Submitted:
28 December 2023
Posted:
29 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
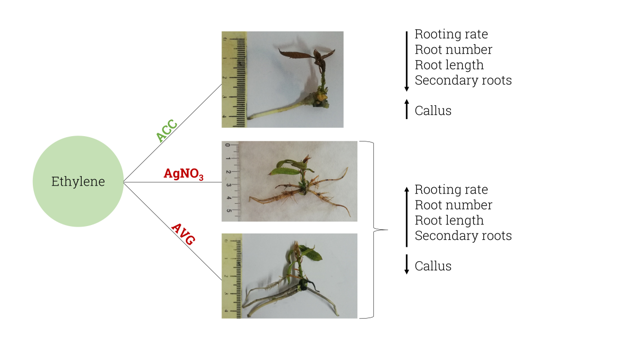
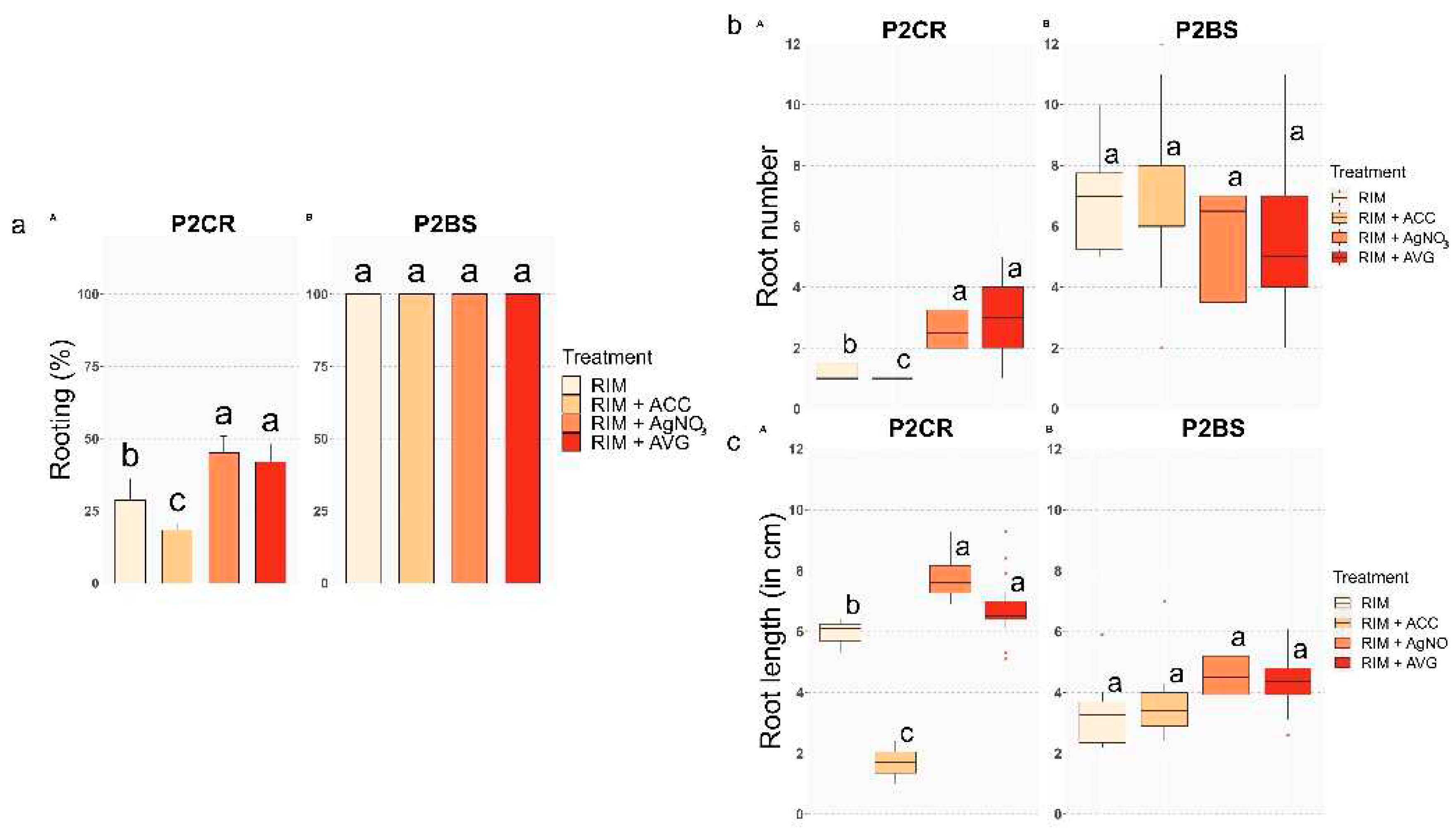
2.1. Rooting experiments
Ethylene negatively modulates adventitious rooting and worsens root system development in mature chestnut
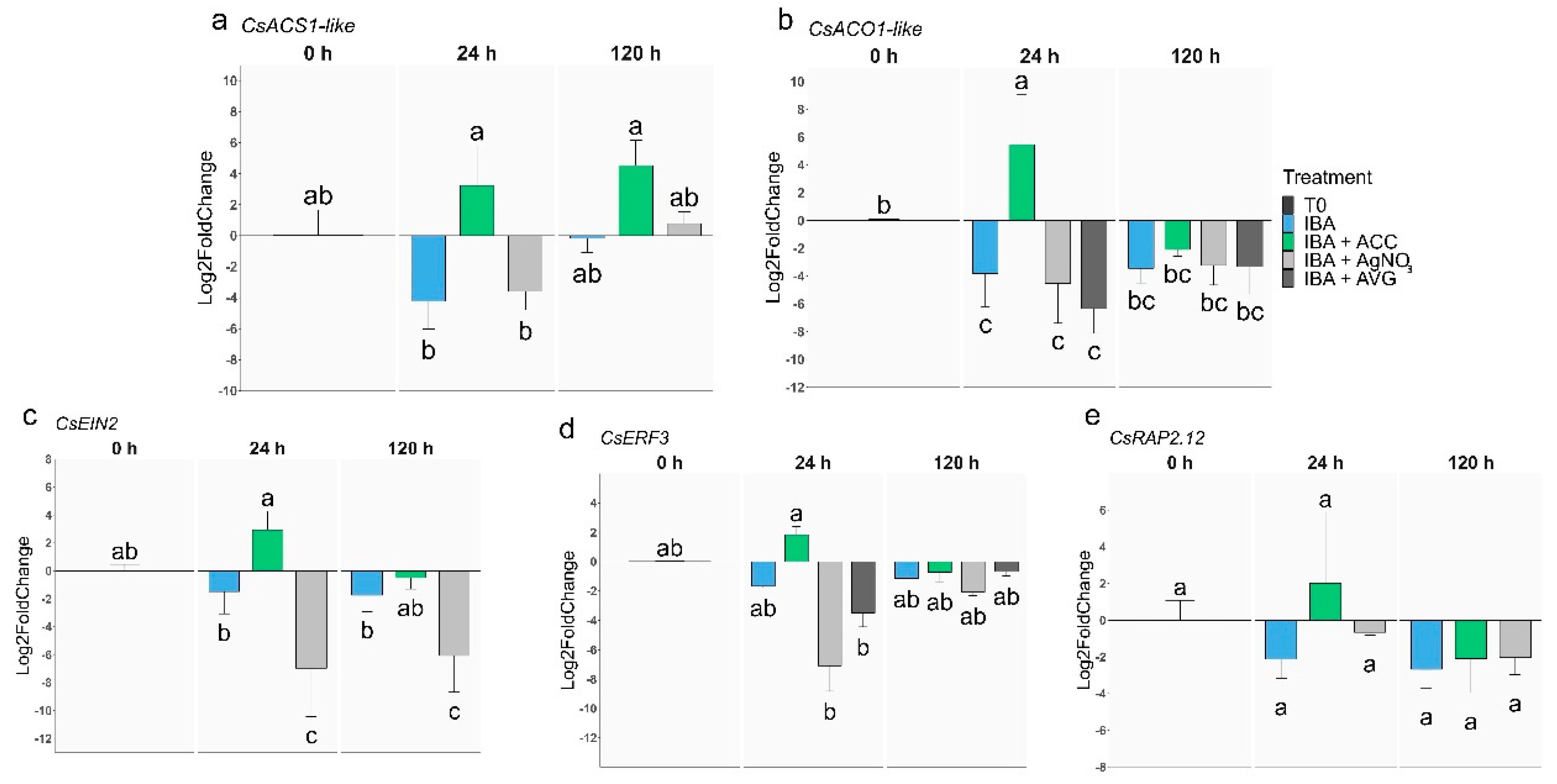
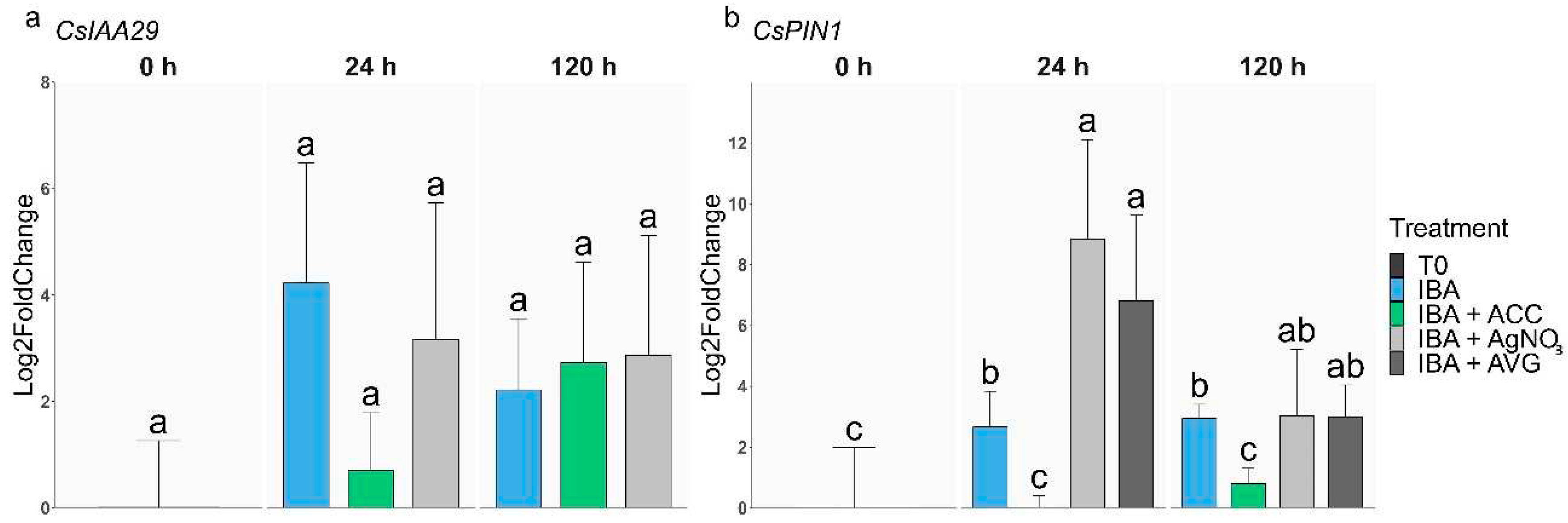
2.2. Analysis of gene expression during AR in rooting-recalcitrant shoots
2.2.1. Ethylene-related genes
2.2.2. Auxin-related genes
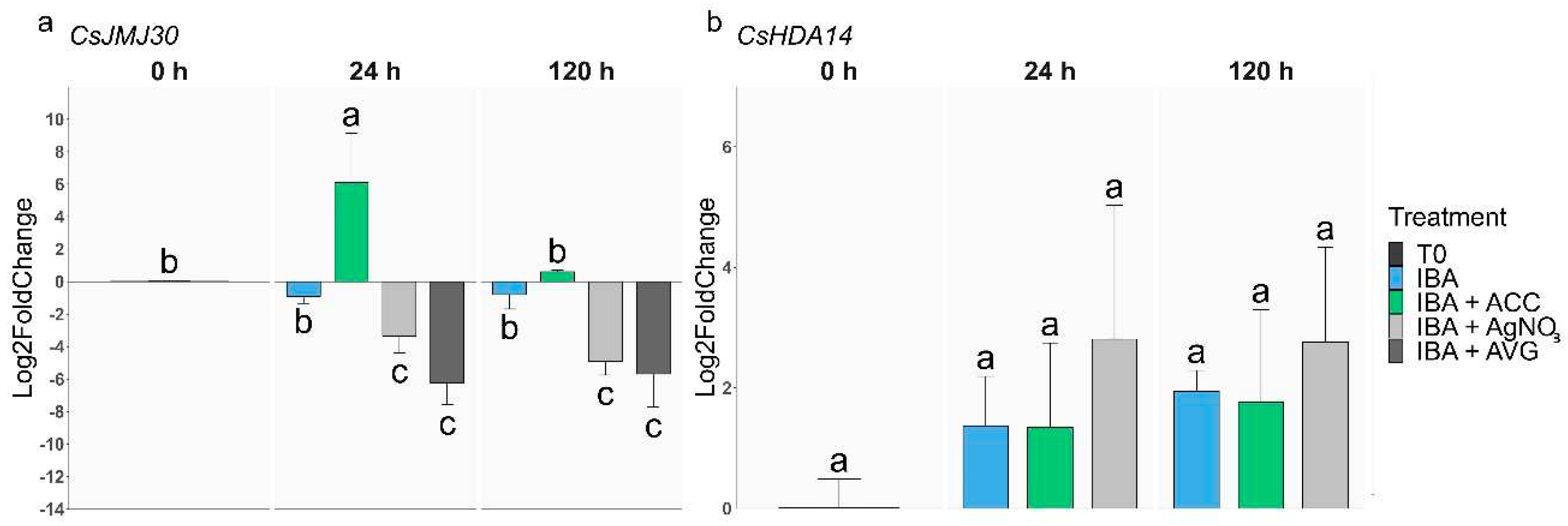
2.2.3. Epigenetic-related genes
3. Discussion
4. Materials and Methods
4.1. Plant material and culture conditions
4.2. Rooting experiments
4.3. RNA extraction and qPCR analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martín, M.A.; Mattioni, C.; Molina, J.R.; Alvarez, J.B.; Cherubini, M.; Herrera, M.A.; Villani, F.; Martín, L.M. Landscape genetic structure of chestnut (Castanea sativa Mill.) in Spain. Tree Genet. Genomes 2012, 8, 127–136. [CrossRef]
- Braga, N.; Rodrigues, F.; P.P. Oliveira, M.B. Castanea sativa by-products: a review on added value and sustainable application. Nat. Prod. Res. 2015, 29, 1–18. [CrossRef]
- Vega, A.; Dieste, A.; Guaita, M.; Majada, J.; Baño, V. Modelling of the mechanical properties of Castanea sativa Mill. structural timber by a combination of non-destructive variables and visual grading parameters. Eur. J. Wood Wood Prod. 2012, 70, 839–844. [CrossRef]
- Nunes, L.; Gower, S.; Monteiro, M.; Lopes, D.; Rego, F. Growth dynamics and productivity of pure and mixed Castanea sativa Mill. and Pseudotsuga menziesii (Mirb.) Franco plantations in northern Portugal. iForest - Biogeosciences For. 2014, 7, 92–102. [CrossRef]
- Guitián, J.; Guitián, P.; Magrach, A.; Docampo, C.; Domínguez, P.; Guitián, L. Effect of management and spatial characteristics on plant species richness of Castanea sativa Mill. woodlots in the NW Iberian Peninsula. J. For. Res. 2012, 17, 98–104. [CrossRef]
- Castaño-Santamaría, J.; Barrio-Anta, M.; Álvarez-Álvarez, P. Potential above ground biomass production and total tree carbon sequestration in the major forest species in NW Spain. Int. For. Rev. 2013, 15, 273–289. [CrossRef]
- Pardo-de-Santayana, M.; Tardío, J.; Blanco, E.; Carvalho, A.M.; Lastra, J.J.; San Miguel, E.; Morales, R. Traditional knowledge of wild edible plants used in the northwest of the Iberian Peninsula (Spain and Portugal): a comparative study. J. Ethnobiol. Ethnomed. 2007, 3, 27. [CrossRef]
- Martínez, S.; Ramil, P.; Chuvieco, E. Monitoring loss of biodiversity in cultural landscapes. New methodology based on satellite data. Landsc. Urban Plan. 2010, 94, 127–140. [CrossRef]
- Vielba; Vidal, N.; San-José, M.C.; Rico, S.; Sánchez, C.; José, M.C.S.; Rico, S.; Sánchez, C. Recent Advances in Adventitious Root Formation in Chestnut. Plants 2020, 9, 1543. [CrossRef]
- da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [CrossRef]
- Rasmussen, A.; Hosseini, S.A.; Hajirezaei, M.-R.; Druege, U.; Geelen, D. Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J. Exp. Bot. 2015, 66, 1437–1452. [CrossRef]
- Christiaens, A.; Gobin, B.; Van Huylenbroeck, J.; Van Labeke, M.-C. Adventitious rooting of Chrysanthemum is stimulated by a low red:far-red ratio. J. Plant Physiol. 2019, 236, 117–123. [CrossRef]
- Steffens, B.; Rasmussen, A. The Physiology of Adventitious Roots. Plant Physiol. 2016, 170, 603–617. [CrossRef]
- Zhang, Y.; Xiao, Z.; Zhan, C.; Liu, M.; Xia, W.; Wang, N. Comprehensive analysis of dynamic gene expression and investigation of the roles of hydrogen peroxide during adventitious rooting in poplar. BMC Plant Biol. 2019, 19, 99. [CrossRef]
- Husen, A.; Pal, M. Variation in Shoot Anatomy and Rooting Behaviour of Stem Cuttings in Relation to Age of Donor Plants in Teak (Tectona grandis Linn. f.). New For. 2006, 31, 57–73. [CrossRef]
- Aumond, M.L.; de Araujo, A.T.; de Oliveira Junkes, C.F.; de Almeida, M.R.; Matsuura, H.N.; de Costa, F.; Fett-Neto, A.G. Events Associated with Early Age-Related Decline in Adventitious Rooting Competence of Eucalyptus globulus Labill. Front. Plant Sci. 2017, 8. [CrossRef]
- Ballester, A.; San-José, M.C.; Vidal, N.; Fernández-Lorenzo, J.L.; Vieitez, A.M. Anatomical and Biochemical Events during in vitro Rooting of Microcuttings from Juvenile and Mature Phases of Chestnut. Ann. Bot. 1999, 83, 619–629. [CrossRef]
- de Almeida, M.R.; de Bastiani, D.; Gaeta, M.L.; de Araújo Mariath, J.E.; de Costa, F.; Retallick, J.; Nolan, L.; Tai, H.H.; Strömvik, M. V.; Fett-Neto, A.G. Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in Eucalyptus. Plant Sci. 2015, 239, 155–165. [CrossRef]
- Vielba, J.M.; Varas, E.; Rico, S.; Covelo, P.; Sánchez, C. Auxin-mediated expression of a GH3 gene in relation to ontogenic state in Chestnut. Trees 2016, 30, 2237–2252. [CrossRef]
- Vielba, J.M.; Rico, S.; Sevgin, N.; Castro-Camba, R.; Covelo, P.; Vidal, N.; Sanchez, C.; Sánchez, C. Transcriptomics Analysis Reveals a Putative Role for Hormone Signaling and MADS-Box Genes in Mature Chestnut Shoots Rooting Recalcitrance. Plants (Basel) 2022, 11, 3486. [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [CrossRef]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J.M. Plant Development and Crop Yield: The Role of Gibberellins. Plants 2022, 11, 2650. [CrossRef]
- Lischweski, S.; Muchow, A.; Guthörl, D.; Hause, B. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 2015, 15, 229. [CrossRef]
- Fattorini, L.; Hause, B.; Gutierrez, L.; Veloccia, A.; Della Rovere, F.; Piacentini, D.; Falasca, G.; Altamura, M.M. Jasmonate promotes auxin-induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 2018, 18, 182. [CrossRef]
- Li, C.; Bian, B.; Gong, T.; Liao, W. Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. J. Plant Physiol. 2018, 229, 185–194. [CrossRef]
- Haffner, V.; Enjalric, F.; Lardet, L.; Carron, M. Maturation of woody plants: a review of metabolic and genomic aspects. Ann. des Sci. For. 1991, 48, 615–630. [CrossRef]
- Schaller, G.E. Ethylene and the regulation of plant development. BMC Biol. 2012, 10, 9. [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 08. [CrossRef]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings. Front. Plant Sci. 2016, 7, 381. [CrossRef]
- Qin, H.; He, L.; Huang, R. The Coordination of Ethylene and Other Hormones in Primary Root Development. Front. Plant Sci. 2019, 10. [CrossRef]
- Neves, M.; Correia, S.; Cavaleiro, C.; Canhoto, J. Modulation of Organogenesis and Somatic Embryogenesis by Ethylene: An Overview. Plants 2021, 10, 1208. [CrossRef]
- Gonin; Bergougnoux; Nguyen; Gantet; Champion What Makes Adventitious Roots? Plants 2019, 8, 240. [CrossRef]
- Maury, S.; Sow, M.D.; Le Gac, A.L.; Genitoni, J.; Lafon-Placette, C.; Mozgova, I. Phytohormone and Chromatin Crosstalk: The Missing Link For Developmental Plasticity? Front Plant Sci 2019, 10, 395. [CrossRef]
- Moore, B.M.; Lee, Y.S.; Wang, P.; Azodi, C.; Grotewold, E.; Shiu, S.-H. Modeling temporal and hormonal regulation of plant transcriptional response to wounding. Plant Cell 2022, 34, 867–888. [CrossRef]
- Park, S.-H.; Elhiti, M.; Wang, H.; Xu, A.; Brown, D.; Wang, A. Adventitious root formation of in vitro peach shoots is regulated by auxin and ethylene. Sci. Hortic. (Amsterdam). 2017, 226, 250–260. [CrossRef]
- Kilkenn, A.J.; Wallace, H.M.; Walton, D.A.; Adkins, M.F.; Trueman, S.J. Improved Root Formation in Eucalypt Cuttings Following Combined Auxin and Anti-ethylene Treatments. J. Plant Sci. 2012, 7, 138–153. [CrossRef]
- Ma, J.-H.; Yao, J.-L.; Cohen, D.; Morris, B. Ethylene inhibitors enhance in vitro root formation from apple shoot cultures. Plant Cell Rep. 1998, 17, 211–214. [CrossRef]
- Druege, U.; Franken, P.; Lischewski, S.; Ahkami, A.H.; Zerche, S.; Hause, B.; Hajirezaei, M.R. Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Front. Plant Sci. 2014, 5. [CrossRef]
- Deng, Y.; Wang, C.; Zhang, M.; Wei, L.; Liao, W. Identification of Key Genes during Ethylene-Induced Adventitious Root Development in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 12981. [CrossRef]
- Jin, X.; Liao, W.-B.; Yu, J.-H.; Ren, P.-J.; Dawuda, M.M.; Wang, M.; Niu, L.-J.; Li, X.-P.; Xu, X.-T. Nitric Oxide Is Involved in Ethylene-Induced Adventitious Rooting in Marigold. Can. J. Plant Sci. 2017, CJPS-2016-0156. [CrossRef]
- Mori, Y.; Miyahara, F.; Tsutsumi, Y.; Kondo, R. Effects of combinational treatment with ethephon and indole-3-butyric acid on adventitious rooting of Pinus thunbergii cuttings. Plant Growth Regul. 2011, 63, 271–278. [CrossRef]
- Mendes, A.F.S.; Cidade, L.C.; Otoni, W.C.; Soares-Filho, W.S.; Costa, M.G.C. Role of auxins, polyamines and ethylene in root formation and growth in sweet orange. Biol. Plant. 2011, 55, 375–378. [CrossRef]
- Li, H.; Yao, L.; Sun, L.; Zhu, Z. ETHYLENE INSENSITIVE 3 suppresses plants de novo root regeneration from leaf explants and mediates age-regulated regeneration decline. Development 2020. [CrossRef]
- Negi, S.; Ivanchenko, M.G.; Muday, G.K. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 2008, 55, 175–187. [CrossRef]
- Park, C.H.; Roh, J.; Youn, J.H.; Son, S.H.; Park, J.H.; Kim, S.Y.; Kim, T.W.; Kim, S.K. Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Mol. Cells 2018, 41, 923–932. [CrossRef]
- Negi, S.; Sukumar, P.; Liu, X.; Cohen, J.D.; Muday, G.K. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 2010, 61, 3–15. [CrossRef]
- Polko, J.K.; Kieber, J.J. 1-Aminocyclopropane 1-Carboxylic Acid and Its Emerging Role as an Ethylene-Independent Growth Regulator. Front. Plant Sci. 2019, 10. [CrossRef]
- Li; Mou, W.; Van de Poel, B.; Chang, C. Something old, something new: Conservation of the ethylene precursor 1-amino-cyclopropane-1-carboxylic acid as a signaling molecule. Curr. Opin. Plant Biol. 2022, 65, 102116. [CrossRef]
- Betti, C.; Della Rovere, F.; Ronzan, M.; Fattorini, L. EIN2 and COI1 control the antagonism between ethylene and jasmonate in adventitious rooting of Arabidopsis thaliana thin cell layers. Plant Cell, Tissue Organ Cult. 2019, 138, 41–51. [CrossRef]
- Zhang; Wang, L.; Qi, B.; Zhao, B.; Ko, E.E.; Riggan, N.D.; Chin, K.; Qiao, H. EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc. Natl. Acad. Sci. 2017, 114, 10274–10279. [CrossRef]
- Trupiano, D.; Yordanov, Y.; Regan, S.; Meilan, R.; Tschaplinski, T.; Scippa, G.S.; Busov, V. Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 2013, 238, 271–282. [CrossRef]
- Wang; Pak, S.; Yang, J.; Wu, Y.; Li, W.; Feng, H.; Yang, J.; Wei, H.; Li, C. Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis. Plant Biotechnol. J. 2022, 20, 1561–1577. [CrossRef]
- Valladares, S.; Varas, E.; Vielba, J.M.; Vidal, N.; Codesido, V.; Castro, R.; Sanchez, C. Expression of a Rap2.12 like-1 ERF gene during adventitious rooting of chestnut and oak microshoots. Isr. J. Plant Sci. 2020, 67, 69–82. [CrossRef]
- Papdi, C.; Pérez-Salamó, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [CrossRef]
- Veloccia, A.; Fattorini, L.; Della Rovere, F.; Sofo, A.; D’Angeli, S.; Betti, C.; Falasca, G.; Altamura, M.M. Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6445–6458. [CrossRef]
- Li Molecular Bases for the Regulation of Adventitious Root Generation in Plants. Front. Plant Sci. 2021, 12. [CrossRef]
- Guan, L.; Li, Y.; Huang, K.; Cheng, Z.-M. Auxin regulation and MdPIN expression during adventitious root initiation in apple cuttings. Hortic. Res. 2020, 7, 143. [CrossRef]
- Hu; Liu, X.; Xuan, W.; Mei, H.; Li, J.; Chen, X.; Zhao, Z.; Zhao, Y.; Jeyaraj, A.; Periakaruppan, R.; et al. Genome-wide identification and characterization of PIN-FORMED (PIN) and PIN-LIKES (PILS) gene family reveals their role in adventitious root development in tea nodal cutting (Camellia Sinensis). Int. J. Biol. Macromol. 2023, 229, 791–802. [CrossRef]
- Neves, M.; Correia, S.; Canhoto, J. Ethylene Inhibition Reduces De Novo Shoot Organogenesis and Subsequent Plant Development from Leaf Explants of Solanum betaceum Cav. Plants 2023, 12, 1854. [CrossRef]
- Li; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z.; Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z.; Li; Xie, Z.-Z.; et al. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [CrossRef]
- Tang, Y.; Wang, L.; Qu, Z.; Huang, C.; Zhao, T.; Li, Y.; Zhang, C. BSISTER transcription factors directly binds to the promoter of IAA19 and IAA29 genes to up-regulate gene expression and promote the root development. Plant Sci. 2022, 321, 111324. [CrossRef]
- Chang, K.N.; Zhong, S.; Weirauch, M.T.; Hon, G.; Pelizzola, M.; Li, H.; Huang, S.C.; Schmitz, R.J.; Urich, M.A.; Kuo, D.; et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2013, 2. [CrossRef]
- Lee, K.; Park, O.; Seo, P.J. JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 2018, 95, 961–975. [CrossRef]
- Wu, J.; Yamaguchi, N.; Ito, T. Histone demethylases control root elongation in response to stress-signaling hormone abscisic acid. Plant Signal. Behav. 2019, 14, 1604019. [CrossRef]
- Sanchez, M.C.; Vieitez, A.M. In vitro morphogenetic competence of basal sprouts and crown branches of mature chestnut. Tree Physiol. 1991, 8, 59–70. [CrossRef]
- Gresshoff, P.M.; Doy, C.H. Development and differentiation of haploid Lycopersicon esculentum (tomato). Planta 1972, 107, 161–170. [CrossRef]
- R Core Team R: A language and environment for statistical computing. 2021.
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




