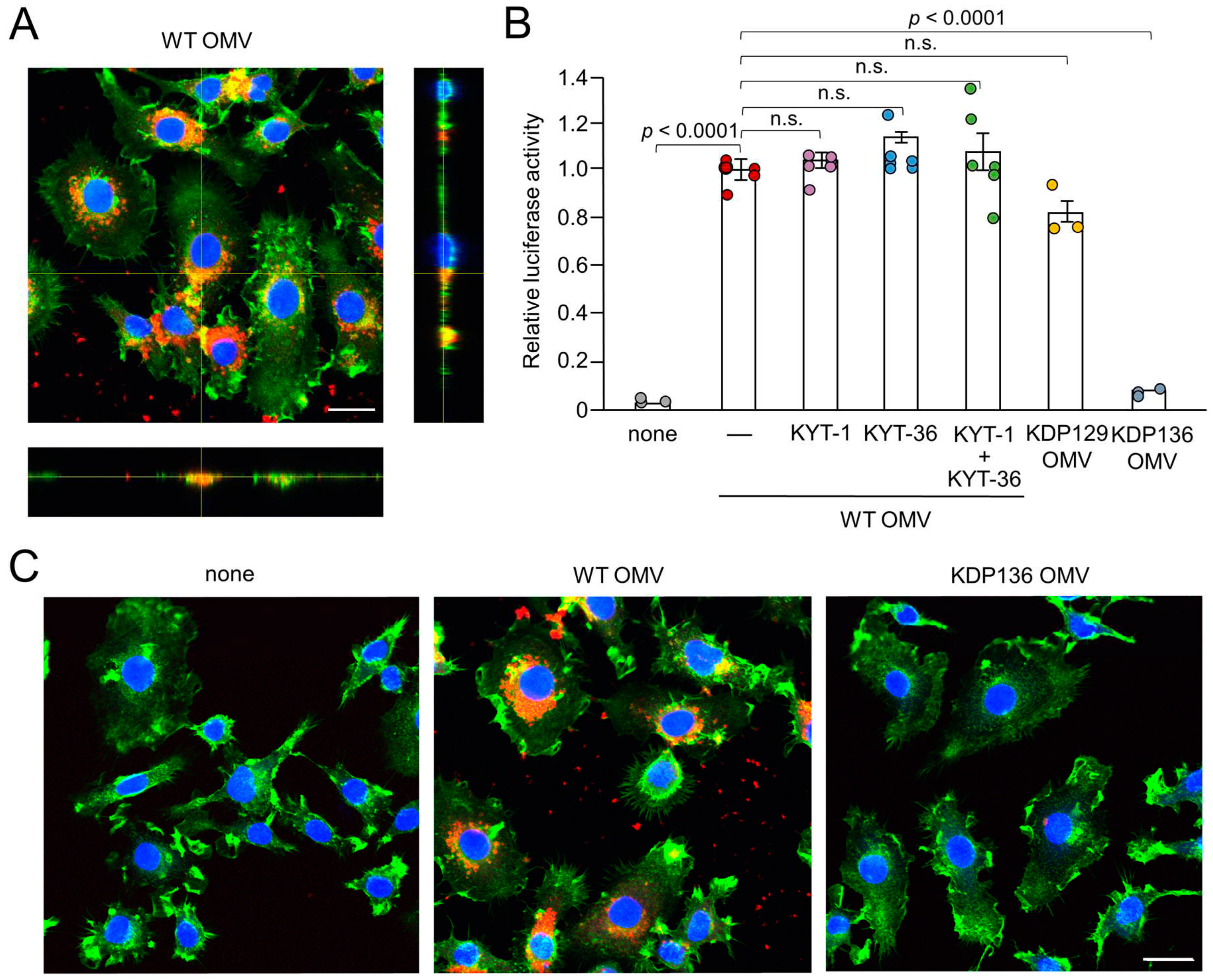
Figure 3.
Effects of pharmacological and genetic inhibition of CatB on IL-1β production by BV-2 microglia following stimulation with Pg LPS and OMVs. (A), (B) The mean relative luciferase activity of the IL-1β probe induced in BV-2 microglia following treatment with Pg LPS (A) and OMVs (B) after treatment with hBD3 (1 μM), CA-074Me (10 μM) or ZRLR (10 μM). The data are presented as the mean ± SE of 3-6 independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p < 0.05 was considered to indicate statistical significance. (C) The mean values of CatB intensity, which were detected by the immunoblot shown, were measured in Nluc reporter BV-2 microglia (Nluc BV-2) and Nluc reporter CatB-knockdown BV-2 microglia (CatB-KD Nluc BV-2) and normalized against the signal of GAPDH. The data are presented as the mean ± SE of three independent experiments, and the p value was calculated using Student’s t-test. (D) The mean luciferase activity (RLU) of the IL-1β probe induced by Pg LPS or OMV in CatB-KD Nuc BV-2 microglia. The data are presented as the mean ± SE of 3 independent experiments.
Figure 3.
Effects of pharmacological and genetic inhibition of CatB on IL-1β production by BV-2 microglia following stimulation with Pg LPS and OMVs. (A), (B) The mean relative luciferase activity of the IL-1β probe induced in BV-2 microglia following treatment with Pg LPS (A) and OMVs (B) after treatment with hBD3 (1 μM), CA-074Me (10 μM) or ZRLR (10 μM). The data are presented as the mean ± SE of 3-6 independent experiments, and p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p < 0.05 was considered to indicate statistical significance. (C) The mean values of CatB intensity, which were detected by the immunoblot shown, were measured in Nluc reporter BV-2 microglia (Nluc BV-2) and Nluc reporter CatB-knockdown BV-2 microglia (CatB-KD Nluc BV-2) and normalized against the signal of GAPDH. The data are presented as the mean ± SE of three independent experiments, and the p value was calculated using Student’s t-test. (D) The mean luciferase activity (RLU) of the IL-1β probe induced by Pg LPS or OMV in CatB-KD Nuc BV-2 microglia. The data are presented as the mean ± SE of 3 independent experiments.
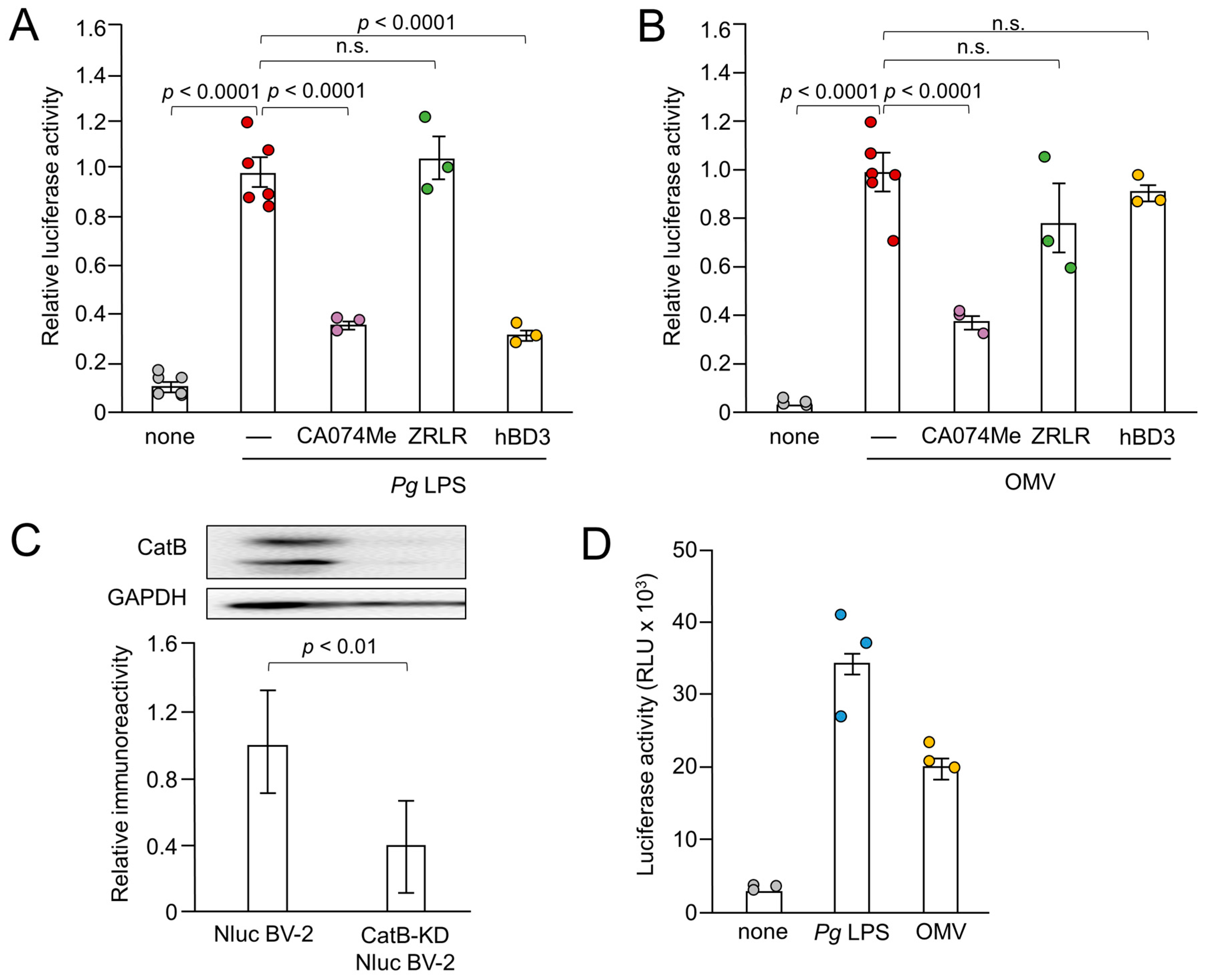
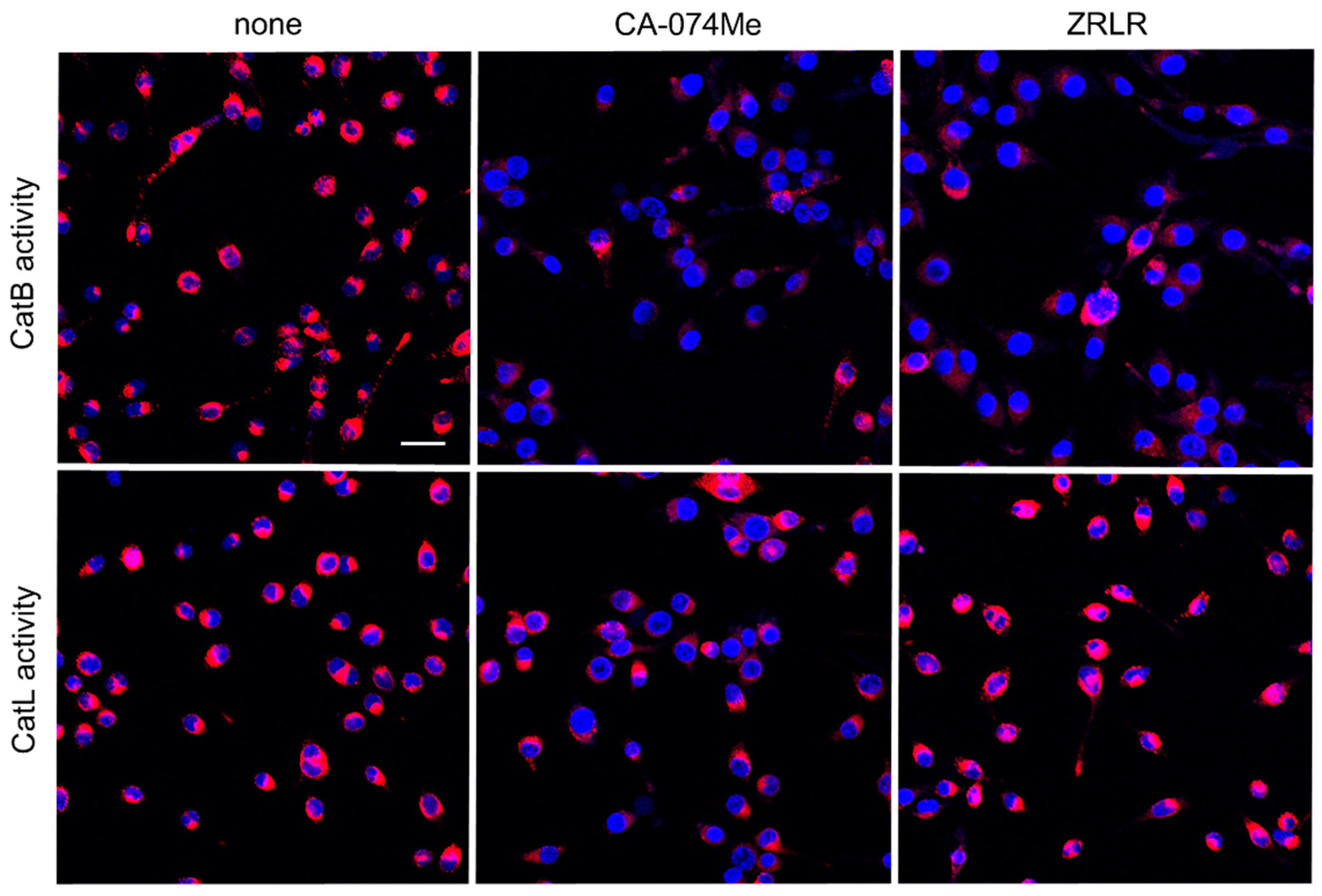
Figure 4.
Enzymatic activities of CatB and CatL visualized using the cell-permeable, fluorescently labeled substrates, z-Arg-Arg-cresyl violet and z-Phe-Arg-cresyl violet, respectively, in the absence (none) and presence of CA-074Me (10 μM) or ZRLR (10 μM). Scale bar = 40 μm.
Figure 4.
Enzymatic activities of CatB and CatL visualized using the cell-permeable, fluorescently labeled substrates, z-Arg-Arg-cresyl violet and z-Phe-Arg-cresyl violet, respectively, in the absence (none) and presence of CA-074Me (10 μM) or ZRLR (10 μM). Scale bar = 40 μm.
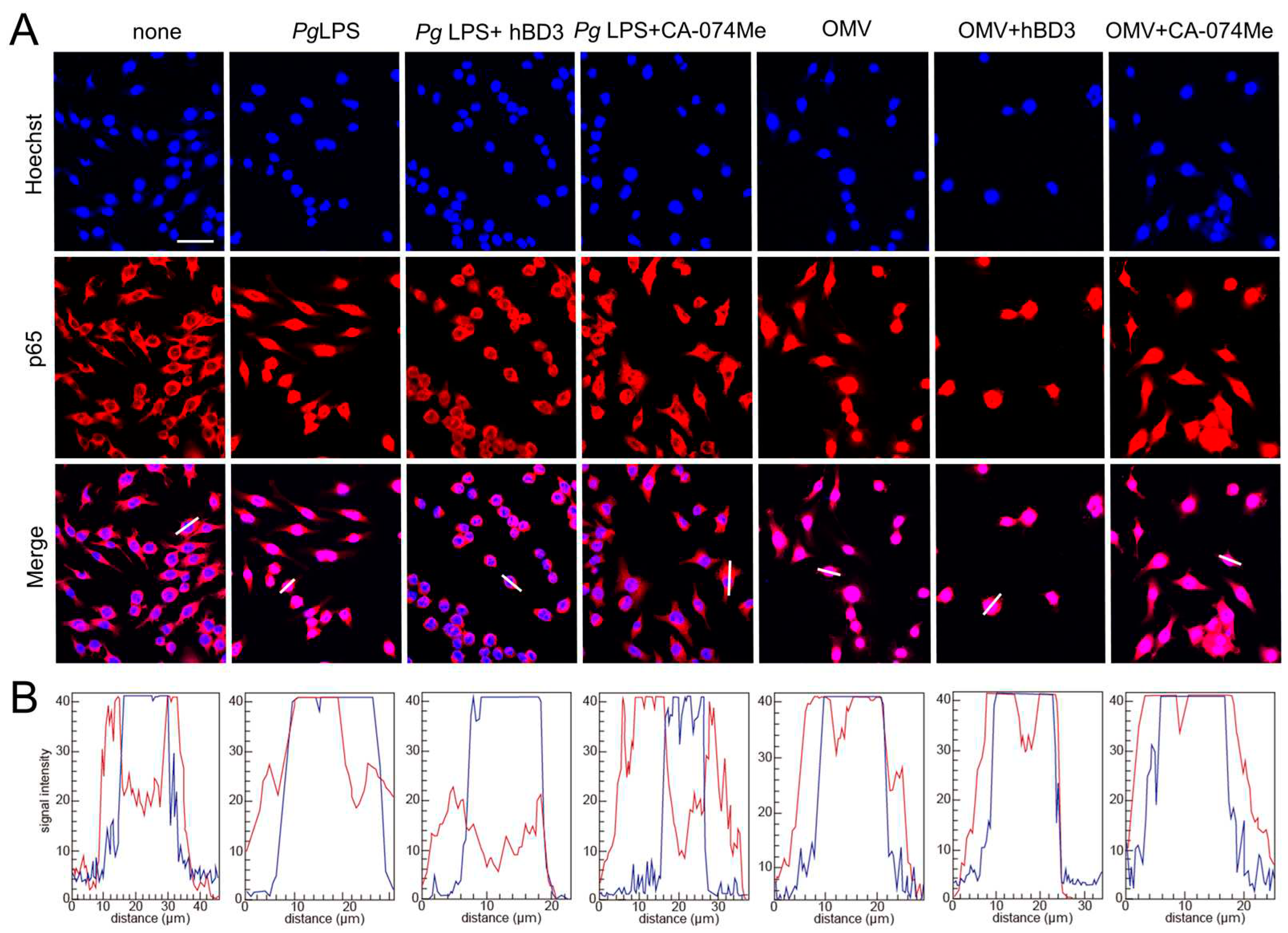
Figure 5.
Effects of hBD3 and CA-074Me on nuclear NF-κB p65 translocation following stimulation with Pg LPS and OMVs. (A) Immunofluorescent CLSM images of BV-2 microglia after treatment with Pg LPS or OMVs in the absence or presence of hBD3 (1 μM) or CA-074Me (10 μM). Nuclear NF-κB p65 translocation was visualized by immunohistochemical staining (red). Nuclei were stained blue by Hoechst 33342 (blue). Scale bar = 40 μm. (B) The typical cells were analyzed by line plot profile to show the cytosol and nuclear NF-κB p65 translocation. indicate The fluorescence intensity of NF-κB p65 and Hoechst 33342 in the cells traversed by white lines in (A) was indicated by red and blue lines, respectively.
Figure 5.
Effects of hBD3 and CA-074Me on nuclear NF-κB p65 translocation following stimulation with Pg LPS and OMVs. (A) Immunofluorescent CLSM images of BV-2 microglia after treatment with Pg LPS or OMVs in the absence or presence of hBD3 (1 μM) or CA-074Me (10 μM). Nuclear NF-κB p65 translocation was visualized by immunohistochemical staining (red). Nuclei were stained blue by Hoechst 33342 (blue). Scale bar = 40 μm. (B) The typical cells were analyzed by line plot profile to show the cytosol and nuclear NF-κB p65 translocation. indicate The fluorescence intensity of NF-κB p65 and Hoechst 33342 in the cells traversed by white lines in (A) was indicated by red and blue lines, respectively.
Figure 6.
Effects of hBD3 and CA-074Me on the degradation of IκBα following treatment with Pg LPS and OMVs. (A) The protein expression of IκBα in BV-2 microglia after stimulation with Pg LPS (10 μg/mL) for 30 min in the presence or absence of hBD3 (1 μM) or CA-074Me (30 μM). (B) The mean values of the IκBα intensity, which were detected by the immunoblots shown in (A), were measured and normalized against the signal of β-actin. The data relative to the values in un treated cells are presented as the mean ± SE of three-five independent experiments. The p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance. (C) The protein expression of IκBα in BV-2 microglia after stimulation with OMVs (150 μg/mL) for 10 min in the presence or absence of hBD3 (1 μM) or CA-074Me (30 μM). (D) The mean values of the IκBα intensity, which were detected by the immunoblots shown in (C), were measured and normalized against the signal of β-actin. The data relative to the values in untreated cells are presented as the mean ± SE of three independent experiments. The p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance.
Figure 6.
Effects of hBD3 and CA-074Me on the degradation of IκBα following treatment with Pg LPS and OMVs. (A) The protein expression of IκBα in BV-2 microglia after stimulation with Pg LPS (10 μg/mL) for 30 min in the presence or absence of hBD3 (1 μM) or CA-074Me (30 μM). (B) The mean values of the IκBα intensity, which were detected by the immunoblots shown in (A), were measured and normalized against the signal of β-actin. The data relative to the values in un treated cells are presented as the mean ± SE of three-five independent experiments. The p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance. (C) The protein expression of IκBα in BV-2 microglia after stimulation with OMVs (150 μg/mL) for 10 min in the presence or absence of hBD3 (1 μM) or CA-074Me (30 μM). (D) The mean values of the IκBα intensity, which were detected by the immunoblots shown in (C), were measured and normalized against the signal of β-actin. The data relative to the values in untreated cells are presented as the mean ± SE of three independent experiments. The p values were calculated using a one-way ANOVA with a post-hoc Tukey’s test. A value of p<0.05 was considered to indicate statistical significance.
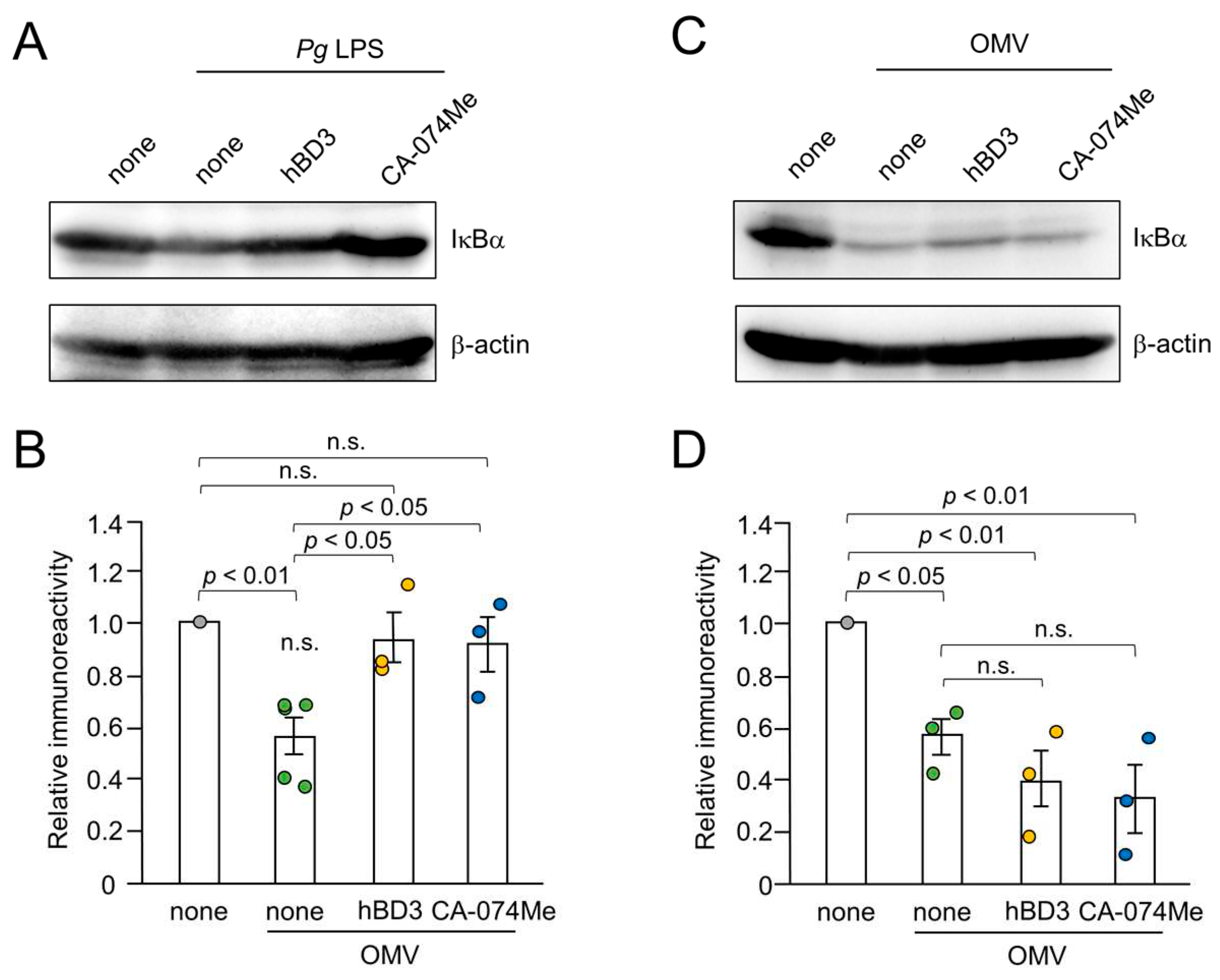
Figure 7.
Prediction of hBD3 binding to CatB. (
A) Structural model of hBD3-bound CatB generated using AlphaFold. The hBD3 model, presented as a ribbon, binds to the molecular surface of CatB. Amino acids are colored based on their pLDDT score. (
B) PAE plots of the hBD3 and CatB complex model. (
C) Binding of hBD3 to the active cleft formed on the molecular surface of CatB. The surface representation of CatB is shown in gray, except for the S1, S3, and S1’ sites, which are shown in light magenta; the S2 and S2’ sites, which are shown in magenta; and Cys29, which is shown in yellow. The hBD3 model, presented as a ribbon, is colored cyan. The figures were drawn using the PyMOL software program [
50].
Figure 7.
Prediction of hBD3 binding to CatB. (
A) Structural model of hBD3-bound CatB generated using AlphaFold. The hBD3 model, presented as a ribbon, binds to the molecular surface of CatB. Amino acids are colored based on their pLDDT score. (
B) PAE plots of the hBD3 and CatB complex model. (
C) Binding of hBD3 to the active cleft formed on the molecular surface of CatB. The surface representation of CatB is shown in gray, except for the S1, S3, and S1’ sites, which are shown in light magenta; the S2 and S2’ sites, which are shown in magenta; and Cys29, which is shown in yellow. The hBD3 model, presented as a ribbon, is colored cyan. The figures were drawn using the PyMOL software program [
50].
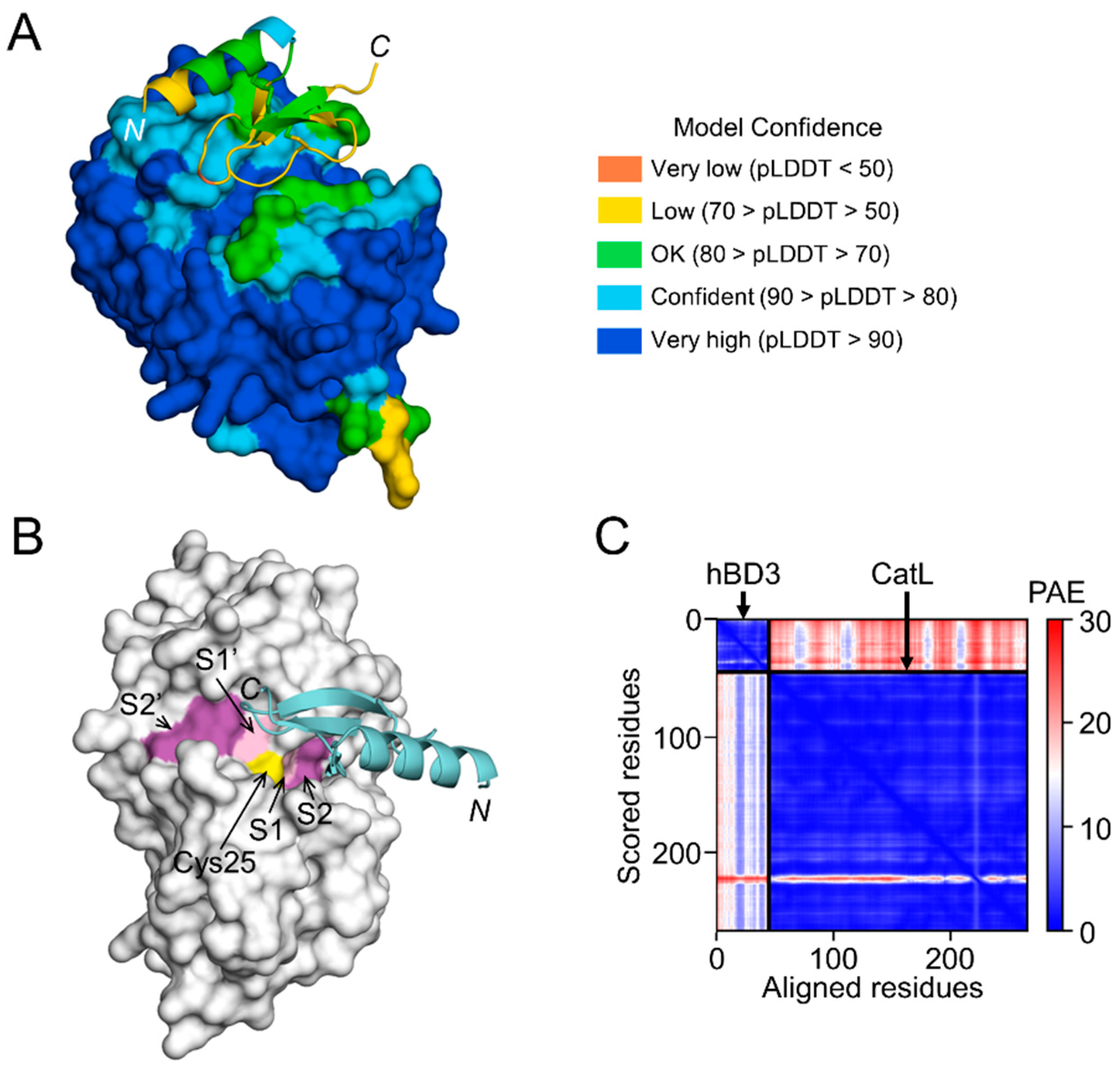
Figure 8.
Prediction of hBD3 binding to CatL. (
A) Structural model of hBD3-bound CatL generated using AlphaFold. hBD3 model, presented as a ribbon, binds to the molecular surface of CatL. Amino acids are colored based on their pLDDT score. (
B) Binding of hBD3 to the active cleft formed on the molecular surface of CatL. The surface representation of CatL is shown in gray, except for S1 and S1’sites, which are shown in light magenta; the S2 and S2’ sites, which are shown in magenta; and Cys25, which is shown in yellow. The hBD3 model, presented as a ribbon, is colored cyan. (
C) PAE plots of the hBD3 and CatL complex model. The figures were drawn using the PyMOL software program [
50].
Figure 8.
Prediction of hBD3 binding to CatL. (
A) Structural model of hBD3-bound CatL generated using AlphaFold. hBD3 model, presented as a ribbon, binds to the molecular surface of CatL. Amino acids are colored based on their pLDDT score. (
B) Binding of hBD3 to the active cleft formed on the molecular surface of CatL. The surface representation of CatL is shown in gray, except for S1 and S1’sites, which are shown in light magenta; the S2 and S2’ sites, which are shown in magenta; and Cys25, which is shown in yellow. The hBD3 model, presented as a ribbon, is colored cyan. (
C) PAE plots of the hBD3 and CatL complex model. The figures were drawn using the PyMOL software program [
50].