Submitted:
26 December 2023
Posted:
27 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
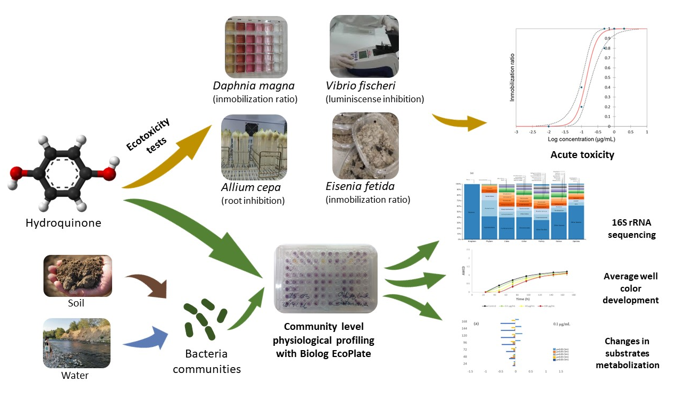
2. Materials and Methods
2.1. Reagents
2.2. Daphnia magna assay
2.3. Vibrio fischeri assay
2.4. Allium cepa assay
2.5. Eisenia fetida assay
2.6. River and soil microorganisms community assay
2.6.1 River samples
2.6.2. Soil samples
2.6.3. Genetic sequencing of river and soil microorganisms
2.6.4. Community-level physiological profiling (CLPP) of river and soil microorganisms
2.7. Statistics and graphic representation
3. Results and Discussion
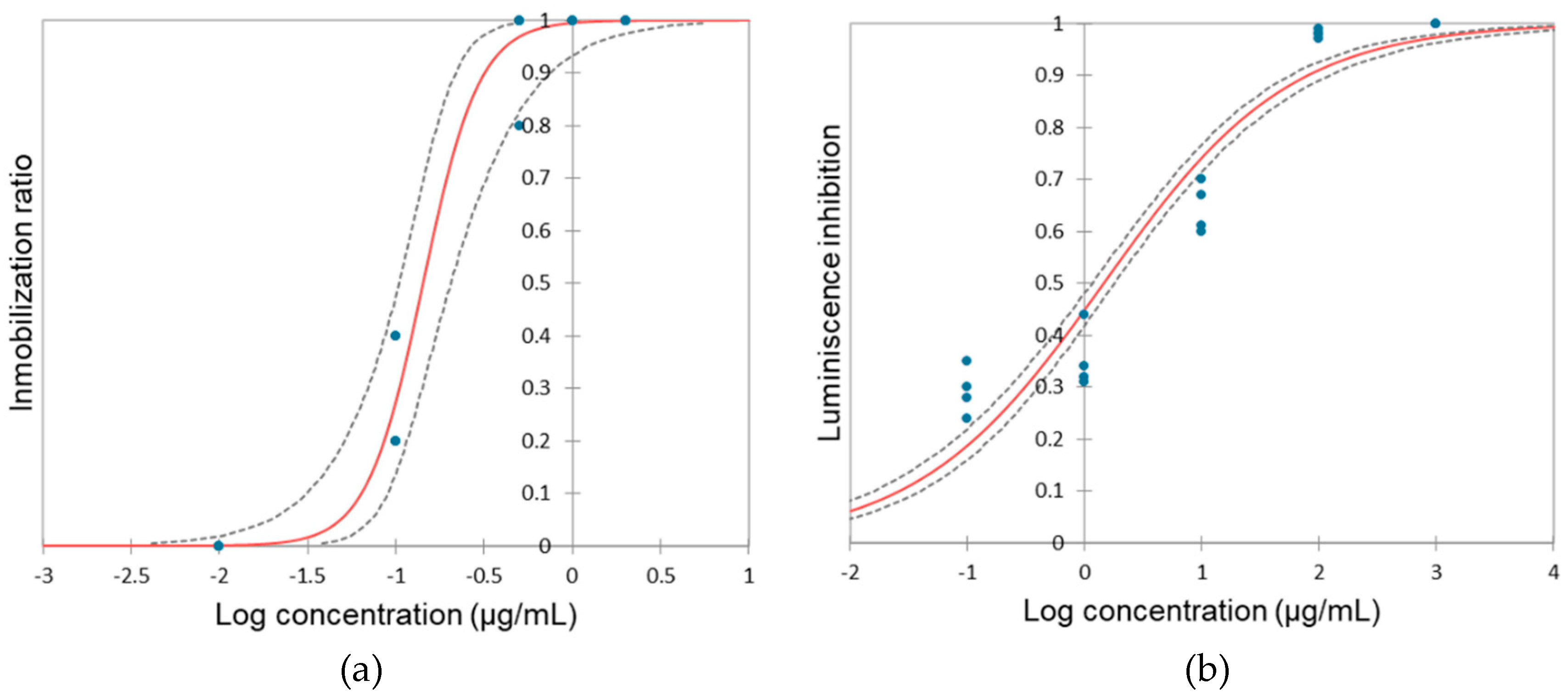
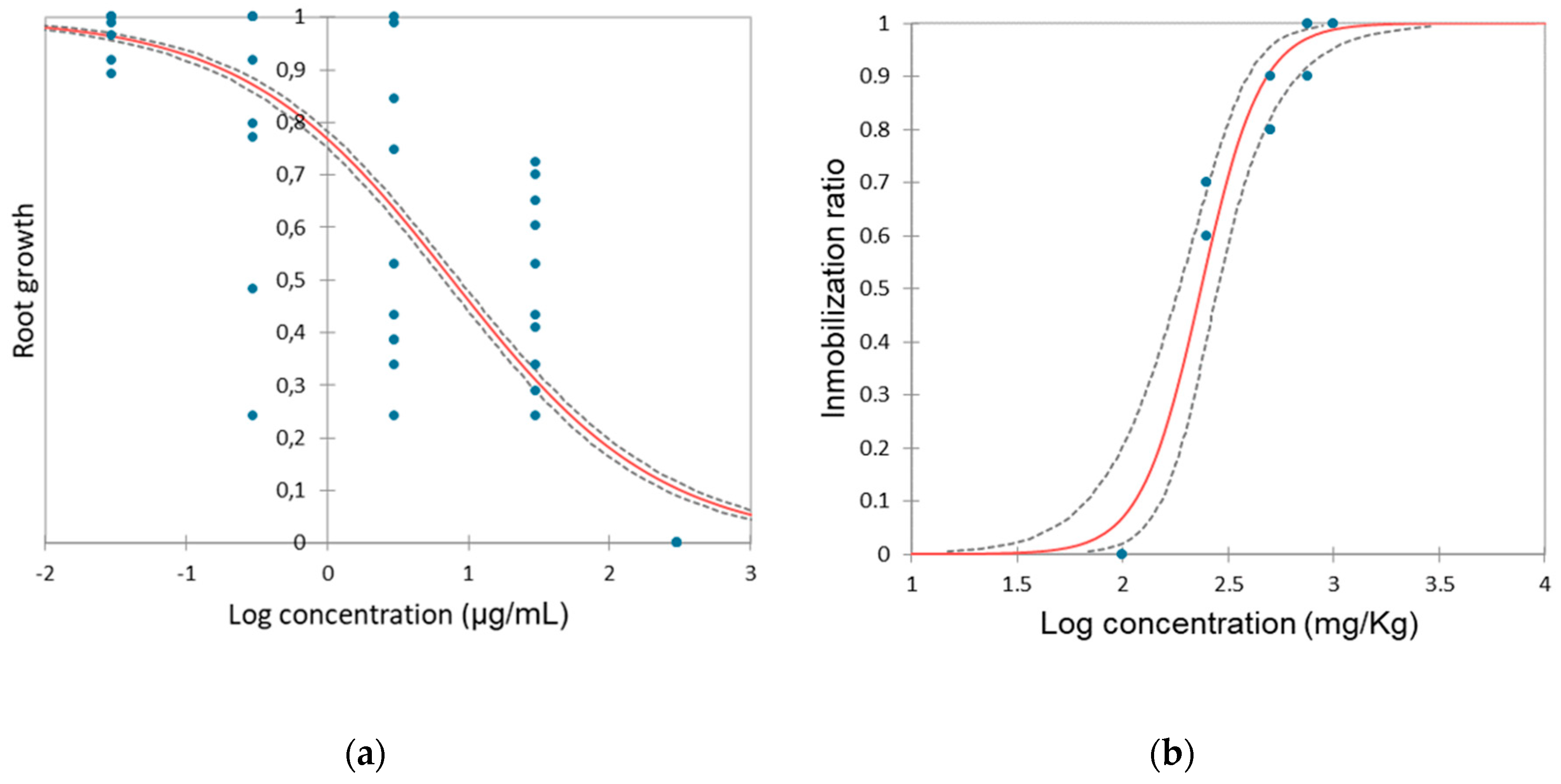
3.1 Impact of hydroquinone on Daphnia magna
3.2. Impact of hydroquinone on V. fisheri
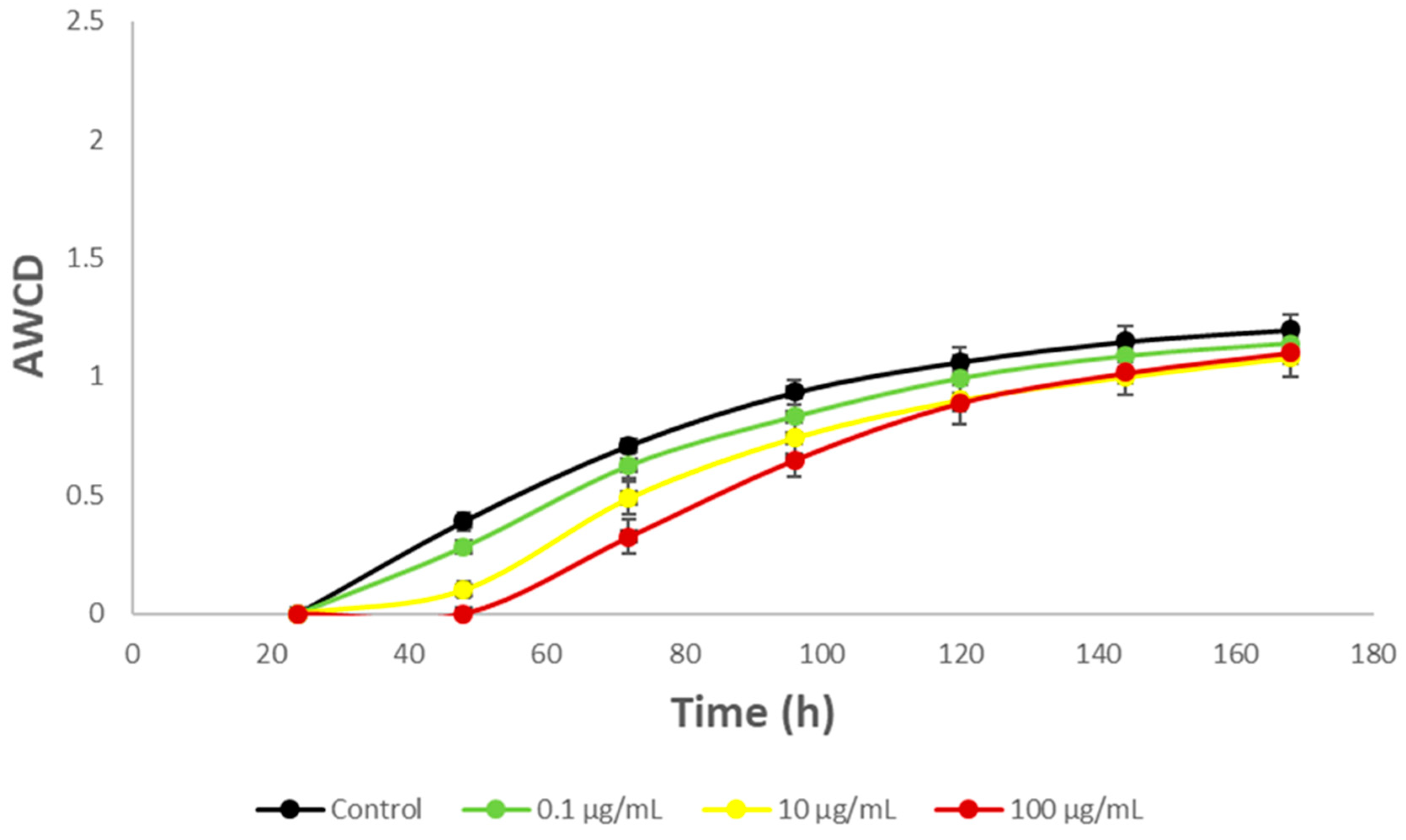
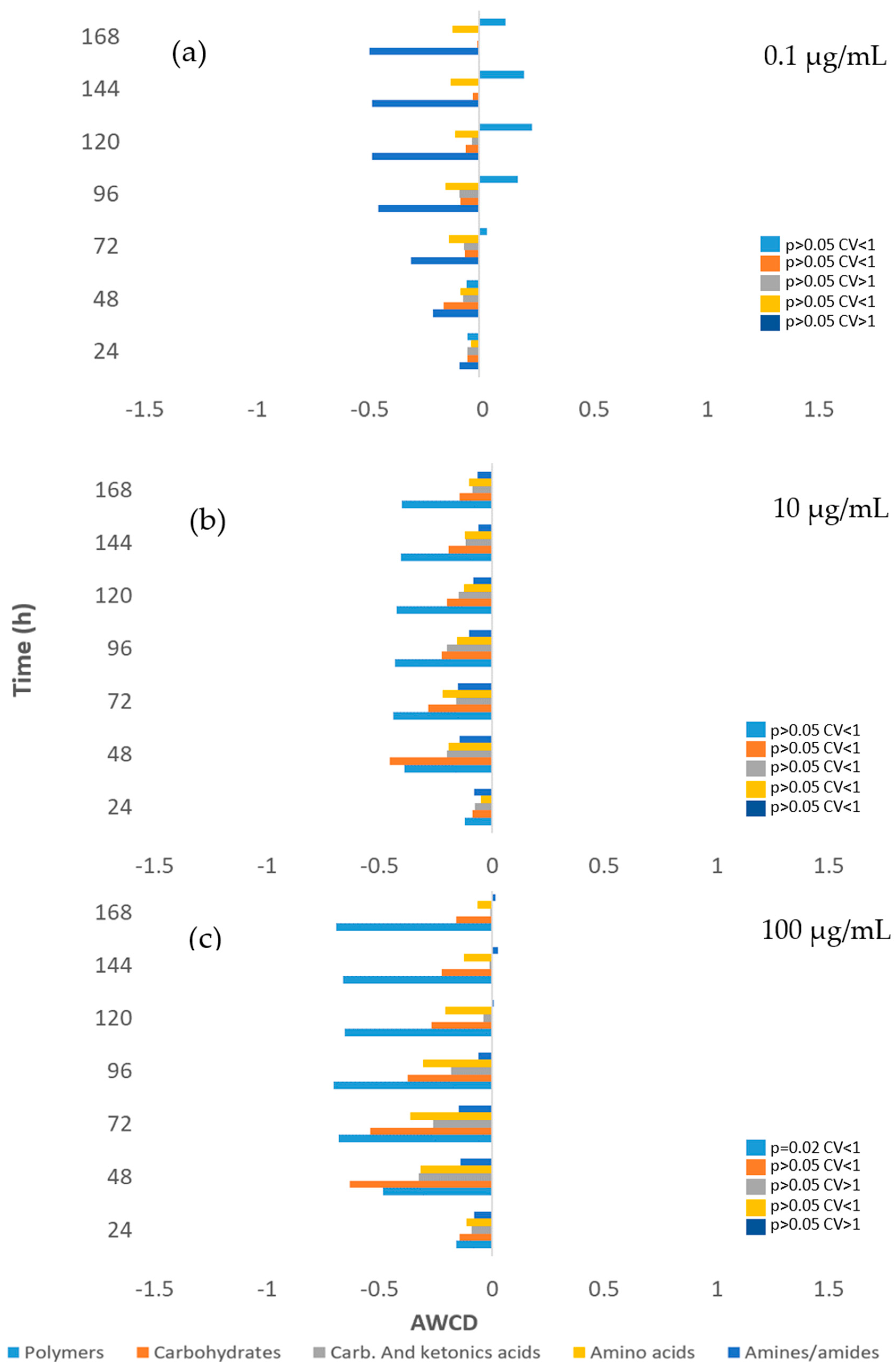
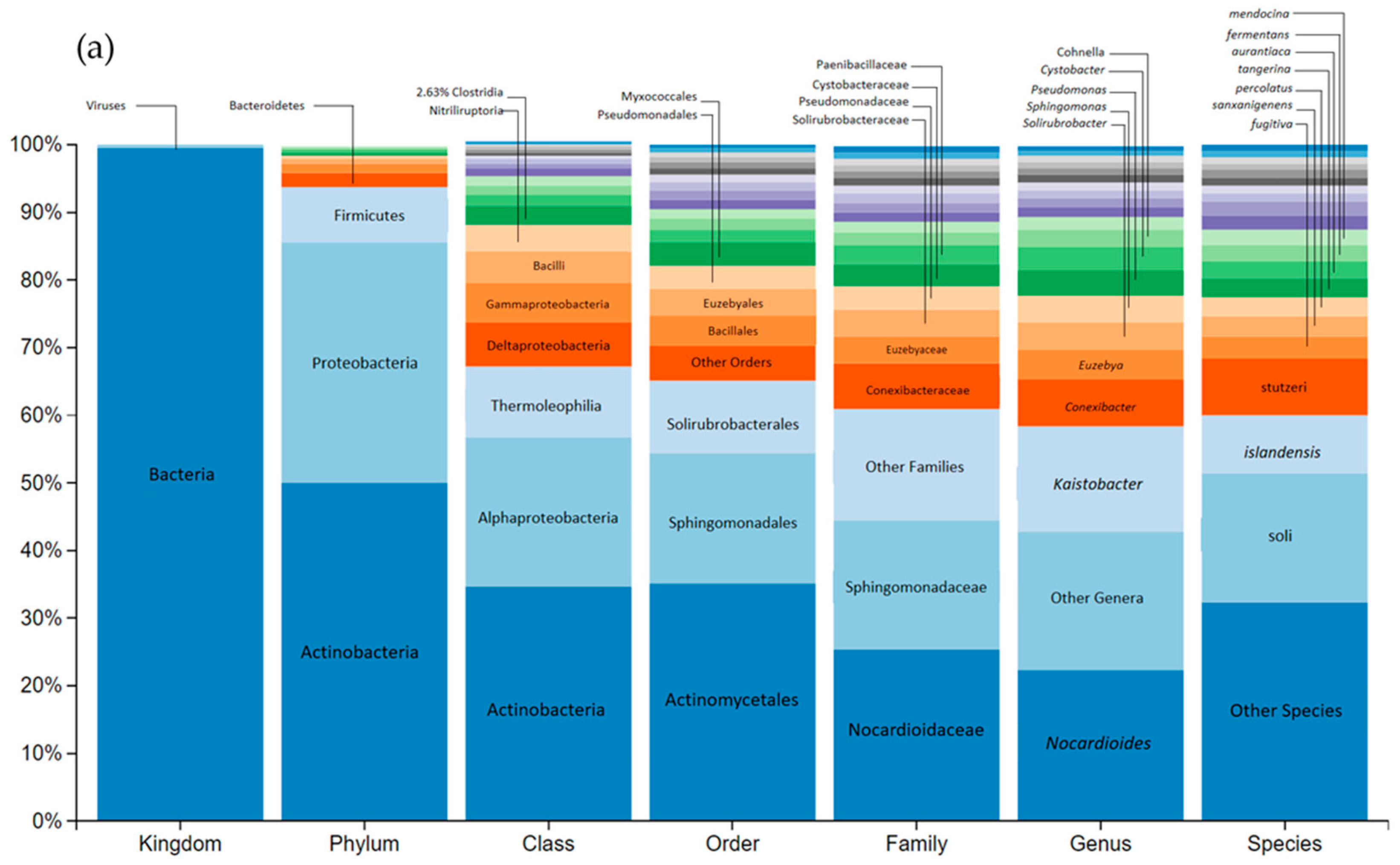
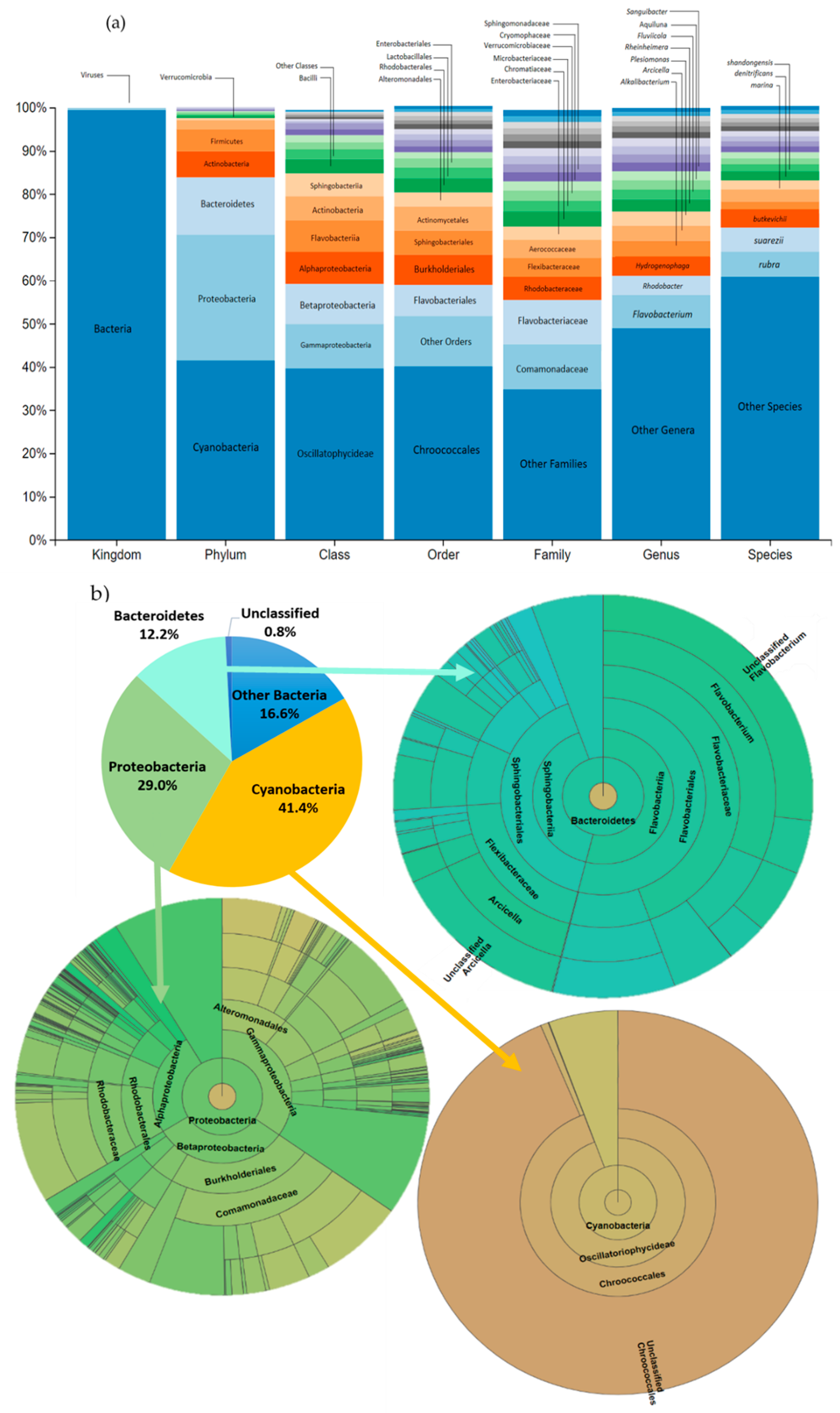
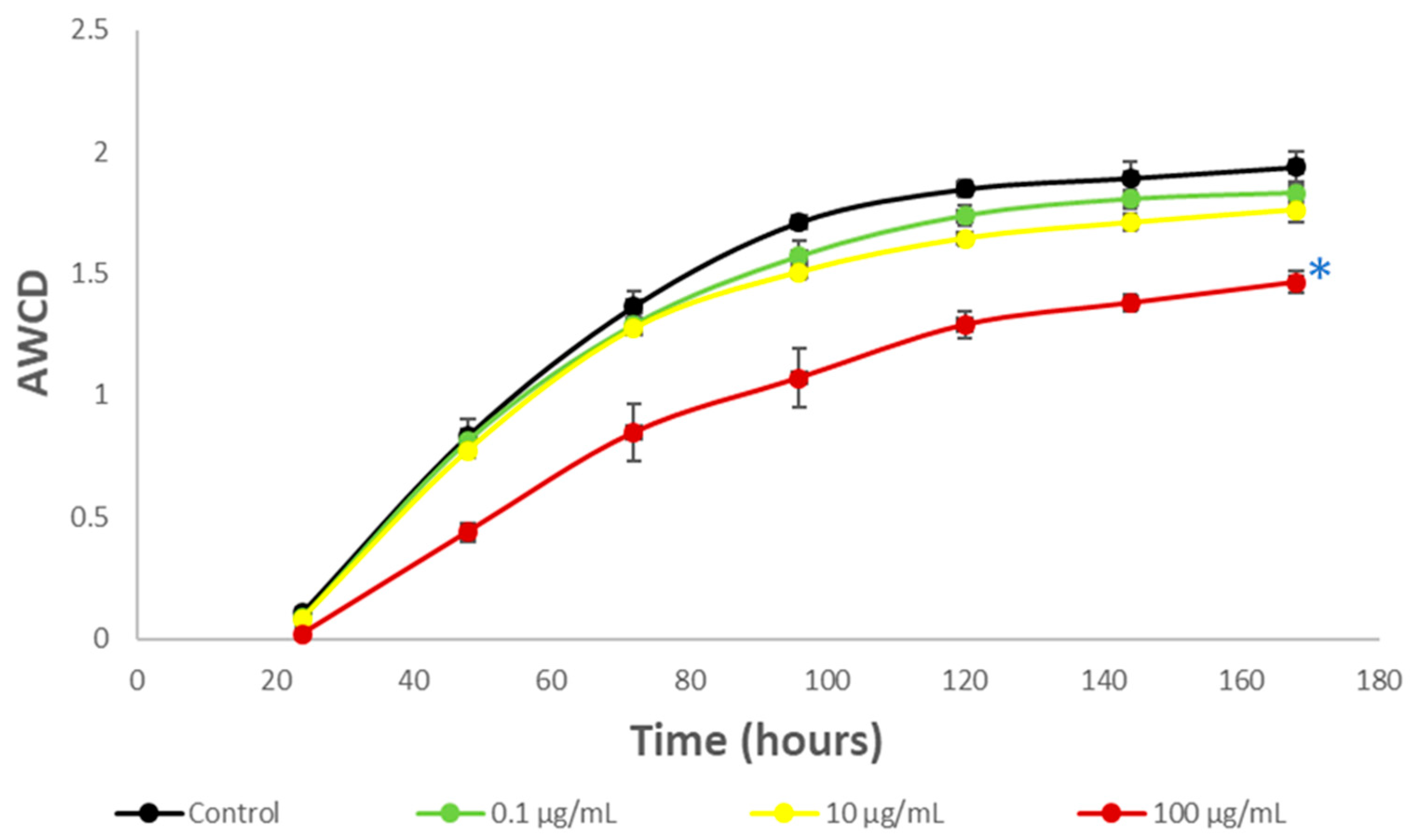
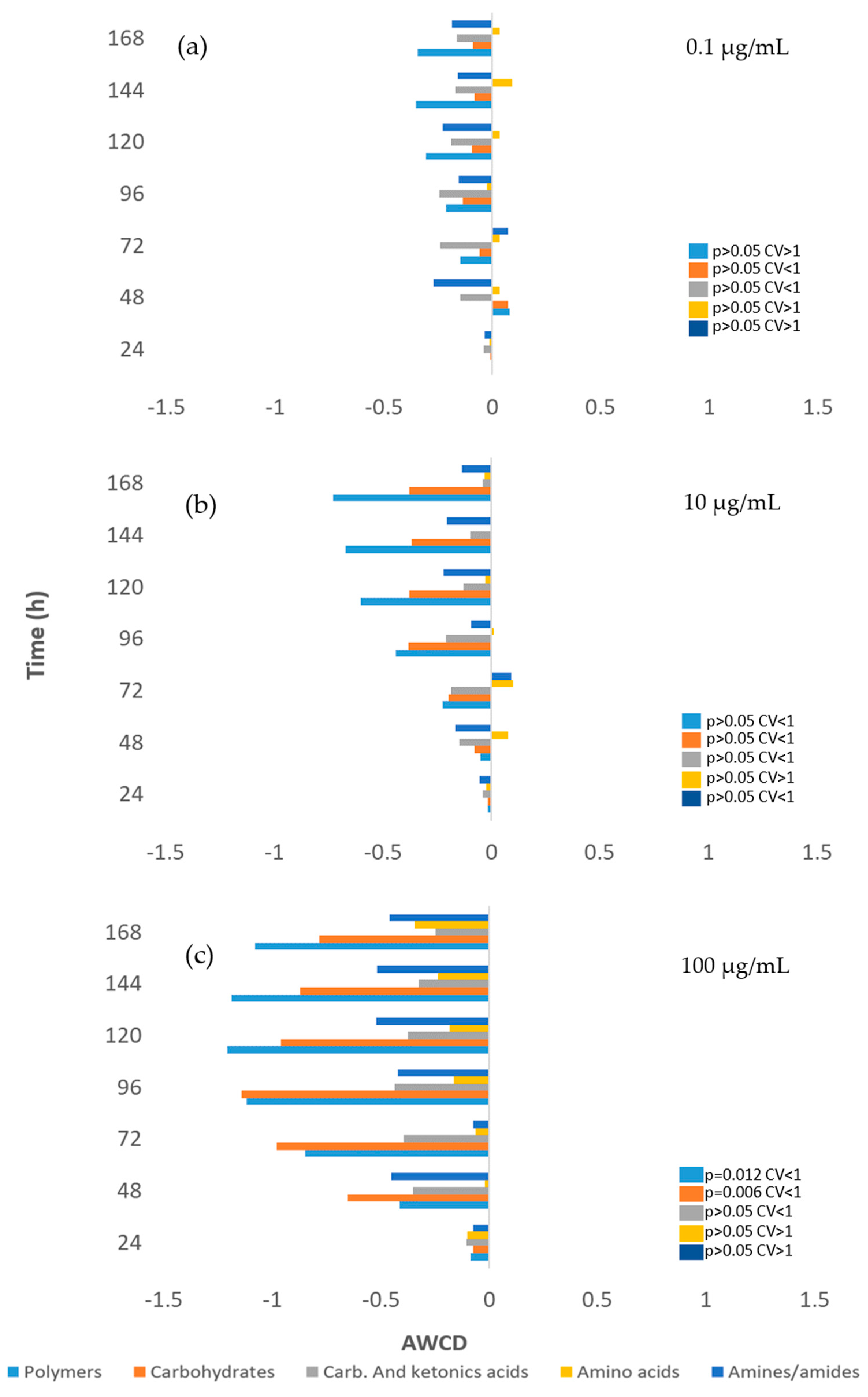
3.3. Impact on river microbial communities: growth and Community-level physiological profiling (CLPP)
3.4. Impact of hydroquinone on Allium cepa
3.5. Impact of Hydroquinone on Eisenia fetida
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix
| Physical-chemical analysis of the river water sample | |
| HCO-3 (mg/L) | 313 |
| TDS (mg/L) | 1925 |
| MES (mg/L) | 6 |
| Cl- (mg/L) | 618±93 |
| SO42- (mg/L) | 415±62 |
| NO3- (mg/L) | 17.7±2.7 |
| NO2- (mg/L) | <0.05 |
| F- (mg/L) | 0.071±0.011 |
| PO43-(mg/L) | 0.6±0.09 |
| NH4+(mg/L) | <0.1 |
| O2 (mg/L) | 2.3 |
| DQO (O2) (mg/L) | <25 |
| DBO5 (mg/L) | <5 |
| Ca (mg/L) | 235±80 |
| Mg (mg/L) | 38.1±13.7 |
| Na (mg/L) | 415±95 |
| K (mg/L) | 6.08±1.95 |
| Soil composition | Surface soil | 30cm deep soil |
| Clay content (%) | 20.98 | 23.61 |
| Sand content (%) | 16.08 | 13.10 |
| Silt content (%) | 62.94 | 63.29 |
| pH | 7.9±0.5 | 8.1±0.5 |
| K (mg/L) | 238±40 | 208±35 |
| Mg (mg/Kg) | 244±39 | 242±39 |
| P Oslen (mg/Kg) | 13±2 | 10±1.7 |
| EC1:5 (dS/m) | 0.6±0.09 | 0.4±0.06 |
| Organic matter (g/100g) | 2.46±0.31 | 2.35±0.30 |
References
- Pandey, D.K.; Mishra, N.; Singh, P. Relative phytotoxicity of hydroquinone on rice (Oryza sativa L.) and associated aquatic weed green musk chary (Chary zeylanica Willd.). Pesticide Biochemistry and Physiology 2005, 83, 82-96. [CrossRef]
- Remberger, M.; Hynning, P.A.; Neilson, A.H. 2,5-dichloro-3,6-dihydroxybenzo-1,4-quinone - identification of a new organochlorine compound in kraft mill bleachery effluents. Environmental Science & Technology 1991, 25, 1903-1907. [CrossRef]
- Neven, L.; Barich, H.; Rutten, R.; De Wael, K. Novel (photo)electrochemical analysis of aqueous industrial samples containing phenols. Microchemical Journal 2022, 181. [CrossRef]
- Choi, Y.; Jeon, J.; Kim, S.D. Identification of biotransformation products of organophosphate ester from various aquatic species by suspect and non-target screening approach. Water Research 2021, 200. [CrossRef]
- Marchlewicz, A.; Guzik, U.; Wojcieszynska, D. Over-the-Counter Monocyclic Non-Steroidal Anti-Inflammatory Drugs in Environment-Sources, Risks, Biodegradation. Water Air and Soil Pollution 2015, 226. [CrossRef]
- Zur, J.; Wojcieszynska, D.; Hupert-Kocurek, K.; Marchlewicz, A.; Guzik, U. Paracetamol - toxicity and microbial utilization. Pseudomonas moorei KB4 as a case study for exploring degradation pathway. Chemosphere 2018, 206, 192-202. [CrossRef]
- Wang, W.; Yu, H.; Qin, H.; Long, Y.; Ye, J.; Qu, Y. Bisphenol A degradation pathway and associated metabolic networks in Escherichia coli harboring the gene encoding CYP450. Journal of Hazardous Materials 2020, 388. [CrossRef]
- Bolobajev, J.; Oncu, N.B.; Viisimaa, M.; Trapido, M.; Balcioglu, I.; Goi, A. Column experiment on activation aids and biosurfactant application to the persulphate treatment of chlorophene-contaminated soil. Environmental Technology 2015, 36, 348-357. [CrossRef]
- Hoang Nhat Phong, V.; Le, G.K.; Thi Minh Hong, N.; Xuan-Thanh, B.; Khanh Hoang, N.; Rene, E.R.; Thi Dieu Hien, V.; Ngoc-Dan Thanh, C.; Mohan, R. Acetaminophen micropollutant: Historical and current occurrences, toxicity, removal strategies and transformation pathways in different environments. Chemosphere 2019, 236. [CrossRef]
- Santos, A.; Yustos, P.; Quintanilla, A.; Garcia-Ochoa, F.; Casas, J.A.; Rodriguez, J.J. Evolution of toxicity upon wet catalytic oxidation of phenol. Environmental Science & Technology 2004, 38, 133-138. [CrossRef]
- Wang, Y.; Li, X.; Sun, X. The transformation mechanism and eco-toxicity evaluation of butylated hydroxyanisole in environment. Ecotoxicology and Environmental Safety 2022, 231. [CrossRef]
- Di Marzio, W.D.; Saenz, M.; Alberdi, J.; Tortorelli, M.; Silvana, G. Risk assessment of domestic and industrial effluents unloaded into a freshwater environment. Ecotoxicology and Environmental Safety 2005, 61, 380-391. [CrossRef]
- Karami-Kolmoti, P.; Beitollahi, H.; Modiri, S. Electrochemical Sensor for Simple and Sensitive Determination of Hydroquinone in Water Samples Using Modified Glassy Carbon Electrode. Biomedicines 2023, 11. [CrossRef]
- Yang, L.; Zhao, H.; Fan, S.; Li, B.; Li, C.-P. A highly sensitive electrochemical sensor for simultaneous determination of hydroquinone and bisphenol A based on the ultrafine Pd nanoparticle@TiO2 functionalized SiC. Analytica Chimica Acta 2014, 852, 28-36. [CrossRef]
- Hernandez, S.R.; Kergaravat, S.V.; Isabel Pividori, M. Enzymatic electrochemical detection coupled to multivariate calibration for the determination of phenolic compounds in environmental samples. Talanta 2013, 106, 399-407. [CrossRef]
- Varela, A.; Martins, C.; Nunez, O.; Martins, I.; Houbraken, J.A.M.P.; Martins, T.M.; Leitao, M.C.; McLellan, I.; Vetter, W.; Galceran, M.T.; et al. Understanding fungal functional biodiversity during the mitigation of environmentally dispersed pentachlorophenol in cork oak forest soils. Environmental Microbiology 2015, 17, 2922-2934. [CrossRef]
- Lin, J.; Chen, J.; Wang, Y.; Cai, X.; Wei, X.; Qiao, X. More toxic and photoresistant products from photodegradation of fenoxaprop-p-ethyl. Journal of Agricultural and Food Chemistry 2008, 56, 8226-8230. [CrossRef]
- Candido, N.R.; Modolo, L.V.; Pasa, V.M.D.; de Fatima, A. Pyroligneous acids of coconut shell, black wattle and eucalyptus: physical-chemical characterization and in vitro evaluation as potential urease inhibitors. Quimica Nova 2023. [CrossRef]
- Baehrs, H.; Putschew, A.; Steinberg, C.E.W. Toxicity of hydroquinone to different freshwater phototrophs is influenced by time of exposure and pH. Environmental Science and Pollution Research 2013, 20, 146-154. [CrossRef]
- Briggs, G.G.; Henderson, I.F. some factors affecting the toxicity of poisons to the slug deroceras-reticulatum (muller) (Pulmonata, limacidae). Crop Protection 1987, 6, 341-346. [CrossRef]
- Lahnsteiner, F. The sensitivity and reproducibility of the zebrafish (Danio rerio) embryo test for the screening of waste water quality and for testing the toxicity of chemicals. Atla-Alternatives to Laboratory Animals 2008, 36, 299-311. [CrossRef]
- Abugazleh, M.K.; Ali, H.M.; Chester, J.A.; Al-Fa'ouri, A.M.; Bouldin, J.L. Aquatic toxicity of hydroquinone and catechol following metal oxide treatment to Ceriodaphnia dubia and Pimephales promelas. Ecotoxicology 2023, 32, 656-665. [CrossRef]
- Degraeve, G.M. Aquatic effects of underground coal gasification wastewater and important constituents; 1979.
- Stadnichenko, A.P.; Pogorelova, N.S.; Rudenko, S.A. The effect of different concentrations of hydroquinone on horn snails (Gastropoda, pulmonata, bulinidae) infected with parthenitae of tylodelphys-excavata (Trematoda, diplostomatidae). Parazitologiya 1991, 25, 462-467.
- Devillers, J.; Boule, P.; Vasseur, P.; Prevot, P.; Steiman, R.; Seiglemurandi, F.; Benoitguyod, J.L.; Nendza, M.; Grioni, C.; Dive, D.; et al. Environmental and health risks of hydroquinone. Ecotoxicology and Environmental Safety 1990, 19, 327-354. [CrossRef]
- Wang, X.D.; Yu, J.Z.; Wang, Y.; Wang, L.S. Mechanism-based quantitative structure-activity relationships for the inhibition of substituted phenols on germination rate of Cucumis sativus. Chemosphere 2002, 46, 241-250. [CrossRef]
- Zhang, X.; Linghu, S.; Chen, Z.; Gu, H.; Chen, X.; Wei, X.; Hu, X.; Yang, Y.; Gao, Y. Bacterial diversity evolution process based on physicochemical characteristics of sludge treating hydroquinone during acclimation. Environmental Science and Pollution Research 2022, 29, 31686-31699. [CrossRef]
- Chen, X.; Hu, X.; Lu, Q.; Yang, Y.; Linghu, S.; Zhang, X. Study on the differences in sludge toxicity and microbial community structure caused by catechol, resorcinol and hydroquinone with metagenomic analysis. Journal of Environmental Management 2022, 302. [CrossRef]
- Wang, W.; Wu, B.; Pan, S.; Yang, K.; Hu, Z.; Yuan, S. Performance robustness of the UASB reactors treating saline phenolic wastewater and analysis of microbial community structure. Journal of Hazardous Materials 2017, 331, 21-27. [CrossRef]
- Tian, H.; Xu, X.; Qu, J.; Li, H.; Hu, Y.; Huang, L.; He, W.; Li, B. Biodegradation of phenolic compounds in high saline wastewater by biofilms adhering on aerated membranes. Journal of Hazardous Materials 2020, 392. [CrossRef]
- Li, W.; Xiao, Q.; Hu, C.; Liu, B.; Sun, R. A comparison of the efficiency of different urease inhibitors and their effects on soil prokaryotic community in a short-term incubation experiment. Geoderma 2019, 354. [CrossRef]
- Dong, D.; Kou, Y.; Yang, W.; Chen, G.; Xu, H. Effects of urease and nitrification inhibitors on nitrous oxide emissions and nitrifying/denitrifying microbial communities in a rainfed maize soil: A 6-year field observation. Soil & Tillage Research 2018, 180, 82-90. [CrossRef]
- CRC handbook of chemistry and physics. 1977.
- ECD; SIDS. “Hydroquinone,” CAS 123-31-9, UNEP Publications. 2012.
- Suresh, S.; Srivastava, V.C.; Mishra, I.M. Adsorption of catechol, resorcinol, hydroquinone, and their derivatives: a review. International Journal of Energy and Environmental Engineering 2012, 3, 1-19. [CrossRef]
- Fiskesjö, G. The allium test in wastewater monitoring. Environmental Toxicology and Water Quality 1993, 8, 291-298. [CrossRef]
- MR, P.; J, V.; AM, M.; E, Z.; C, E.; E, L. Acute toxicological effects on the earthworm Eisenia fetida of 18 common pharmaceuticals in artificial soil. The Science of the total environment 2015, 518-519. [CrossRef]
- MR, P.-O.; S, M.; J, V.; E, N. Effects of 18 pharmaceuticals on the physiological diversity of edaphic microorganisms. The Science of the total environment 2017, 595. [CrossRef]
- Pohland, B., Owen, B. TAS. technical bulletin Biolog. 2009, 1 pp. 1–3.
- JL, G.; AL, M. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Applied and environmental microbiology 1991, 57. [CrossRef]
- Pino-Otin, M.R.; Langa, E.; Val, J.; Mainar, A.M.; Ballestero, D. Impact of citronellol on river and soil environments using non-target model organisms and natural populations. Journal of Environmental Management 2021, 287. [CrossRef]
- Pino-Otín, M.R.; Ballestero, D.; Navarro, E.; González-Coloma, A.; Val, J.; Mainar, A.M. Ecotoxicity of a novel biopesticide from Artemisia absinthium on non-target aquatic organisms. Chemosphere 2019, 216, 131-146. [CrossRef]
- Lu, N.; Lu, Y.; Liu, F.; Zhao, K.; Yuan, X.; Zhao, Y.; Li, Y.; Qin, H.; Zhu, J. H3PW12O40/TiO2 catalyst-induced photodegradation of bisphenol A (BPA): Kinetics, toxicity and degradation pathways. Chemosphere 2013, 91, 1266-1272. [CrossRef]
- Tissot, A.; Boule, P.; Lemaire, J.; Lambert, S.; Palla, J.C. Photochemistry and environment .10. evaluation of the toxicity of phototransformation products of hydroquinone and chlorophenols in aqueous-media. Chemosphere 1985, 14, 1221-1230. [CrossRef]
- Crisinel, A.; Delaunay, L.; Rossel, D.; Tarradellas, J.; Meyer, H.; Saiah, H.; Vogel, P.; Delisle, C.; Blaise, C. cyst-based ecotoxicological tests using anostracans - comparison of 2 species of Streptocephalus. Environmental Toxicology and Water Quality 1994, 9, 317-326. [CrossRef]
- Kuznetsova, T.V.; Sladkova, G.V.; Kholodkevich, S.V. Evaluation of functional state of crayfish Pontastacus leptodactylus in normal and toxic environment by characteristics of their cardiac activity and hemolymph biochemical parameters. Journal of Evolutionary Biochemistry and Physiology 2010, 46, 241-250. [CrossRef]
- Prosser, L., Temperature,. Sravnitel 'naya fiziologiya zhivotnykh (Comparative Animal Physiology). 1977, 2, 84-209.
- IPCS. Hydroquinone. Environmental Health Criteria 1994, 157.
- Sladkova, S.V.; Kholodkevich, S.V. Total protein in hemolymph of crawfish Pontastacus leptodactylus as a parameter of the functional state of animals and a biomarker of quality of habitat. Journal of Evolutionary Biochemistry and Physiology 2011, 47, 160-167. [CrossRef]
- Mondrala, S.; Eastmond, D.A. Topoisomerase II inhibition by the bioactivated benzene metabolite hydroquinone involves multiple mechanisms. Chemico-Biological Interactions 2010, 184, 259-268. [CrossRef]
- El Najjar, N.H.; Touffet, A.; Deborde, M.; Journel, R.; Leitner, N.K.V. Kinetics of paracetamol oxidation by ozone and hydroxyl radicals, formation of transformation products and toxicity. Separation and Purification Technology 2014, 136, 137-143. [CrossRef]
- Muneer, M.; Singh, H.K.; Bahnemann, D. Semiconductor-mediated photocatalysed degradation of two selected priority organic pollutants, benzidine and 1,2-diphenylhydrazine, in aqueous suspension. Chemosphere 2002, 49, 193-203. [CrossRef]
- Turkay, O.; Barisci, S.; Ozturk, H.; Ozturk, B.; Seker, M.G. Toxicological Profile of 1,4-Benzoquinone and Its Degradation By-Products during Electro-Fenton, Electrocoagulation, and Electrosynthesized Fe(VI) Oxidation. Journal of Environmental Engineering 2018, 144. [CrossRef]
- Guo, Q.; Zhou, Y.; Pang, S.-Y.; Gao, Y.; Duan, J.; Li, J.; Jiang, J. Transformation and detoxification of sulfamethoxazole by permanganate (Mn(VII)) in the presence of phenolic humic constituents. Chemical Engineering Journal 2021, 413. [CrossRef]
- El-Ghenymy, A.; Maria Rodriguez, R.; Brillas, E.; Oturan, N.; Oturan, M.A. Electro-Fenton degradation of the antibiotic sulfanilamide with Pt/carbon-felt and BDD/carbon-felt cells. Kinetics, reaction intermediates, and toxicity assessment. Environmental Science and Pollution Research 2014, 21, 8368-8378. [CrossRef]
- Rosal, R.; Gonzalo, M.S.; Boltes, K.; Leton, P.; Vaquero, J.J.; Garcia-Calvo, E. Identification of intermediates and assessment of ecotoxicity in the oxidation products generated during the ozonation of clofibric acid. Journal of Hazardous Materials 2009, 172, 1061-1068. [CrossRef]
- Calza, R.; Massolino, C.; Pelizzetti, E. Photo-induced transformation of hexaconazole and dimethomorph over TiO2 suspension. Journal of Photochemistry and Photobiology a-Chemistry 2008, 200, 356-363. [CrossRef]
- Jeyanthi, V.; Anbu, P.; Vairamani, M.; Velusamy, P. Isolation of hydroquinone (benzene-1,4-diol) metabolite from halotolerant <i>Bacillus methylotrophicus</i> MHC10 and its inhibitory activity towards bacterial pathogens. Bioprocess and Biosystems Engineering 2016, 39, 429-439. [CrossRef]
- Jurica, K.; Gobin, I.; Kremer, D.; Cepo, D.V.; Grubesic, R.J.; Karaconji, I.B.; Kosalec, I. Arbutin and its metabolite hydroquinone as the main factors in the antimicrobial effect of strawberry tree (<i>Arbutus unedo</i> L.) leaves. Journal of Herbal Medicine 2017, 8, 17-23. [CrossRef]
- Bikowska, B.Z.; Franiczek, R.; Sowa, A.; Polukord, G.; Krzyzanowska, B.; Sroka, Z. Antimicrobial and Antiradical Activity of Extracts Obtained from Leaves of Five Species of the Genus <i>Bergenia</i>: Identification of Antimicrobial Compounds. Microbial Drug Resistance 2017, 23, 771-780. [CrossRef]
- Sathiyamoorthi, E.; Faleye, O.S.; Lee, J.-H.; Lee, J. Hydroquinone derivatives attenuate biofilm formation and virulence factor production in<i> Vibrio</i> spp. International Journal of Food Microbiology 2023, 384. [CrossRef]
- Mol, V.P.L.; Abdulaziz, A.; Sneha, K.G.; Praveen, P.J.; Raveendran, T.V.; Parameswaran, P.S. Inhibition of pathogenic <i>Vibrio harveyi</i> using calamenene, derived from the Indian gorgonian <i>Subergorgia reticulata</i>, and its synthetic analog. 3 Biotech 2020, 10. [CrossRef]
- Genuario, D.B.; Vaz, M.; de Vielo, I.S. Phylogenetic insights into the diversity of homocytous cyanobacteria from Amazonian rivers. Molecular Phylogenetics and Evolution 2017, 116, 120-135. [CrossRef]
- Sun, F.L.; Wang, Y.S.; Wu, M.L.; Sun, C.C. Cyanobacterial community diversity in the sediments of the Pearl River Estuary in China. Scientia Marina 2017, 81, 477-485. [CrossRef]
- McGregor, G.B.; Fabbro, L.D.; Lobegeiger, J.S. Freshwater planktic Chroococcales (Cyanoprokaryota) from North-Eastern Australia: a morphological evaluation. Nova Hedwigia 2007, 84, 299-331. [CrossRef]
- Battistuzzi, F.U.; Hedges, S.B. A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land. Molecular Biology and Evolution 2009, 26, 335-343. [CrossRef]
- Xia, N.; Xia, X.H.; Zhu, B.T.; Zheng, S.K.; Zhuang, J. Bacterial diversity and community structure in the sediment of the middle and lower reaches of the Yellow River, the largest turbid river in the world. Aquatic Microbial Ecology 2013, 71, 43-U168. [CrossRef]
- Narciso-da-Rocha, C.; Manaia, C.M. Multidrug resistance phenotypes are widespread over different bacterial taxonomic groups thriving in surface water. Science of the Total Environment 2016, 563, 1-9. [CrossRef]
- Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the <i>para</i>-nitrophenol catabolic pathway, and characterization of three enzymes involved in the hydroquinone pathway, in <i>pseudomonas</i> sp 1-7. Bmc Microbiology 2012, 12. [CrossRef]
- Spain, J.C.; Gibson, D.T. Pathway for biodegradation of para-nitrophenol in a Moraxella sp. Applied and Environmental Microbiology 1991, 57, 812-819. [CrossRef]
- Madigan, T.M.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, S.A. Brock. Biología de los microorganismos, 14th ed.; Pearson Education: Madrid, 2015.
- Liu, S.; Wang, P.; Wang, C.; Chen, J.; Wang, X.; Hu, B.; Yuan, Q. Ecological insights into the disturbances in bacterioplankton communities due to emerging organic pollutants from different anthropogenic activities along an urban river. Science of the Total Environment 2021, 796. [CrossRef]
- Zhang, M.; Sun, Q.; Chen, P.; Wei, X.; Wang, B. How microorganisms tell the truth of potentially toxic elements pollution in environment. Journal of Hazardous Materials 2022, 431. [CrossRef]
- Christensen, A.M.; Ingerslev, F.; Baun, A. Ecotoxicity of mixtures of antibiotics used in aquacultures. Environmental Toxicology and Chemistry 2006, 25, 2208-2215. [CrossRef]
- Schoffelen, N.J.; Mohr, W.; Ferdelman, T.G.; Duerschlag, J.; Littmann, S.; Ploug, H.; Kuypers, M.M.M. Phosphate availability affects fixed nitrogen transfer from diazotrophs to their epibionts. Isme Journal 2019, 13, 2701-2713. [CrossRef]
- Wang, Z.; Han, S.; Cai, M.; Du, P.; Zhang, Z.; Li, X. Environmental behavior of methamphetamine and ketamine in aquatic ecosystem: Degradation, bioaccumulation, distribution, and associated shift in toxicity and bacterial community. Water Research 2020, 174. [CrossRef]
- Zhang, C.; Li, J.; Cheng, F.; Liu, Y. Enhanced phenol removal in an innovative lignite activated coke-assisted biological process. Bioresource Technology 2018, 260, 357-363. [CrossRef]
- Zhao, J.; Chen, X.; Bao, L.; Bao, Z.; He, Y.; Zhang, Y.; Li, J. Correlation between microbial diversity and toxicity of sludge treating synthetic wastewater containing 4-chlorophenol in sequencing batch reactors. Chemosphere 2016, 153, 138-145. [CrossRef]
- Ansola, G.; Arroyo, P.; Saenz de Miera, L.E. Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Science of the Total Environment 2014, 473, 63-71. [CrossRef]
- Rios-Miguel, A.B.; Smith, G.J.; Cremers, G.; van Alen, T.; Jetten, M.S.M.; Camp, H.J.M.O.d.; Welte, C.U. Microbial paracetamol degradation involves a high diversity of novel amidase enzyme candidates. Water Research X 2022, 16. [CrossRef]
- Park, S.; Oh, S. Activated sludge-degrading analgesic drug acetaminophen: Acclimation, microbial community dynamics, degradation characteristics, and bioaugmentation potential. Water Research 2020, 182. [CrossRef]
- Traversi, D.; Villa, S.; Lorenzi, E.; Degan, R.; Gilli, G. Application of a real-time qPCR method to measure the methanogen concentration during anaerobic digestion as an indicator of biogas production capacity. Journal of Environmental Management 2012, 111, 173-177. [CrossRef]
- Villegas, L.G.C.; Mashhadi, N.; Chen, M.; Mukherjee, D.; Taylor, K.E.; Biswas, N. A Short Review of Techniques for Phenol Removal from Wastewater. Current Pollution Reports 2016, 2, 157-167. [CrossRef]
- Westman, C.A. The Effect Hydroquinone on Mitosis in <em>Allium cepa</em>; 1949.
- Shettel, N.L.; Balke, N.E. Plant-growth response to several allelopathic chemicals. Weed Science 1983, 31, 293-298. [CrossRef]
- Keller, C.P.; Barkosky, R.R.; Seil, J.E.; Mazurek, S.A.; Grundstad, M.L. The electrical response of Phaseolus vulgaris roots to abrupt exposure to hydroquinone. Plant signaling & behavior 2008, 3, 633-640. [CrossRef]
- Bouknana, D.; Jodeh, S.; Sbaa, M.; Hammouti, B.; Arabi, M.; Darmous, A.; Slamini, M.; Haboubi, K. A phytotoxic impact of phenolic compounds in olive oil mill wastewater on fenugreek "Trigonella foenum-graecum". Environmental Monitoring and Assessment 2019, 191. [CrossRef]
- Pino-Otín, M.R.; Lorca, G.; Val, J.; Ferrando, N.; Ballestero, D.; Langa, E. Ecotoxicological Study of Tannic Acid on Soil and Water Non-Target Indicators and Its Impact on Fluvial and Edaphic Communities. Plants 2023, 12, 4041. [CrossRef]
- Wang, H.; Cheng, Z.; Djouonkep, L.D.W.; Wang, L.; Cai, S.; Gauthier, M. Synthesis and properties of biodegradable aliphatic-aromatic polyesters derived from 4-hydroxybenzaldehyde. Journal of Applied Polymer Science 2023, 140. [CrossRef]
- Osman, A.M.; Den Besten, P.J.; van Noort, P.C.M. Menadione enhances oxyradical formation in earthworm extracts: vulnerability of earthworms to quinone toxicity. Aquatic Toxicology 2003, 65, 101-109. [CrossRef]
- Suthar, S.; Singh, S.; Dhawan, S. Earthworms as bioindicator of metals (Zn, Fe, Mn, Cu, Pb and Cd) in soils: Is metal bioaccumulation affected by their ecological category? Ecological Engineering 2008, 32, 99-107. [CrossRef]
- Saxena, P.N.; Gupta, S.K.; Murthy, R.C. Comparative toxicity of carbaryl, carbofuran, cypermethrin and fenvalerate in <i>Metaphire posthuma</i> and <i>Eisenia fetida</i>-A possible mechanism. Ecotoxicology and Environmental Safety 2014, 100, 218-225. [CrossRef]
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Waomar, M. Biological effects of essential oils - A review. Food and Chemical Toxicology 2008, 46, 446-475. [CrossRef]
- Ahmed, N.; Al-Mutairi, K.A. Earthworms Effect on Microbial Population and Soil Fertility as Well as Their Interaction with Agriculture Practices. Sustainability 2022, 14. [CrossRef]
- Edwards, C.A.; Fletcher, K.E. Interactions between earthworms and microorganisms in organic-matter breakdown. Agriculture Ecosystems & Environment 1988, 24, 235-247. [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Applied and Environmental Microbiology 2006, 72, 1719-1728. [CrossRef]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. Isme Journal 2009, 3, 992-1000. [CrossRef]
- Zhang, L.; Xu, Z.H. Assessing bacterial diversity in soil. Journal of Soils and Sediments 2008, 8, 379-388. [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity (vol 180, pg 4765, 1998). Journal of Bacteriology 1998, 180, 6793-6793. [CrossRef]
- Wang, Y.J.; Liu, L.; Yang, J.F.; Duan, Y.M.; Luo, Y.; Taherzadeh, M.J.; Li, Y.F.; Li, H.K.; Awasthi, M.K.; Zhao, Z.Y. The diversity of microbial community and function varied in response to different agricultural residues composting. Science of the Total Environment 2020, 715. [CrossRef]
- Zhang, S.Y.; Fan, C.; Wang, Y.X.; Xia, Y.S.; Xiao, W.; Cui, X.L. Salt-tolerant and plant-growth-promoting bacteria isolated from high-yield paddy soil. Canadian Journal of Microbiology 2018, 64, 968-978. [CrossRef]
- Blain, N.P.; Helgason, B.L.; Germida, J.J. Endophytic root bacteria associated with the natural vegetation growing at the hydrocarbon-contaminated Bitumount Provincial Historic site. Canadian Journal of Microbiology 2017, 63, 502-515. [CrossRef]
- Zhang, W.; Chen, L.; Zhang, R.; Lin, K. High throughput sequencing analysis of the joint effects of BDE209-Pb on soil bacterial community structure. Journal of Hazardous Materials 2016, 301, 1-7. [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. Bmc Microbiology 2018, 18. [CrossRef]
- Pino-Otin, M.R.; Gan, C.; Terrado, E.; Sanz, M.A.; Ballestero, D.; Langa, E. Antibiotic properties of Satureja montana L. hydrolate in bacteria and fungus of clinical interest and its impact in non-target environmental microorganisms. Scientific Reports 2022, 12. [CrossRef]
- Chen, H.; Yao, J.; Wang, F.; Choi, M.M.F.; Bramanti, E.; Zaray, G. Study on the toxic effects of diphenol compounds on soil microbial activity by a combination of methods. Journal of Hazardous Materials 2009, 167, 846-851. [CrossRef]
- Chien, S.H.; Prochnow, L.I.; Cantarella, H. Chapter 8 Recent Developments of Fertilizer Production and Use to Improve Nutrient Efficiency and Minimize Environmental Impacts. In Advances in Agronomy; Academic Press: 2009; Volume 102, pp. 267-322.
- Bremner, J.M.; Chai, H.S. effects of phosphoroamides on ammonia volatilization and nitrite accumulation in soils treated with urea. Biology and Fertility of Soils 1989, 8, 227-230.
- Zaman, M.; Nguyen, M.L.; Blennerhassett, J.D.; Quin, B.F. Reducing NH, NO and NO3 losses from a pasture soil with urease or nitrification inhibitors and elemental S-amended nitrogenous fertilizers. Biology and Fertility of Soils 2008, 44, 693-705. [CrossRef]
- Luisa Castrejon-Godinez, M.; Tovar-Sanchez, E.; Ortiz-Hernandez, M.L.; Encarnacion-Guevara, S.; Gabriel Martinez-Batallar, A.; Hernandez-Ortiz, M.; Sanchez-Salinas, E.; Rodriguez, A.; Mussali-Galante, P. Proteomic analysis of <i>Burkholderia zhejiangensis</i> CEIB S4-3 during the methyl parathion degradation process. Pesticide Biochemistry and Physiology 2022, 187. [CrossRef]
- Jia, Y.H. Diversity of utilizing substrates of strain dyella sp. la-4 and its application in soi remediation. 2009.
- Flood, J.J.; Copley, S.D. Genome-Wide Analysis of Transcriptional Changes and Genes That Contribute to Fitness during Degradation of the Anthropogenic Pollutant Pentachlorophenol by Sphingobium chlorophenolicum. Msystems 2018, 3. [CrossRef]
- Jain, R.K.; Dreisbach, J.H.; Spain, J.C. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Applied and Environmental Microbiology 1994, 60, 3030-3032. [CrossRef]
- Cejková, A.; Masák, J.; Jirku, V.; Vesely, M.; Pátek, M.; Nesvera, J. Potential of <i>Rhodococcus erythropolis</i> as a bioremediation organism. World Journal of Microbiology & Biotechnology 2005, 21, 317-321. [CrossRef]
- Organisation for Economic Co-operation and Development (OECD). Test No. 202: Daphnia sp. Acute Immobilisation Test. In OECD Guidelines for the Testing of Chemicals, Section 2; OECD: Paris, France, 2004.
- UNE-EN ISO 6341; Water Quality—Determination of the Inhibition of the Mobility of Daphnia magna Straus (Cladocera, Crustacea)—Acute Toxicity Test. AENOR: Madrid, Spain, 2012.
- ISO 11348; Water Quality—Determination of the Inhibitory Effect of Water Samples on the Light Emission of Vibrio fischeri (Luminescent Bacteria Test). International Organization for Standardization: Geneva, Switzerland, 2007.
- Organisation for Economic Co-operation and Development (OECD). Test No. 207: Earthworm, Acute Toxicity Tests. In OECD Guidelines for the Testing of Chemicals, Section 2; OECD: Paris, France, 1984.
- UNE-EN ISO 19458:2007; Water Quality—Sampling for Microbiological Analysis (ISO 19458:2006). AENOR: Madrid, Spain, 2007.
- Commission Implementing Decision (EU) 2018/840 of 5 June 2018 establishing a watch list of substances for Union-wide monitoring in the field of water policy pursuant to Directive 2008/105/EC of the European Parliament and of the Council and repealing Commission Implementing Decision (EU) 2015/495 (notified under document C(2018) 3362). Official Journal of the European Union. L 141/9.
| Hydroquinone properties | |
| Molecular weight | 110.11 g/mol[33] |
| Water solubility | 73 g/L at 25 °C[33] |
| Melting point | 170-172 °C[33] |
| Boiling point | 287 °C[33] |
| Dipole moment | 1.4-2.4D[33] |
| Density | 1341 kg/m[34] |
| Vapour pressure | 2.34x10-3 Pa at 25 °C[34] |
| pH | 4.0-7.0[34] |
| Partition coefficient (log pow) | 0.59[35] |
| pKa | pK1=9.9 pK2=11.6[35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





