Submitted:
28 December 2023
Posted:
29 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
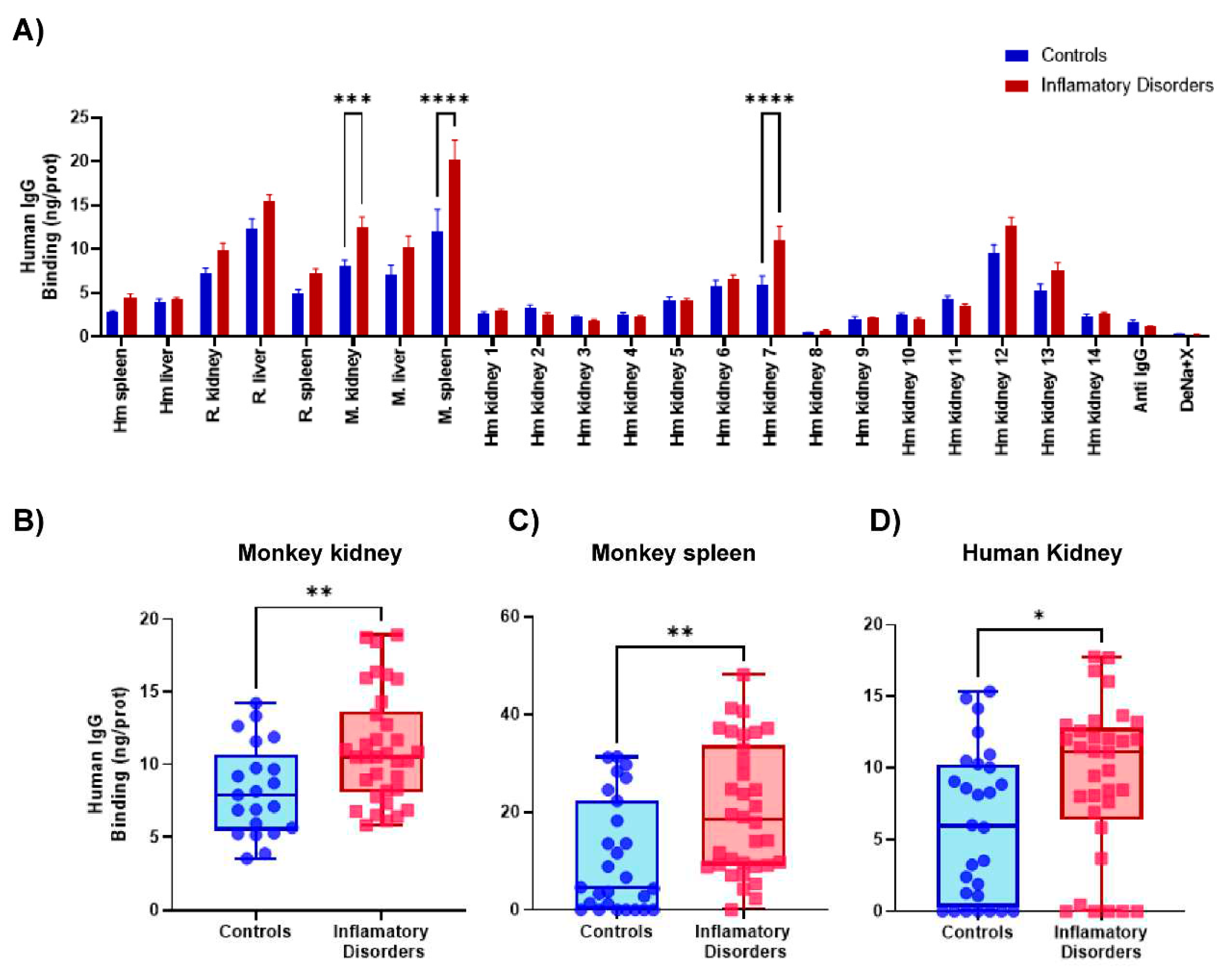
2.1. Detection of self-reactive antibodies in sera from patients with autoimmune disorders
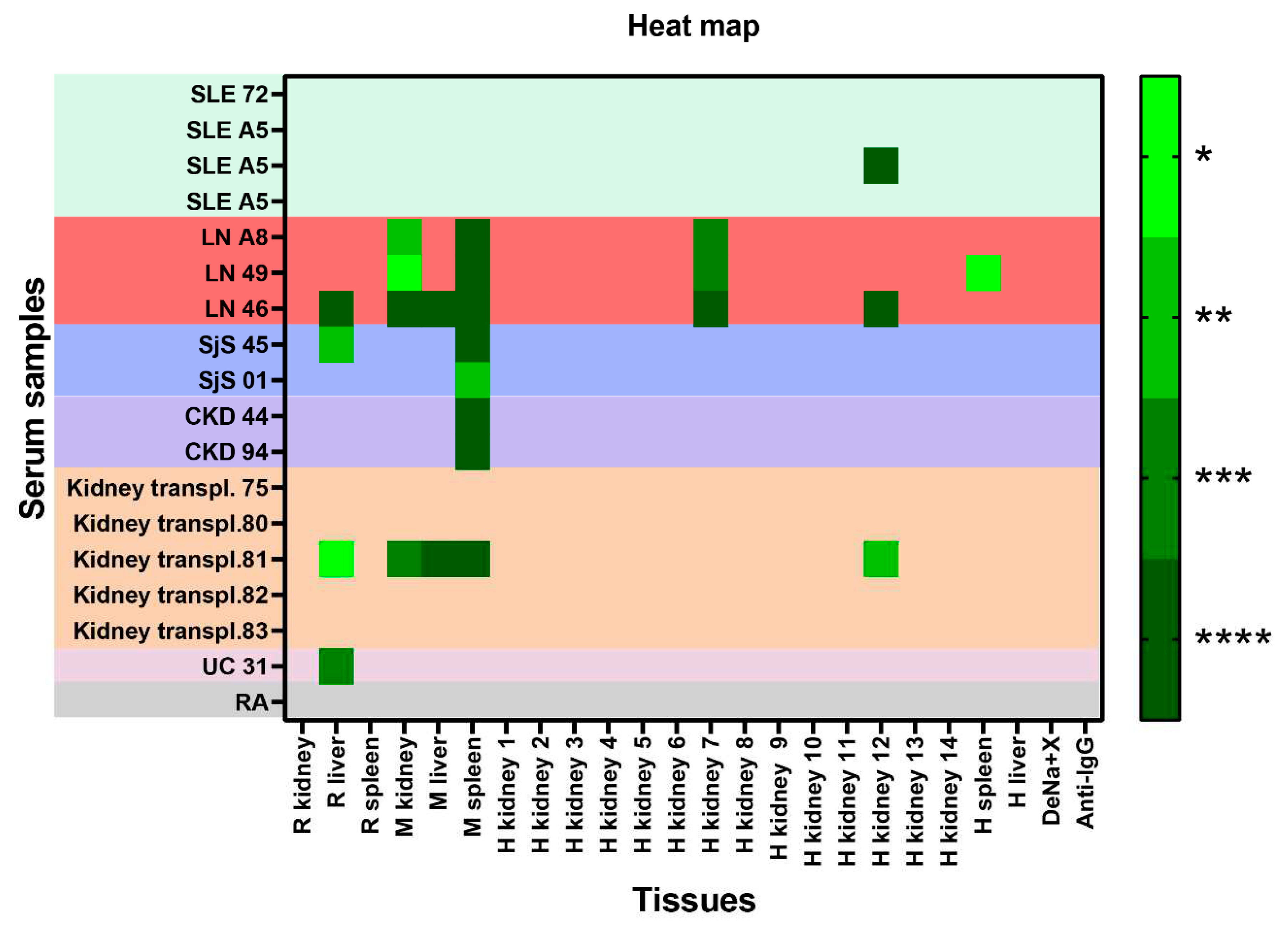
2.2. Reactivity of sera to the panel of samples immobilized in CMMAs
2.3. Evaluation of the test performance in the detection of reactive antibodies
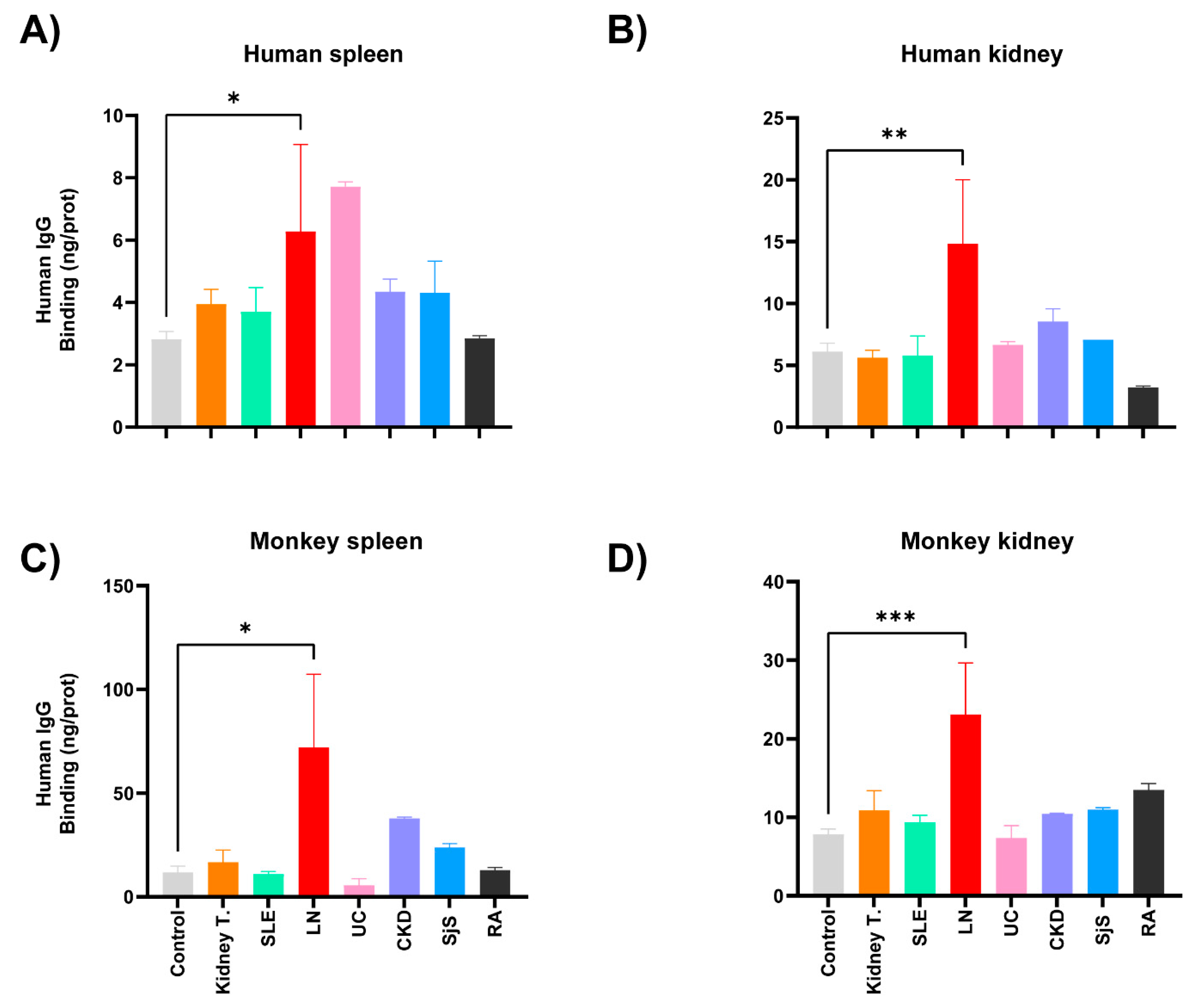
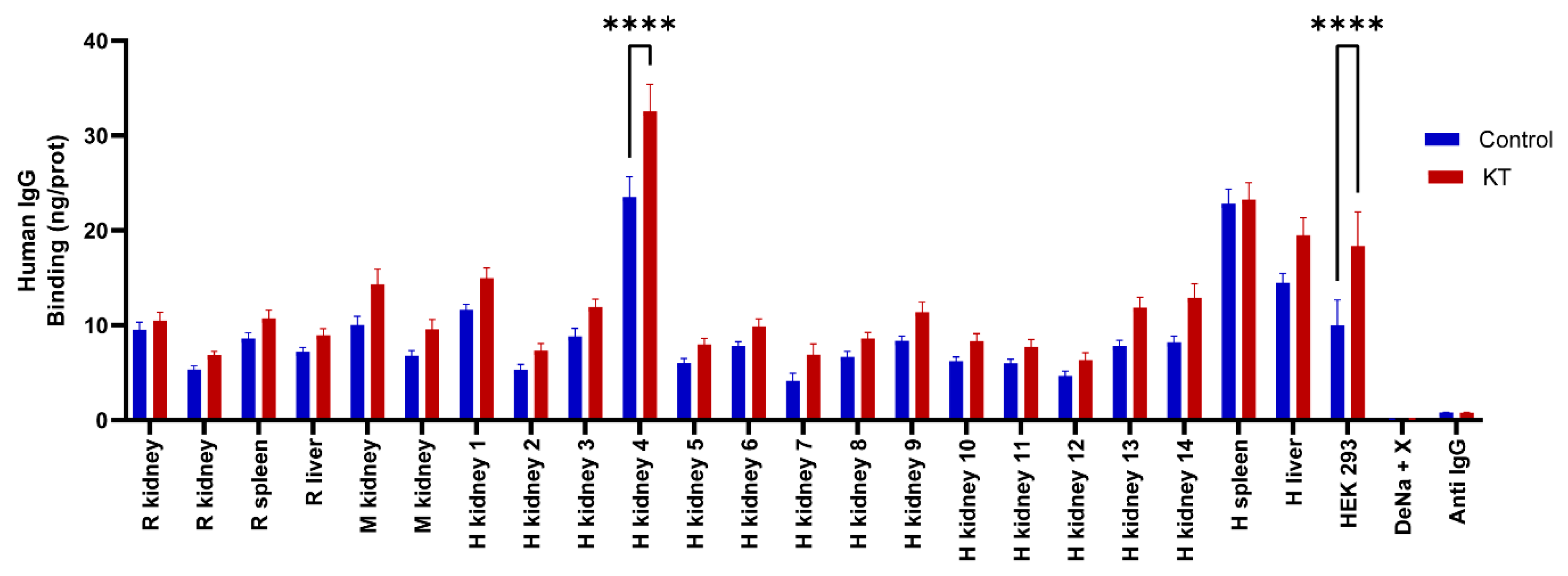
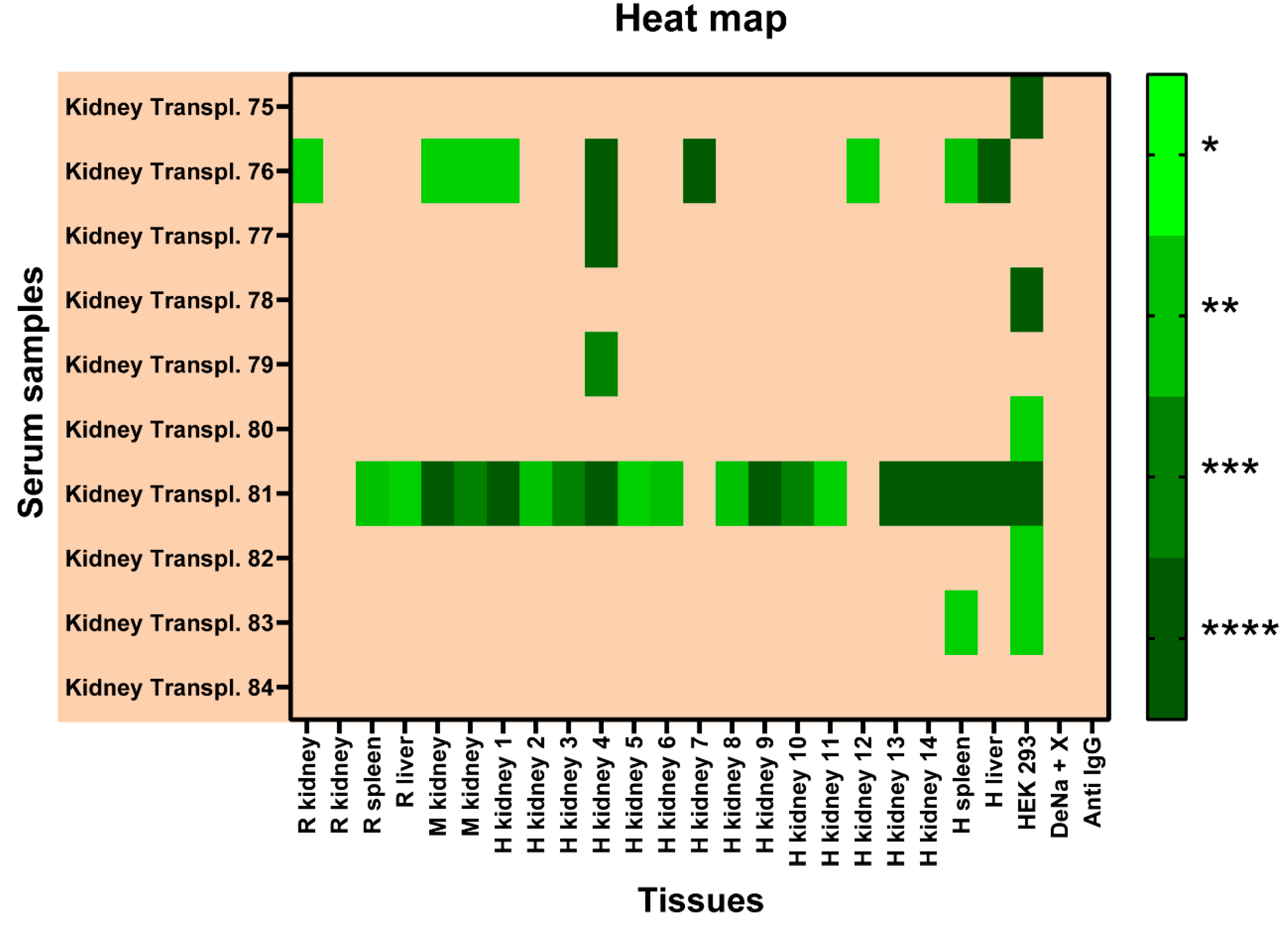
2.4. Detection of self-reactive antibodies in sera from patients with KT
2.5. Reactivity of renal transplant patients´ sera to the CMMAs antigenic panel
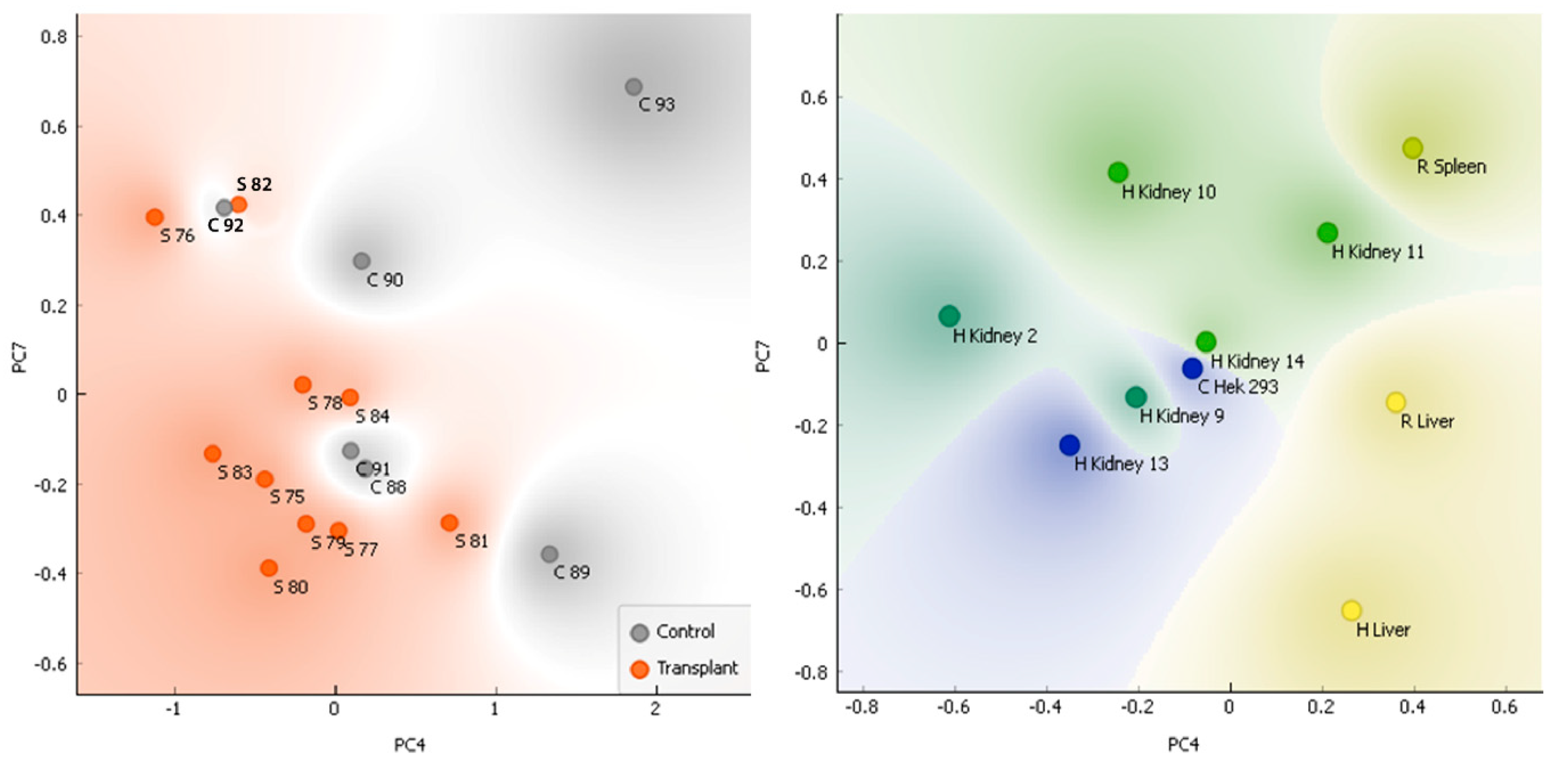
2.6. Principal components analysis (PCA)
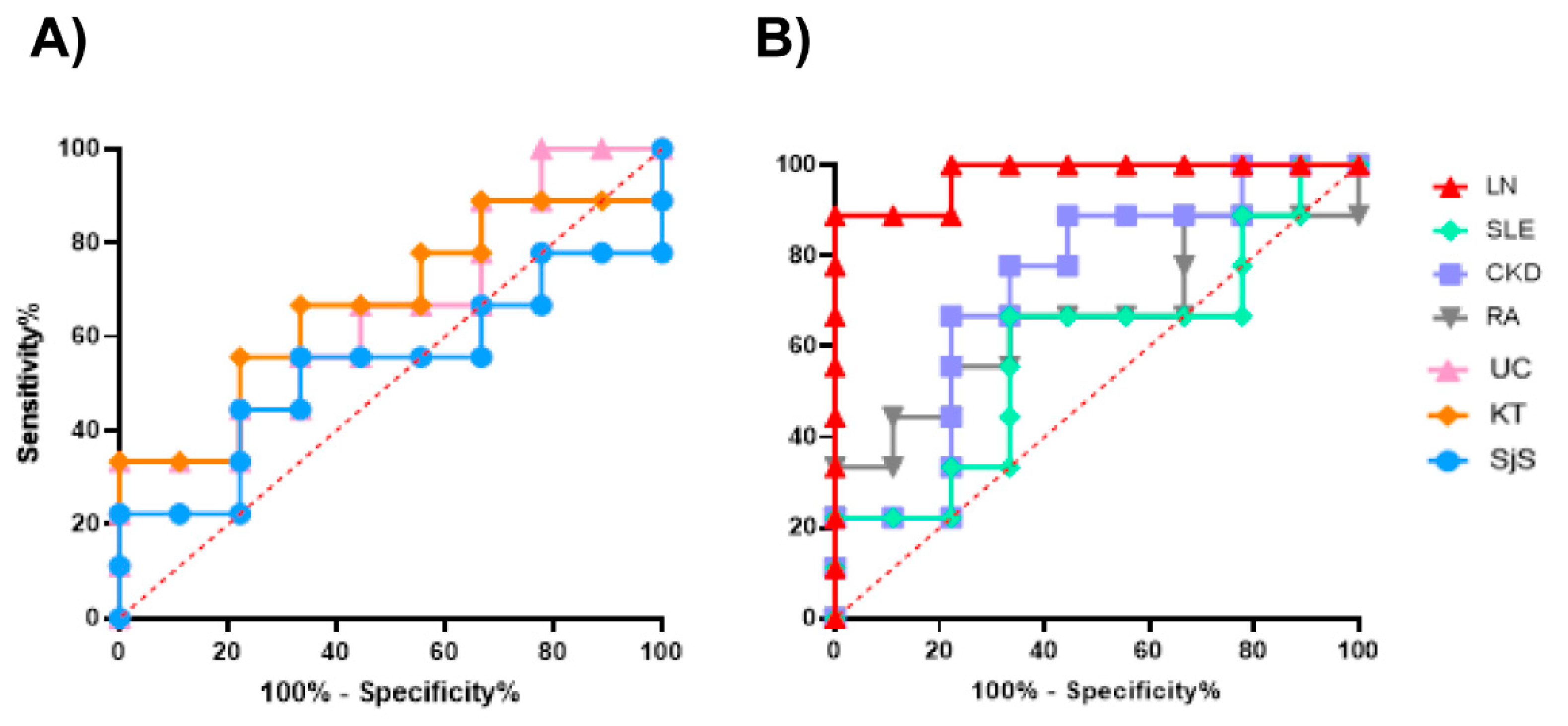
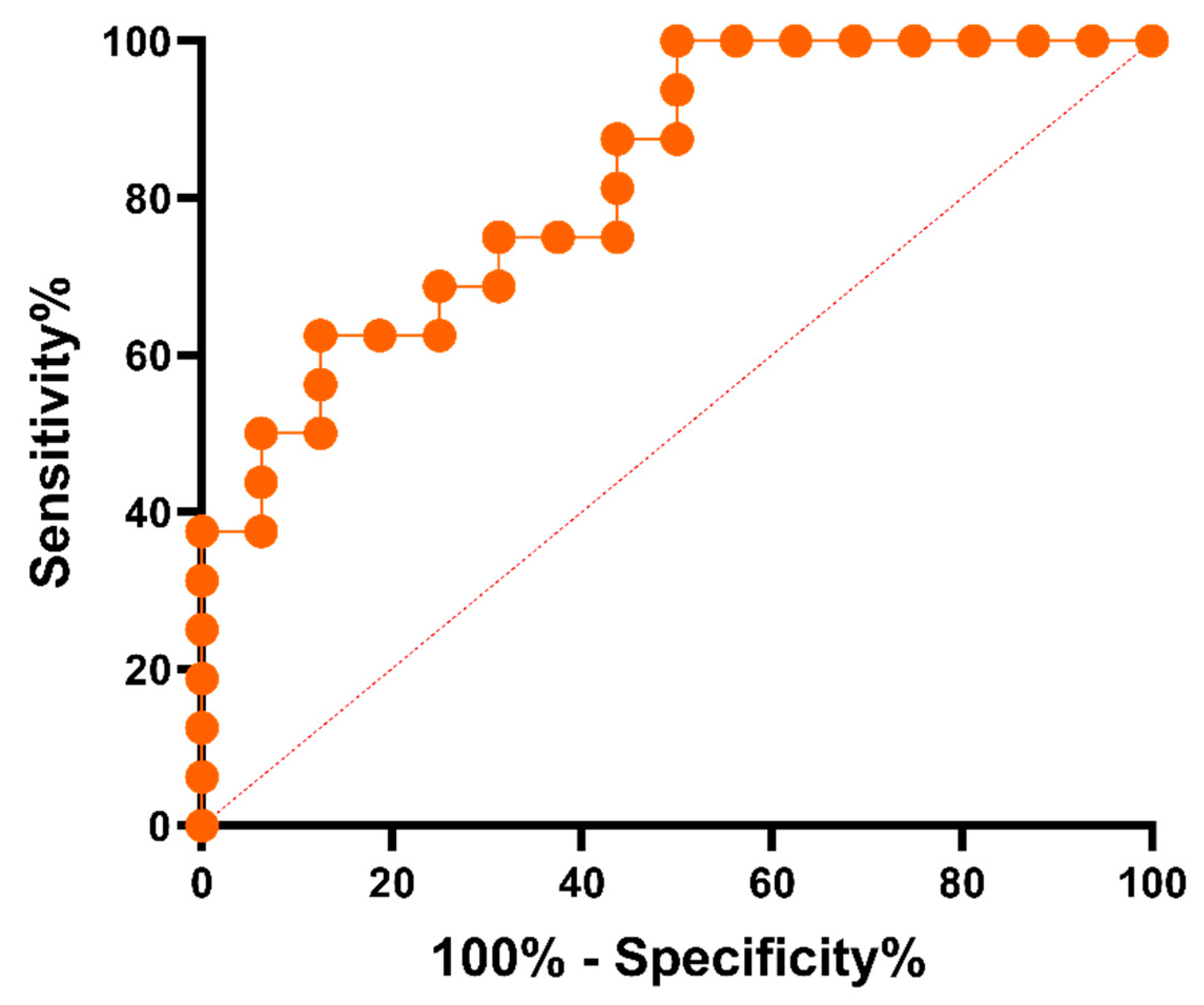
2.7. Evaluation of the test performance. Receiver Operating Characteristic curve
3. Discussion
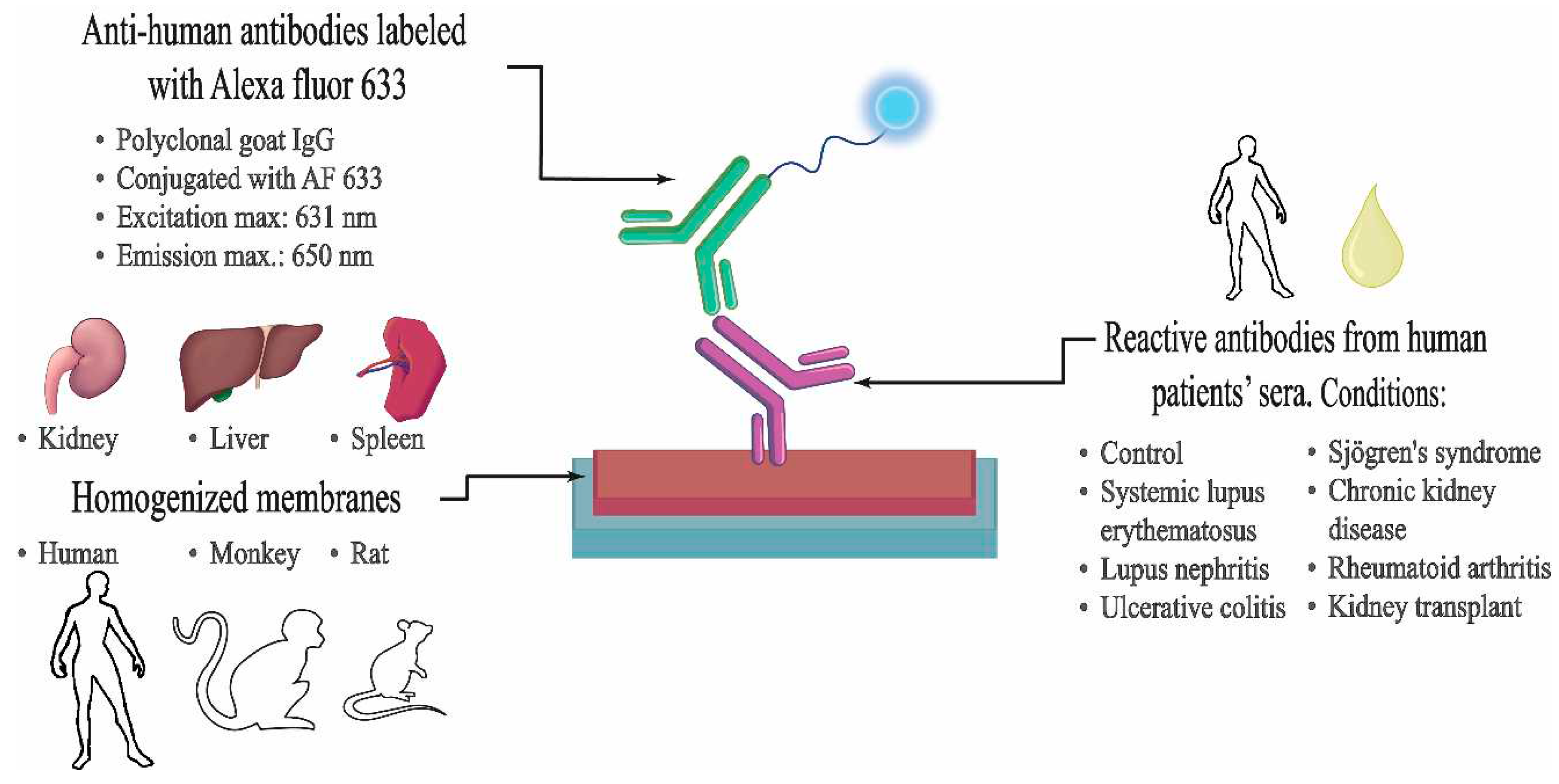
4. Materials and Methods
4.1. Serum samples
| ID | Age At Excision | Sex | Primary Diagnosis | Supplier |
|---|---|---|---|---|
| 248900A8 | 45 | Female | Lupus nephritis | BioIVT |
| 286224A5 | 51 | Female | Systemic lupus erythematosus | BioIVT |
| HMN645046 | 36 | Female | Lupus nephritis | BioIVT |
| HUMANSRM-875072 | 55 | Female | Systemic lupus erythematosus | BioIVT |
| HMN773201 | 54 | Female | Sjogren syndrome | BioIVT |
| HMN700845 | 55 | Female | Sjogren syndrome | BioIVT |
| HMN711044 | 54 | Female | Chronic kidney disease | BioIVT |
| HMN776494 | 54 | Female | Chronic kidney disease | BioIVT |
| HUMANSRM-838031 | 60 | Male | Ulcerative colitis | BioIVT |
| HMN1073049 | 32 | Female | Lupus nephritis | BioIVT |
| HMN869646 | 72 | Male | Rheumatoid Arthritis | BioIVT |
| HMN410375 | 68 | Male | Kidney transplant | BioIVT |
| HMN410376 | 49 | Male | Kidney transplant | BioIVT |
| HMN410377 | 64 | Female | Kidney transplant | BioIVT |
| HMN410378 | 54 | Female | Kidney transplant | BioIVT |
| HMN410379 | 25 | Male | Kidney transplant | BioIVT |
| HMN410380 | 66 | Male | Kidney transplant | BioIVT |
| HMN410381 | 60 | Female | Kidney transplant | BioIVT |
| HMN410382 | 68 | Male | Kidney transplant | BioIVT |
| HMN410383 | 70 | Male | Kidney transplant | BioIVT |
| HMN410384 | 58 | Male | Kidney transplant | BioIVT |
| HMN410388 | 83 | Female | Control | BioIVT |
| HMN410389 | 25 | Male | Control | BioIVT |
| HMN410390 | 60 | Female | Control | BioIVT |
| HMN410391 | 19 | Male | Control | BioIVT |
| HMN410392 | 29 | Female | Control | BioIVT |
| HMN410393 | 30 | Female | Control | BioIVT |
| AAA8816702 | 45 | Male | Control | Biobank of Aragon Health System |
| AAA8825493 | 46 | Male | Control | Biobank of Aragon Health System |
| AAA8825485 | 45 | Male | Control | Biobank of Aragon Health System |
| AAA7371433 | 36 | Female | Control | Biobank of Aragon Health System |
| AAA8825480 | 59 | Female | Control | Biobank of Aragon Health System |
| AAA8825474 | 49 | Male | Control | Biobank of Aragon Health System |
| AAA8825470 | 42 | Male | Control | Biobank of Aragon Health System |
| AAA8825467 | 67 | Male | Control | Biobank of Aragon Health System |
| AAA8825462 | 20 | Female | Control | Biobank of Aragon Health System |
| AAA8825459 | 54 | Male | Control | Biobank of Aragon Health System |
| AAA8825453 | 20 | Female | Control | Biobank of Aragon Health System |
| AAA8825408 | 48 | Male | Control | Biobank of Aragon Health System |
4.2. Tissue samples
4.3. Cell Membrane Microarray Fabrication
4.4. Immunoassay Procedure
4.5. Data processing and Normalization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pahwa, R.; Goyal, A.; Jialal, I. Chronic Inflammation. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2023.
- Michels da Silva, D.; Langer, H.; Graf, T. Inflammatory and Molecular Pathways in Heart Failure-Ischemia, HFpEF and Transthyretin Cardiac Amyloidosis. Int J Mol Sci 2019, 20, 2322. [CrossRef]
- Etxebarria, A.; Díez-Martín, E.; Astigarraga, E.; Barreda-Gómez, G. Role of the Immune System in Renal Transplantation, Types of Response, Technical Approaches and Current Challenges. Immuno 2022, 2, 548–570. [CrossRef]
- Domínguez-Fernández, C.; Egiguren-Ortiz, J.; Razquin, J.; Gómez-Galán, M.; De Las Heras-García, L.; Paredes-Rodríguez, E.; Astigarraga, E.; Miguélez, C.; Barreda-Gómez, G. Review of Technological Challenges in Personalised Medicine and Early Diagnosis of Neurodegenerative Disorders. Int J Mol Sci 2023, 24, 3321. [CrossRef]
- Pontrelli, P.; Grandaliano, G.; Van Kooten, C. Editorial: Kidney Transplantation and Innate Immunity. Front Immunol 2020, 11, 603982. [CrossRef]
- Varela, M.L.; Mogildea, M.; Moreno, I.; Lopes, A. Acute Inflammation and Metabolism. Inflammation 2018, 41, 1115–1127. [CrossRef]
- Nicholson, L.B. The Immune System. Essays Biochem 2016, 60, 275–301. [CrossRef]
- Ferrero-Miliani, L.; Nielsen, O.H.; Andersen, P.S.; Girardin, S.E. Chronic Inflammation: Importance of NOD2 and NALP3 in Interleukin-1beta Generation. Clin Exp Immunol 2007, 147, 227–235. [CrossRef]
- Siu, J.H.Y.; Surendrakumar, V.; Richards, J.A.; Pettigrew, G.J. T Cell Allorecognition Pathways in Solid Organ Transplantation. Front Immunol 2018, 9, 2548. [CrossRef]
- Tsai, D.-H.; Riediker, M.; Berchet, A.; Paccaud, F.; Waeber, G.; Vollenweider, P.; Bochud, M. Effects of Short- and Long-Term Exposures to Particulate Matter on Inflammatory Marker Levels in the General Population. Environ Sci Pollut Res Int 2019, 26, 19697–19704. [CrossRef]
- Siebert, S.; Tsoukas, A.; Robertson, J.; McInnes, I. Cytokines as Therapeutic Targets in Rheumatoid Arthritis and Other Inflammatory Diseases. Pharmacol Rev 2015, 67, 280–309. [CrossRef]
- Richard-Eaglin, A.; Smallheer, B.A. Immunosuppressive/Autoimmune Disorders. Nurs Clin North Am 2018, 53, 319–334. [CrossRef]
- Ilchmann-Diounou, H.; Menard, S. Psychological Stress, Intestinal Barrier Dysfunctions, and Autoimmune Disorders: An Overview. Front Immunol 2020, 11, 1823. [CrossRef]
- Prideaux, L.; De Cruz, P.; Ng, S.C.; Kamm, M.A. Serological Antibodies in Inflammatory Bowel Disease: A Systematic Review. Inflamm Bowel Dis 2012, 18, 1340–1355. [CrossRef]
- Xiao, Z.X.; Miller, J.S.; Zheng, S.G. An Updated Advance of Autoantibodies in Autoimmune Diseases. Autoimmun Rev 2021, 20, 102743. [CrossRef]
- Cusick, M.F.; Libbey, J.E.; Fujinami, R.S. Molecular Mimicry as a Mechanism of Autoimmune Disease. Clin Rev Allergy Immunol 2012, 42, 102–111. [CrossRef]
- Gao, Z.; Feng, Y.; Xu, J.; Liang, J. T-Cell Exhaustion in Immune-Mediated Inflammatory Diseases: New Implications for Immunotherapy. Front Immunol 2022, 13, 977394. [CrossRef]
- Ho, J.; Wiebe, C.; Gibson, I.W.; Rush, D.N.; Nickerson, P.W. Immune Monitoring of Kidney Allografts. Am J Kidney Dis 2012, 60, 629–640. [CrossRef]
- Rienda, B.; Elexpe, A.; Tolentino-Cortez, T.; Gulak, M.; Bruzos-Cidón, C.; Torrecilla, M.; Astigarraga, E.; Barreda-Gómez, G. Analysis of Acetylcholinesterase Activity in Cell Membrane Microarrays of Brain Areas as a Screening Tool to Identify Tissue Specific Inhibitors. Analytica 2021, 2, 25–36. [CrossRef]
- Fernández, R.; Garate, J.; Tolentino-Cortez, T.; Herraiz, A.; Lombardero, L.; Ducrocq, F.; Rodríguez-Puertas, R.; Trifilieff, P.; Astigarraga, E.; Barreda-Gómez, G.; et al. Microarray and Mass Spectrometry-Based Methodology for Lipid Profiling of Tissues and Cell Cultures. Anal Chem 2019, 91, 15967–15973. [CrossRef]
- de la Fuente, M.; Rodríguez-Agirretxe, I.; Vecino, E.; Astigarraga, E.; Acera, A.; Barreda-Gómez, G. Elevation of Tear MMP-9 Concentration as a Biomarker of Inflammation in Ocular Pathology by Antibody Microarray Immunodetection Assays. Int J Mol Sci 2022, 23, 5639. [CrossRef]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognition Letters 2006, 27, 861–874. [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J Intern Med 2013, 4, 627–635.
- Castro, C.; Gourley, M. Diagnostic Testing and Interpretation of Tests for Autoimmunity. J Allergy Clin Immunol 2010, 125, S238–S247. [CrossRef]
- Pisetsky, D.S. Pathogenesis of Autoimmune Disease. Nat Rev Nephrol 2023, 19, 509–524. [CrossRef]
- Rahman, A.; Manson, J.J.; Isenberg, D.A. Autoantibodies and Lupus Nephritis. In Lupus Nephritis; Lewis, E.J., Schwartz, M.M., Korbet, S.M., Chan, D.T.M., Eds.; Oxford University Press, 2010; p. 0 ISBN 978-0-19-956805-5.
- Ungaro, R.; Colombel, J.-F.; Lissoos, T.; Peyrin-Biroulet, L. A Treat-to-Target Update in Ulcerative Colitis: A Systematic Review. Am J Gastroenterol 2019, 114, 874–883. [CrossRef]
- Mitsuyama, K.; Niwa, M.; Takedatsu, H.; Yamasaki, H.; Kuwaki, K.; Yoshioka, S.; Yamauchi, R.; Fukunaga, S.; Torimura, T. Antibody Markers in the Diagnosis of Inflammatory Bowel Disease. World J Gastroenterol 2016, 22, 1304–1310. [CrossRef]
- Anders, H.-J.; Saxena, R.; Zhao, M.; Parodis, I.; Salmon, J.E.; Mohan, C. Lupus Nephritis. Nat Rev Dis Primers 2020, 6, 1–25. [CrossRef]
- Schaub, J.A.; Kretzler, M. Chapter 33 - Systems Biology in Diagnosis and Treatment of Kidney Disease. In Regenerative Nephrology (Second Edition); Goligorsky, M.S., Ed.; Academic Press, 2022; pp. 465–479 ISBN 978-0-12-823318-4.
- Borchers, A.T.; Leibushor, N.; Naguwa, S.M.; Cheema, G.S.; Shoenfeld, Y.; Gershwin, M.E. Lupus Nephritis: A Critical Review. Autoimmunity Reviews 2012, 12, 174–194. [CrossRef]
- Mejia-Vilet, J.M.; Rovin, B.H. 59 - Epidemiology and Management of Lupus Nephritis. In Dubois’ Lupus Erythematosus and Related Syndromes (Ninth Edition); Wallace, D.J., Hahn, B.H., Eds.; Elsevier: London, 2019; pp. 727–744 ISBN 978-0-323-47927-1.
- Musa, R.; Brent, L.H.; Qurie, A. Lupus Nephritis. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2023.
- Alarcón-Riquelme, M.E. The heterogeneity of systemic lupus erythematosus: Looking for a molecular answer. Rev Colomb Reumatol 2021, 28, 31–38. [CrossRef]
- Stinton, L.; Barr, S.; Tibbles, L.A.; Yilmaz, S.; Sar, A.; Benediktsson, H.; Fritzler, M. Autoantibodies in Lupus Nephritis Patients Requiring Renal Transplantation. Lupus 2007, 16, 394–400. [CrossRef]
- Elsayed, S.A.; Mohafez, O.M.M. Autoantibodies Spectrum in Lupus Nephritis in a Cohort of Egyptian Patients: Relation to Disease Activity and Prognostic Value. Egyptian Rheumatology and Rehabilitation 2020, 47, 39. [CrossRef]
- Chen, M.; Wang, Y.-Y.; Zhao, M.-H.; Zhang, Y.-K.; Wang, H.-Y. Autoantibodies against Glomerular Mesangial Cells and Their Target Antigens in Lupus Nephritis. Ren Fail 2005, 27, 507–513. [CrossRef]
- Lewis, E.J.; Schwartz, M.M. Pathology of Lupus Nephritis. Lupus 2005, 14, 31–38. [CrossRef]
- Carsons, S.E.; Patel, B.C. Sjogren Syndrome. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2023.
- Aiyegbusi, O.; McGregor, L.; McGeoch, L.; Kipgen, D.; Geddes, C.C.; Stevens, K.I. Renal Disease in Primary Sjögren’s Syndrome. Rheumatol Ther 2021, 8, 63–80. [CrossRef]
- Icardi, A.; Araghi, P.; Ciabattoni, M.; Romano, U.; Lazzarini, P.; Bianchi, G. [Kidney involvement in rheumatoid arthritis]. Reumatismo 2003, 55, 76–85. [CrossRef]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure-Function of the Immune System in the Spleen. Sci Immunol 2019, 4, eaau6085. [CrossRef]
- Tipton, C.M.; Fucile, C.F.; Darce, J.; Chida, A.; Ichikawa, T.; Gregoretti, I.; Schieferl, S.; Hom, J.; Jenks, S.; Feldman, R.J.; et al. Diversity, Cellular Origin and Autoreactivity of Antibody-Secreting Cell Expansions in Acute Systemic Lupus Erythematosus. Nat Immunol 2015, 16, 755–765. [CrossRef]
- Ehrenstein, M.R.; Katz, D.R.; Griffiths, M.H.; Papadaki, L.; Winkler, T.H.; Kalden, J.R.; Isenberg, D.A. Human IgG Anti-DNA Antibodies Deposit in Kidneys and Induce Proteinuria in SCID Mice. Kidney Int 1995, 48, 705–711. [CrossRef]
- Leadbetter, E.A.; Rifkin, I.R.; Hohlbaum, A.M.; Beaudette, B.C.; Shlomchik, M.J.; Marshak-Rothstein, A. Chromatin-IgG Complexes Activate B Cells by Dual Engagement of IgM and Toll-like Receptors. Nature 2002, 416, 603–607. [CrossRef]
- Hakkim, A.; Fürnrohr, B.G.; Amann, K.; Laube, B.; Abed, U.A.; Brinkmann, V.; Herrmann, M.; Voll, R.E.; Zychlinsky, A. Impairment of Neutrophil Extracellular Trap Degradation Is Associated with Lupus Nephritis. Proc Natl Acad Sci U S A 2010, 107, 9813–9818. [CrossRef]
- Santambrogio, L.; Marrack, P. The Broad Spectrum of Pathogenic Autoreactivity. Nat Rev Immunol 2023, 23, 69–70. [CrossRef]
- Trier, N.H.; Houen, G. Antibody Cross-Reactivity in Auto-Immune Diseases. International Journal of Molecular Sciences 2023, 24, 13609. [CrossRef]
- Lin, Y.-C.; Boone, M.; Meuris, L.; Lemmens, I.; Van Roy, N.; Soete, A.; Reumers, J.; Moisse, M.; Plaisance, S.; Drmanac, R.; et al. Genome Dynamics of the Human Embryonic Kidney 293 Lineage in Response to Cell Biology Manipulations. Nat Commun 2014, 5, 4767. [CrossRef]
- Jeon, J.-H.; Baek, I.-C.; Hong, C.-H.; Park, K.H.; Lee, H.; Oh, E.-J.; Kim, T.-G. Establishment of HLA Class I and MICA/B Null HEK-293T Panel Expressing Single MICA Alleles to Detect Anti-MICA Antibodies. Sci Rep 2021, 11, 15716. [CrossRef]
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential Transformation of Human Neuronal Cells by Human Adenoviruses and the Origin of HEK 293 Cells. FASEB J 2002, 16, 869–871. [CrossRef]
- Oka, Y.; Nakajima, K.; Nagao, K.; Miura, K.; Ishii, N.; Kobayashi, H. 293FT Cells Transduced with Four Transcription Factors (OCT4, SOX2, NANOG, and LIN28) Generate Aberrant ES-like Cells. J Stem Cells Regen Med 2010, 6, 149–156. [CrossRef]
- Chae, S.W.; Cho, E.-Y.; Park, M.S.; Lee, K.-B.; Kim, H.; Kim, U. Polycystin-1 Expression in Fetal, Adult and Autosomal Dominant Polycystic Kidney. J Korean Med Sci 2006, 21, 425–429. [CrossRef]
- Larrea, A.; Elexpe, A.; Díez-Martín, E.; Torrecilla, M.; Astigarraga, E.; Barreda-Gómez, G. Neuroinflammation in the Evolution of Motor Function in Stroke and Trauma Patients: Treatment and Potential Biomarkers. Current Issues in Molecular Biology 2023, 45, 8552–8585. [CrossRef]
- Mishra, M.N.; Baliga, K.V. Significance of Panel Reactive Antibodies in Patients Requiring Kidney Transplantation. Saudi J Kidney Dis Transpl 2013, 24, 495–499. [CrossRef]
- Friedewald, J.J.; Kurian, S.M.; Heilman, R.L.; Whisenant, T.C.; Poggio, E.D.; Marsh, C.; Baliga, P.; Odim, J.; Brown, M.M.; Ikle, D.N.; et al. Development and Clinical Validity of a Novel Blood-Based Molecular Biomarker for Subclinical Acute Rejection Following Kidney Transplant. Am J Transplant 2019, 19, 98–109. [CrossRef]
- Naesens, M.; Khatri, P.; Li, L.; Sigdel, T.K.; Vitalone, M.J.; Chen, R.; Butte, A.J.; Salvatierra, O.; Sarwal, M.M. Progressive Histological Damage in Renal Allografts Is Associated with Expression of Innate and Adaptive Immunity Genes. Kidney Int 2011, 80, 1364–1376. [CrossRef]
- Gibney, E.M.; Cagle, L.R.; Freed, B.; Warnell, S.E.; Chan, L.; Wiseman, A.C. Detection of Donor-Specific Antibodies Using HLA-Coated Microspheres: Another Tool for Kidney Transplant Risk Stratification **Portions of This Study Were Delivered as an Oral Presentation at the American Transplant Congress, Seattle, WA, 20–25 May 2005. Nephrology Dialysis Transplantation 2006, 21, 2625–2629. [CrossRef]
- Zhang, Q.; Reed, E.F. The Importance of Non-HLA Antibodies in Transplantation. Nat Rev Nephrol 2016, 12, 484–495. [CrossRef]
- Josephson, M.A. Monitoring and Managing Graft Health in the Kidney Transplant Recipient. Clin J Am Soc Nephrol 2011, 6, 1774–1780. [CrossRef]
- Han, H.S.; Lubetzky, M.L. Immune Monitoring of Allograft Status in Kidney Transplant Recipients. Frontiers in Nephrology 2023, 3.
- Guerra, G.; Srinivas, T.R.; Meier-Kriesche, H.-U. Calcineurin Inhibitor-Free Immunosuppression in Kidney Transplantation. Transplant International 2007, 20, 813–827. [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-Linked Immunosorbent Assay for the Quantitative/Qualitative Analysis of Plant Secondary Metabolites. J Nat Med 2018, 72, 32–42. [CrossRef]
- Elexpe, A.; Nieto, N.; Fernández-Cuétara, C.; Domínguez-Fernández, C.; Morera-Herreras, T.; Torrecilla, M.; Miguélez, C.; Laso, A.; Ochoa, E.; Bailen, M.; et al. Study of Tissue-Specific Reactive Oxygen Species Formation by Cell Membrane Microarrays for the Characterization of Bioactive Compounds. Membranes (Basel) 2021, 11, 943. [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem 1976, 72, 248–254. [CrossRef]
- Hebert-Chatelain, E.; Desprez, T.; Serrat, R.; Bellocchio, L.; Soria-Gomez, E.; Busquets-Garcia, A.; Pagano Zottola, A.C.; Delamarre, A.; Cannich, A.; Vincent, P.; et al. A Cannabinoid Link between Mitochondria and Memory. Nature 2016, 539, 555–559. [CrossRef]
- Manuel, I.; Barreda-Gómez, G.; González de San Román, E.; Veloso, A.; Fernández, J.A.; Giralt, M.T.; Rodríguez-Puertas, R. Neurotransmitter Receptor Localization: From Autoradiography to Imaging Mass Spectrometry. ACS Chem Neurosci 2015, 6, 362–373. [CrossRef]
| ID | Category | Anatomical Site | Sex | Age | Race | Procurement Type | Procurement Date |
|---|---|---|---|---|---|---|---|
| S6-32 Liver | Normal tissues | Liver | Male | 46 | Caucasian (White) | Autopsy | 18/03/2011 |
| S13-41 Spleen | Normal tissues | Spleen | Male | 48 | Caucasian (White) | Autopsy | 27/02/2012 |
| 90-M-13-28 Kidney, Cortex | Normal tissues | Kidney, Cortex and Medulla | Male | 44 | Caucasian (White) | Autopsy | 12/01/2015 |
| 4650955 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Female | 69 | Caucasian (White) | Autopsy | 02/05/2017 |
| 4641480 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Female | 70 | Caucasian (White) | Autopsy | 28/04/2017 |
| 4647875 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Female | 68 | Caucasian (White) | Autopsy | 27/04/2017 |
| 4638771 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Male | 60 | Caucasian (White) | Autopsy | 19/04/2017 |
| 4698698 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Female | 52 | Caucasian (White) | Autopsy | 17/08/2017 |
| 4645480 Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Male | 48 | Caucasian (White) | Autopsy | 18/04/2017 |
| 113-AFJF Kidney CM | Normal tissues | Kidney, Cortex and Medulla | Male | 60 | Caucasian (White) | Organ recovery | 08/10/2018 |
| A27 Kidney Medulla | Normal tissues | Kidney, Medulla | Male | 50 | Caucasian (White) | Autopsy | 14/04/2007 |
| S1-32 Kidney Medulla | Normal tissues | Kidney, Medulla | Male | 38 | Caucasian (White) | Autopsy | 18/02/2015 |
| S2-32 Kidney Medulla | Normal tissues | Kidney, Medulla | Female | 52 | Caucasian (White) | Autopsy | 24/02/2015 |
| 113-AGET292 Kidney Cortex | Normal tissues | Kidney, Cortex | Male | 41 | Caucasian (White) | Organ recovery | 24/05/2019 |
| 113-AHAM474 Kidney Cortex | Normal tissues | Kidney, Cortex | Male | 64 | Caucasian (White) | Organ recovery | 13/01/2020 |
| 4616286 Kidney CM | Normal tissues | Kidney, Cortex | Female | 46 | Caucasian (White) | Organ recovery | 24/02/2016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





