Introduction
Brucella belongs to the Brucellaceae family, the orders Rhizobiales, Daeguia, Crabtreella, Mycoplana, Pseudochrobactrum, and Paenochrobactrum are all members of the Alpha proteobacteria class1. It is heat sensitive and can survive for several weeks in water. The survival of Brucella abortus in the environment is influenced by sunlight and temperature2. Bacterial development and diversity are linked to organic matter; thus, microbial numbers are greatest at the soil's surface (10 cm) and decrease with depth3. Identify different ecotypes and the potential for new strains, giving them insight into the environmental maintenance and transmission of emerging and re-emerging disease risks in Iraq. The presence of Brucella species in soil is one of the risk factors for the disease's spread to animals and humans4. Brucellosis is a zoonotic infection that can affect both animals and humans and it is still prevalent in the United States5. Brucella species are pathogenic bacteria that adapt to new hosts and are naturally transmitted to their primary hosts by direct or indirect contact, as well as to other vulnerable hosts unwittingly6. The use of cows, buffaloes, sheep, and goats in mixed farming has increased the risk of brucellosis with small ruminants acting as primary hosts and cattle acting as an overflow host for B. melitensis7. The current study investigates the association between soil risk variables (physical and chemical) and the occurrence of Brucella species. the epidemiological relevance of brucellosis in terms of risk of transmission to humans and cattle8. Seiler & Berendonk, (2012)9 showed that the risk of metal resistance in the environment was assessed based on heavy metal concentrations, Analyses of the data indicate that agricultural and aquacultural practices represent major sources of soil and water contamination with moderately to highly toxic metals such as mercury (Hg), cadmium (Cd), copper (Cu), and zinc (Zn). If those metals reach the environment and accumulate to critical concentrations, they can trigger co-selection of antibiotic resistance. Li et al.,(2013)10 found that incidence of brucellosis were strongly associated with lower temperatures and less sunshine in the winter and spring, climatic factors likely influence the ecology of brucellosis both directly and indirectly by affecting several parameters, including the growth and reproduction dynamics of domestic animals, interactions between sheep, goats and humans, pathogen replication, and population immunity11. Ahmed et al., (2017)12 discovered DNA of Brucella species in soil samples (n=1280) from nine districts in Punjab.
Material and methods
The Soil Samples Collection
The animals soil samples collected seasonally from February to November, 2021 were taken the samples from the upper layer for bacteriological study and from the deepest layer (25-30 cm in depth) for environmental study, dried by air and sieved (to ~ 2 mm particle size) during the collecting the samples measured air temperature and soil temperature and determination the sites by GPS.
Total Organic Carbon is the wet oxidation technique was utilized exothermic heating and oxidation of organic carbon of the sample with potassium dichromate by the titration13, Heavy Metals include soils samples for heavy metals determination were digested according to the procedure described Sharidah (1999)14. One gram of dried soil samples were digested and determined with by Flame Atomic absorption - spectrophotometer (Type Aa 7000),Shimdzu /Japan was used to determine the concentrations (mg/L) of the elements15.
Preparation of Culture Media:
Culture media used in this study were prepared according to the manufacturer’s instructions:
Brucella Agar Base
It was prepared according to Himedia manufacturing company (India) to Isolation and detection of Brucella spp: suspend(21.55gm) in (500ml) D.W,heating to boiling to dissolve the medium completely. sterilize by autoclaving at (150C) Ibs pressure (1210C) for 15 minutes, Cool to (45-500C) and aseptically add sterile (5%) v\v inactivated horse serum (RM 1239, inactivated by heating at (560C) for (30 minutes) and then add the antibiotics (Polymxin B sulphate, Vancomycin, Bacitracin, Nystatin, Nalidixic acid, Cycloheximide) as supplement.
Brain Heart Infusion Broth (BHI)
This media were prepared according to the manufacturer’s instruction. It was used for preservation of bacterial isolates as stock for long time16.
Brucella isolation from soil samples
After samples collection in sterile bags from sites taken (1gm) from soil's sample and put in plain tube completed the size to (5ml) by normal saline then mixed by vortex and leave to settle after that incubation for (15) minute in Incubator. Two hundred (200 μl) were taken from mixing by micropipette and published in petridishs where all samples subcultured in selective Brucella agar base media with Polymyxin B (2,500IU), Vancomycin(10.0mg), Bacitracin (12,500IU), Cycloheximide(50.0mg), Nystatin(50,000 IU) and Nalidixic acid (2.5mg) and then5% of inactivated horse serum. Bacterial cultures were incubated for 14 days at (37°C) and 10% carbon dioxide until appearance of growth17,18,19.
Molecular diagnostic methods
Methods are also currently being used for the detection of Brucella spp. in various samples20.
Bacterial DNA Extraction:
Genomic DNA extracted from bacterial isolates cultured from the soil according to the manufacturer's protocol FavorPrepTM Blood/ Cultured Cells Genomic DNA Extraction Mini Kit.
Conventional PCR for Bacteria Extracted from Soil
Conventional PCR were used to amplify the target bacterial DNA using specific primer pairs. It includes three consecutive steps that repeated for specific number of cycles to get PCR product which can be finally visualized after agarose gel electrophoresis. The primer sequence, PCR product size and thermal cycling conditions mentioned in (table 1-1) and (table 1-2).
Table 1-1.
The Sequence of Primers.
Table 1-1.
The Sequence of Primers.
| Primer |
Sequence |
Primer sequence |
Tm
(ᵒC)
|
GC% |
Size of Product (bp) |
16s rRNA
bacterial primers
|
27F
|
5'- AGAGTTTGATCCTGGCTCAG- 3' |
54.3 |
50.0 |
1250
Srinivasan et al., (2015)21
|
| 1392R |
5'- GGTTACCTTGTTACGACTT- 3’ |
49.4 |
42.1 |
Table 1-2.
The Optimum Conditions for Detection the Bacterial Isolates from Soil (Stages and Temperature of PCR for 16s rRNA gene).
Table 1-2.
The Optimum Conditions for Detection the Bacterial Isolates from Soil (Stages and Temperature of PCR for 16s rRNA gene).
| |
|
Temperature 0C |
Time |
cycle |
| Stage 1 |
Initial Denaturation |
95ºC |
5 min |
1 |
| Stage 2 |
Denaturation |
95ºC |
45 sec |
|
| |
annealing |
56ºC |
45 sec |
35 |
| |
Extension |
72ºC |
1 min |
|
| Stage 3 |
Final Extension |
72ºC |
5 min |
1 |
16S rRNA Sequence Analysis and Phylogenetic Tree
Sequencing method was performed for study of genetic changes and phylogenetic tree draw of 16SrRNA gene in some local Brucella isolates by comparing with NCBI-GenBank Brucella isolates. The sequencing of the 16SrRNA gene were done after assurance in presence amplification of PCR products for required volume; These products were sent to company (Macrogen) in Korea for performing Sanger sequence, after getting on nitrogenous bases sequence for 16S rRNA gene amplified products of Brucella isolates. This sequence analyzed by NCBI-blast programme for purpose compared homology or diversity degree to local Brucella isolates with the world isolates recorded in NCBI-GenBank. Also phylogenetic analysis for draw of phylogenetic tree and determination phylogenetic relationship used (MEGA 6) programme to compare local one strain with strains all of the world states.
Recording of Iraqi Brucella isolates in gene bank –NCBI
Sequences of Brucella isolates were isolated from animals soils in Al-Hilla city\Iraq and each sequences have variations.
Results
Soil Samples
The current study results showed that Brucella spp. was found in all seasons,
Genetic Detection
Conventional PCR for Bacterial Isolates and 16SrRNA Sequencing
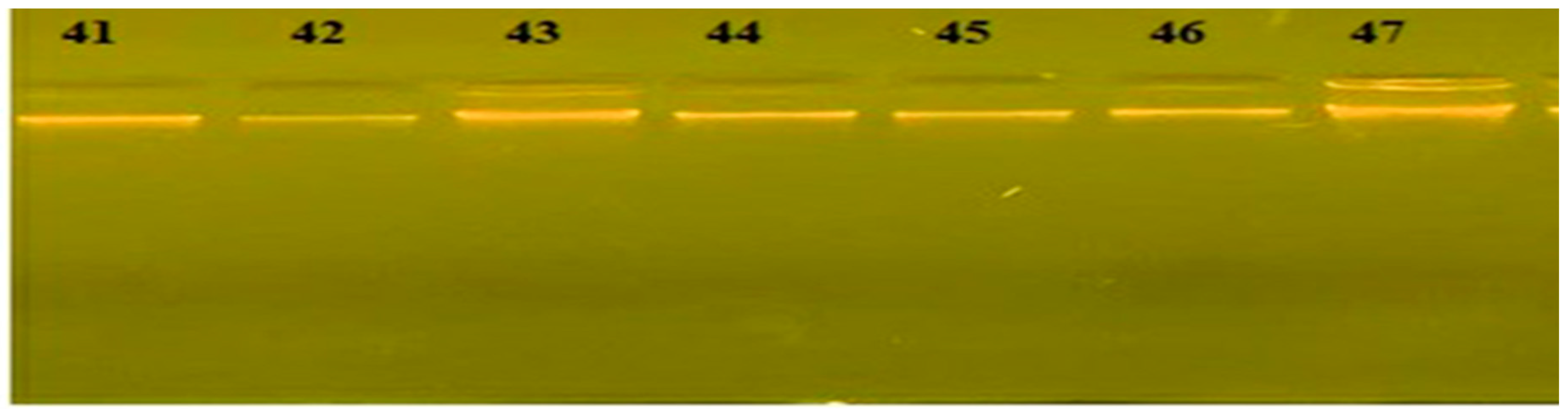
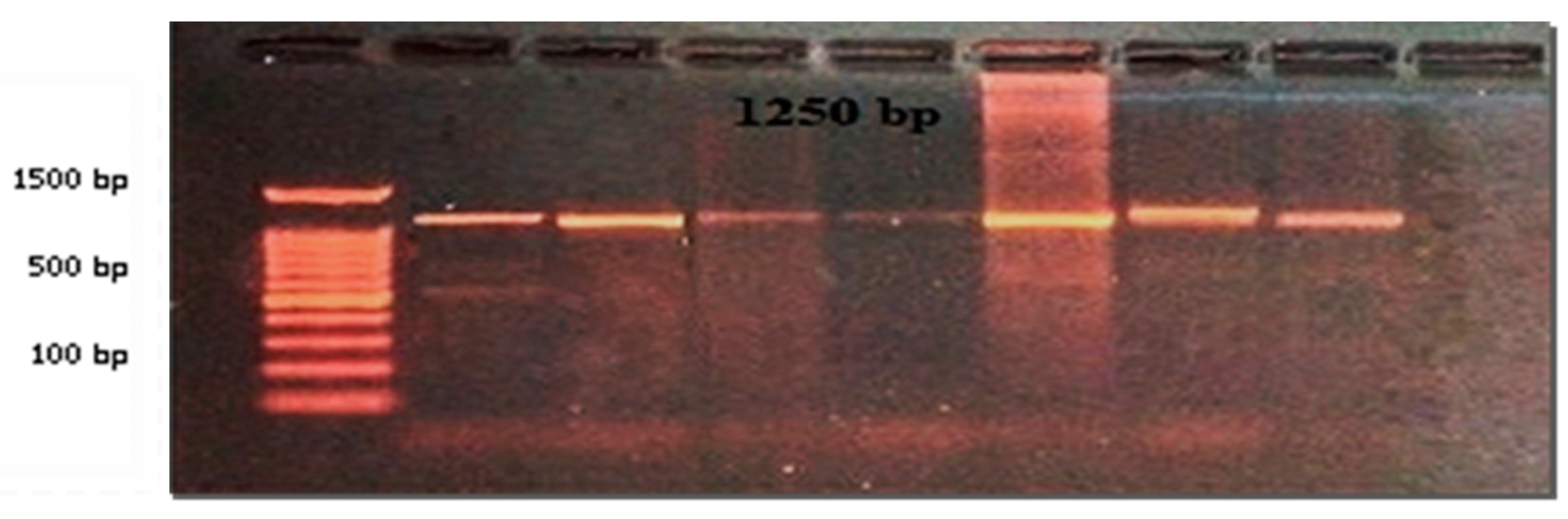
After the culture for both samples from the soil on Brucella agar base media and DNA extraction from bacterial isolates and electrophoresis of DNA as in Figure (1-1) after that, identified the positive samples by conventional PCR to knowledge Brucella sp., as in Figure (1-2).
Figure (1-1).
Gel electrophoresis for Extracted DNA from soil bacterial isolates, (Agarose 1%, at 70 volts, 60 min). Visualized after staining with ethidium bromide stain.
Figure (1-1).
Gel electrophoresis for Extracted DNA from soil bacterial isolates, (Agarose 1%, at 70 volts, 60 min). Visualized after staining with ethidium bromide stain.
Figure (1-2).
Agarose gel electrophoresis (1%) for 16s-rRNA bacterial primer-bacteria isolated from the soil samples (1250 bp), Primer Ta at (560C ), at (65Amp ,70 volts, 60min). It was visualized under U.V light after staining with Eco Safe dye , Lane M 100 bp DNA Ladder.
Figure (1-2).
Agarose gel electrophoresis (1%) for 16s-rRNA bacterial primer-bacteria isolated from the soil samples (1250 bp), Primer Ta at (560C ), at (65Amp ,70 volts, 60min). It was visualized under U.V light after staining with Eco Safe dye , Lane M 100 bp DNA Ladder.
After Sequencing analysis for both bacteria isolated from soil and blood samples found that Brucella isolates from soil samples for the four seasons and sented for sequencing was 6 new strains recorded in GenBank mentioned in table (1-3). These samples divided to (5) new strains for Brucella intermedia in Summer and in Autumn, one species of Brucella rhizosphaerae in Spring.
Table 1-3.
Brucella isolates recorded in GenBank for Brucella spp. isolated from soil.
Table 1-3.
Brucella isolates recorded in GenBank for Brucella spp. isolated from soil.
The results of 16SrRNA sequencing revealed the presence variation for Brucella isolates from soil established (6 new strains) for two species, indicating the presence of genetic diversity among the isolates.
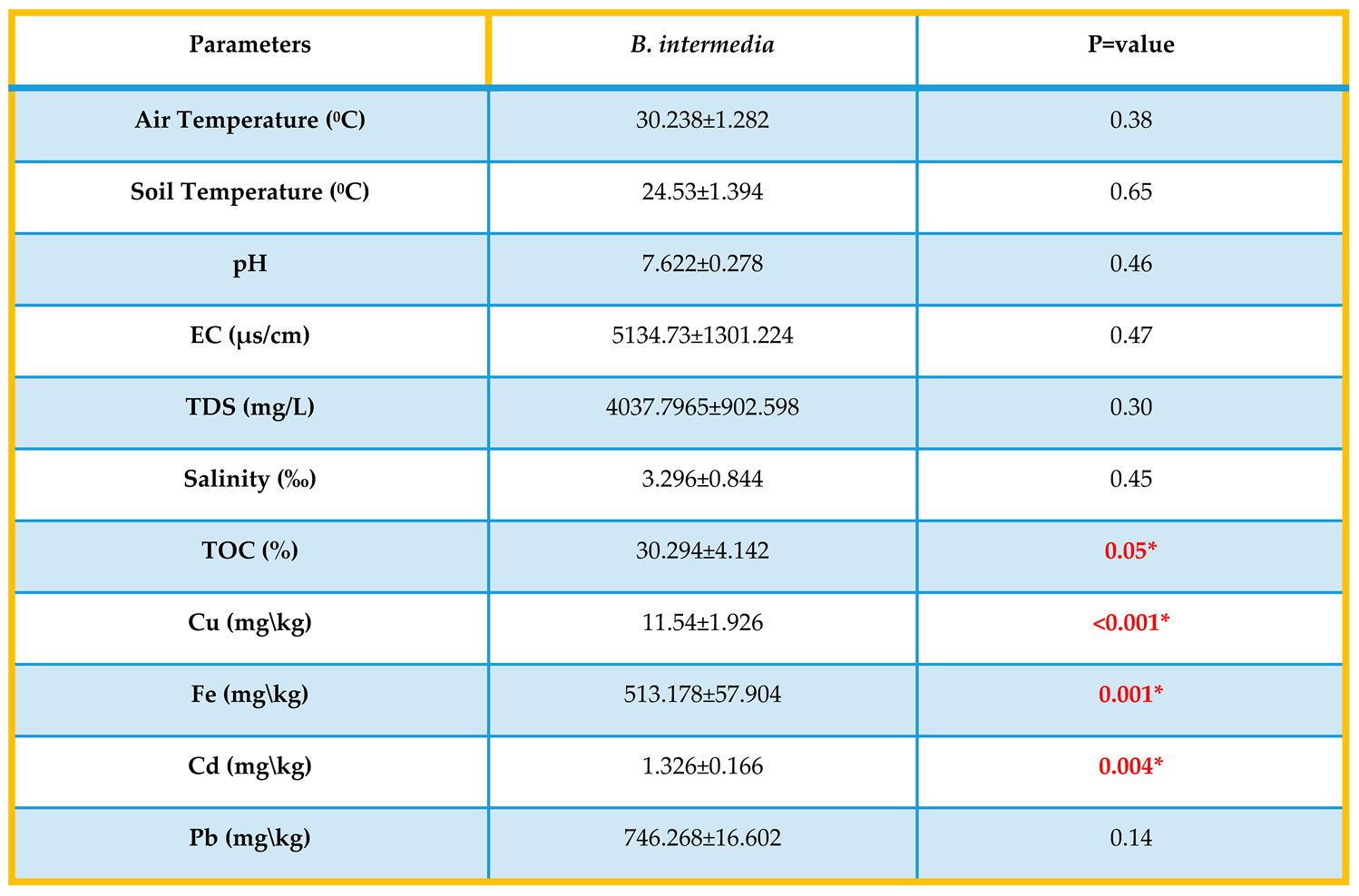
Table (1-4).
The relationship between B.intermedia and environmental factors, Mean ±S.E. p≤0.05.
Table (1-4).
The relationship between B.intermedia and environmental factors, Mean ±S.E. p≤0.05.
This table (1-4) showed that all isolates have significant differences (p<0.05) between genotype and environmental factors that included physical and chemical parameters (air and soil temperature, pH, EC, TDS, salinity, TOC) and heavy metals (Cu, Fe, Cd, Pb) according to statistical analysis.
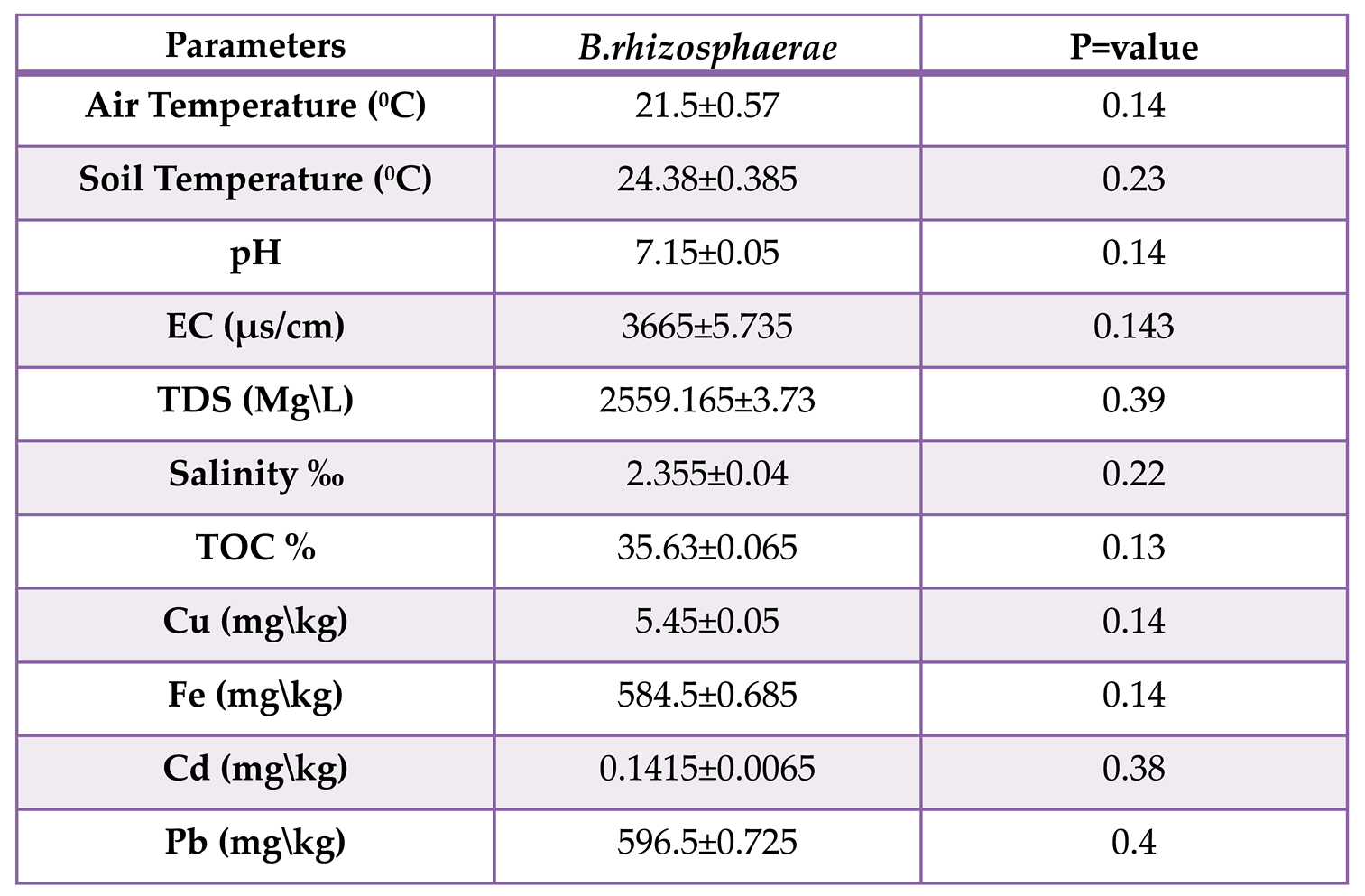
Table (1-5).
The relationship between B.rhizosphaerae and environmental factors, Mean ±S.E. p≤0.05.
Table (1-5).
The relationship between B.rhizosphaerae and environmental factors, Mean ±S.E. p≤0.05.
While Brucella rhizosphaerae mentioned in tables (1-5) do not exhibit significant differences (p>0.05) between genotype and environmental parameters, this suggests that genetic diversity is present but has no impact on the makeup of the bacterium. B.rhizosphaerae founded one in the spring season.
Phylogenetic Tree Draw
Sanger sequencing was successful for soil samples with an expected PCR product size (1250 bp) by 16S rRNA bacterial primers. The sequences were identified as belonging to Brucella spp. following similarity searched by blast (sequence identity of 99%). 16S rRNA sequencing method was performed for study of genetic changes and phylogenetic tree draw of 16SrRNA gene in some local Brucella species isolates by compared with NCBI-GenBank Brucella species. The sequences were deposited in GenBank with accession numbers as mentioned in table (1-6).
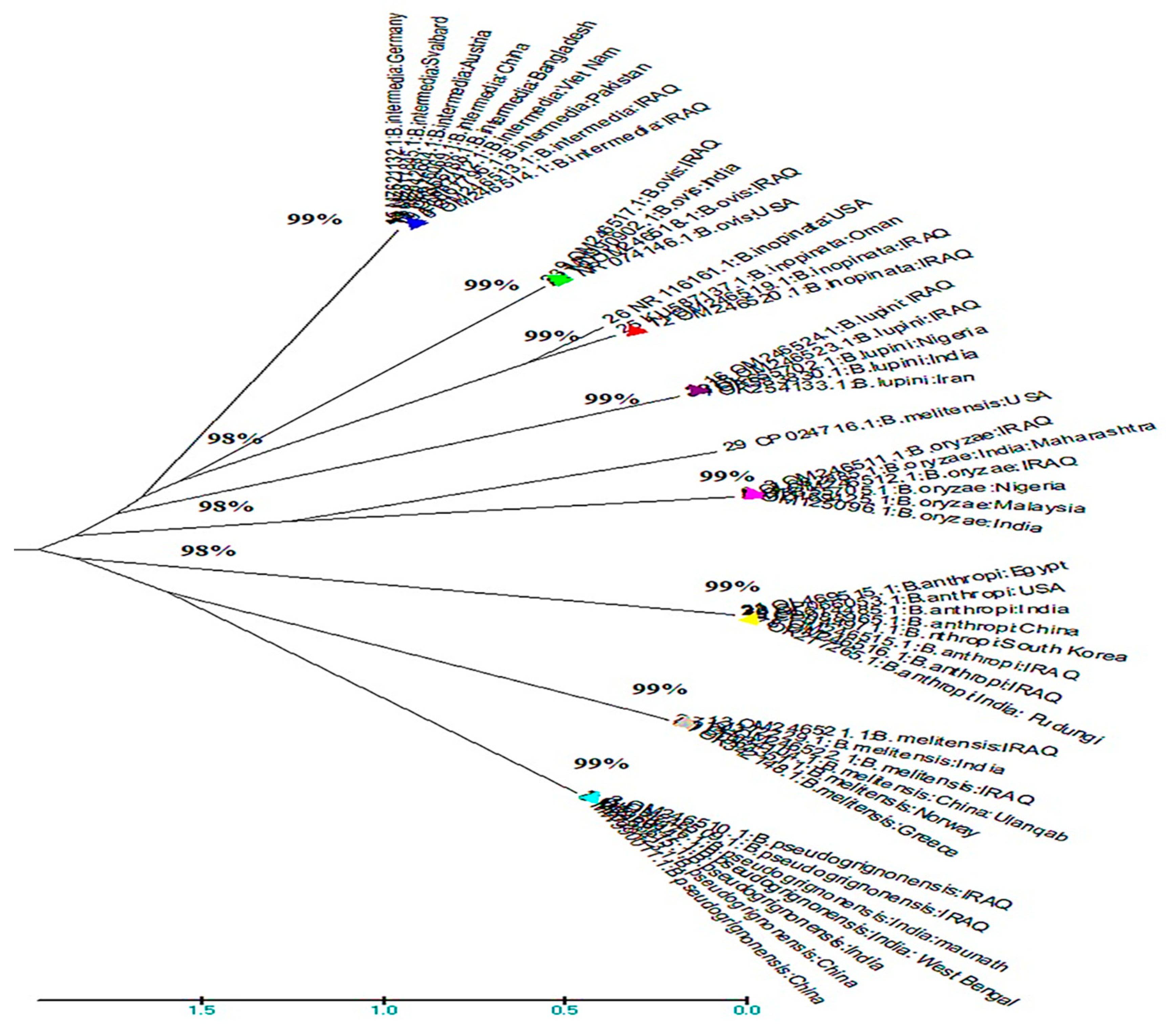
Figure 1-3.
Evolutionary analysis (phylogenetic tree of Brucella isolates isolated from soil)) of 16S rRNA gene sequences at compared of B.intermedia with different states. This figure(1-3) found that B.intermedia was nearest to Pakistan.
Figure 1-3.
Evolutionary analysis (phylogenetic tree of Brucella isolates isolated from soil)) of 16S rRNA gene sequences at compared of B.intermedia with different states. This figure(1-3) found that B.intermedia was nearest to Pakistan.
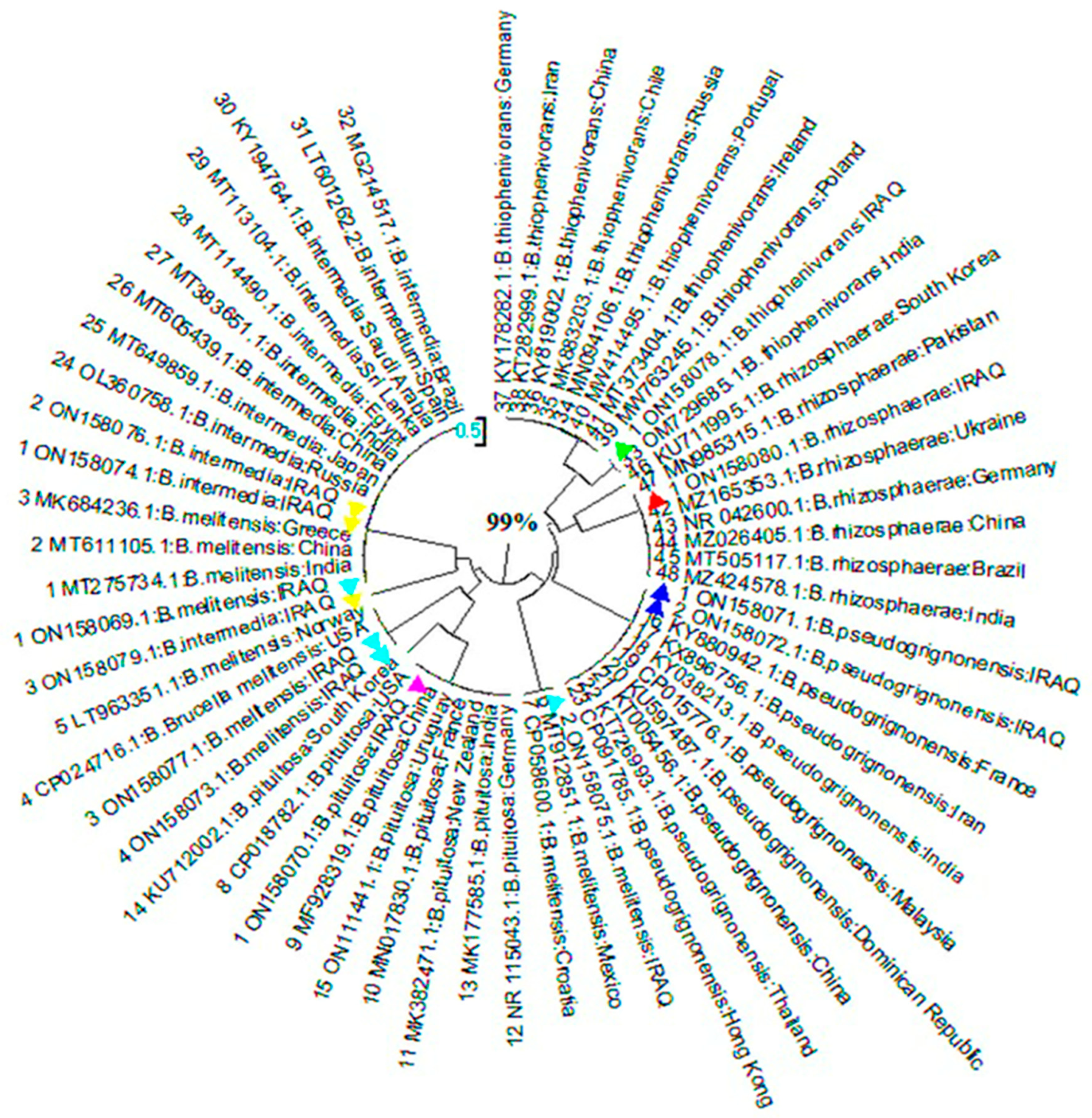
Figure (1-4).
Evolutionary analysis (phylogenetic tree of Brucella isolates isolated from soil) of 16S rRNA gene sequences of B.intermedi and B.rhizosphaerae at compared with different state. This figure(1-4) found that B.intermedia was nearest to Russia while B.rhizosphaerae was nearest to Ukraine and Pakistan.
Figure (1-4).
Evolutionary analysis (phylogenetic tree of Brucella isolates isolated from soil) of 16S rRNA gene sequences of B.intermedi and B.rhizosphaerae at compared with different state. This figure(1-4) found that B.intermedia was nearest to Russia while B.rhizosphaerae was nearest to Ukraine and Pakistan.
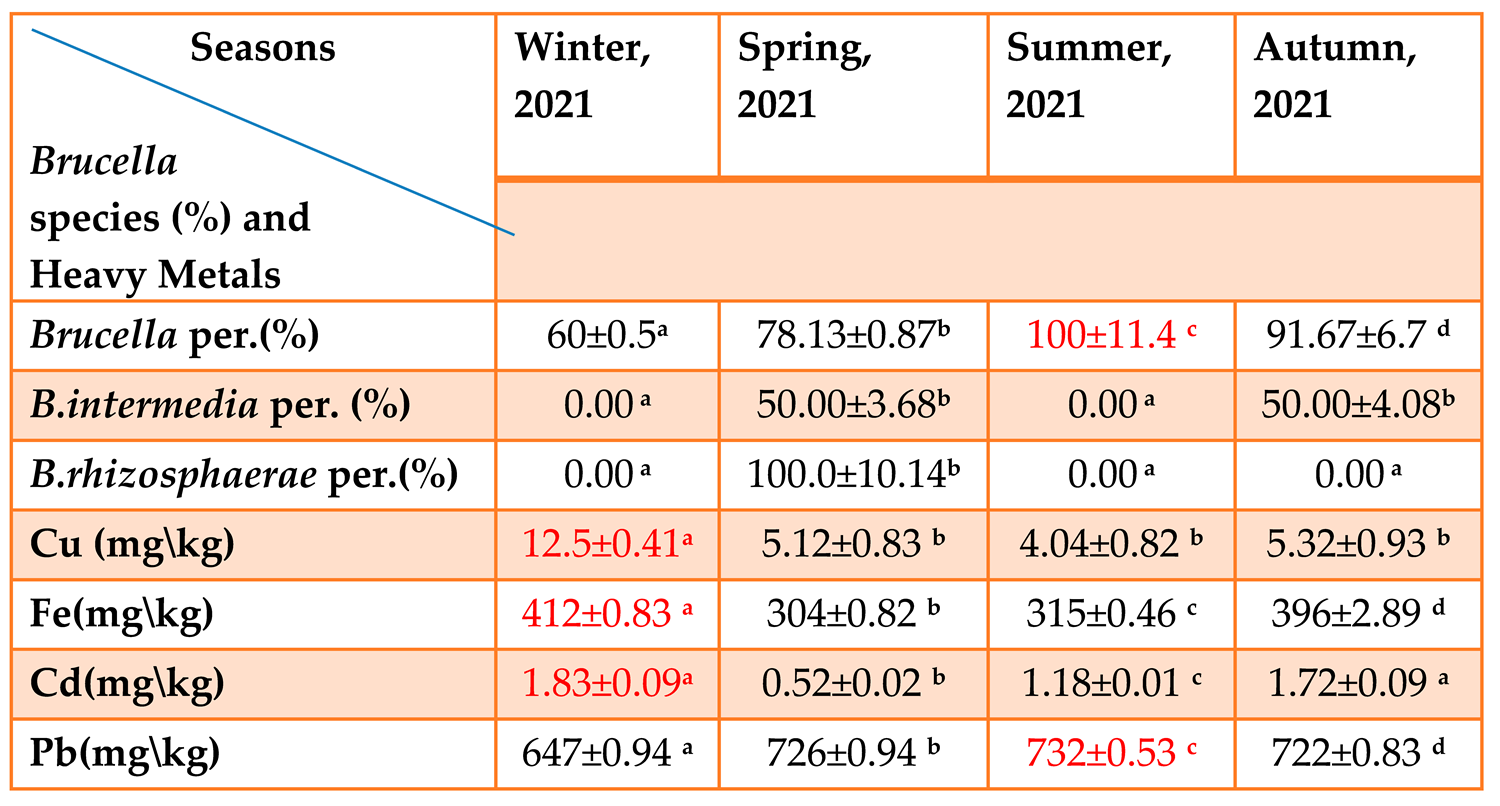
The Relationship between the Environmental Factors and Brucella species
Our results proved existence relation between the presence of Brucella and the physical, chemical properties.
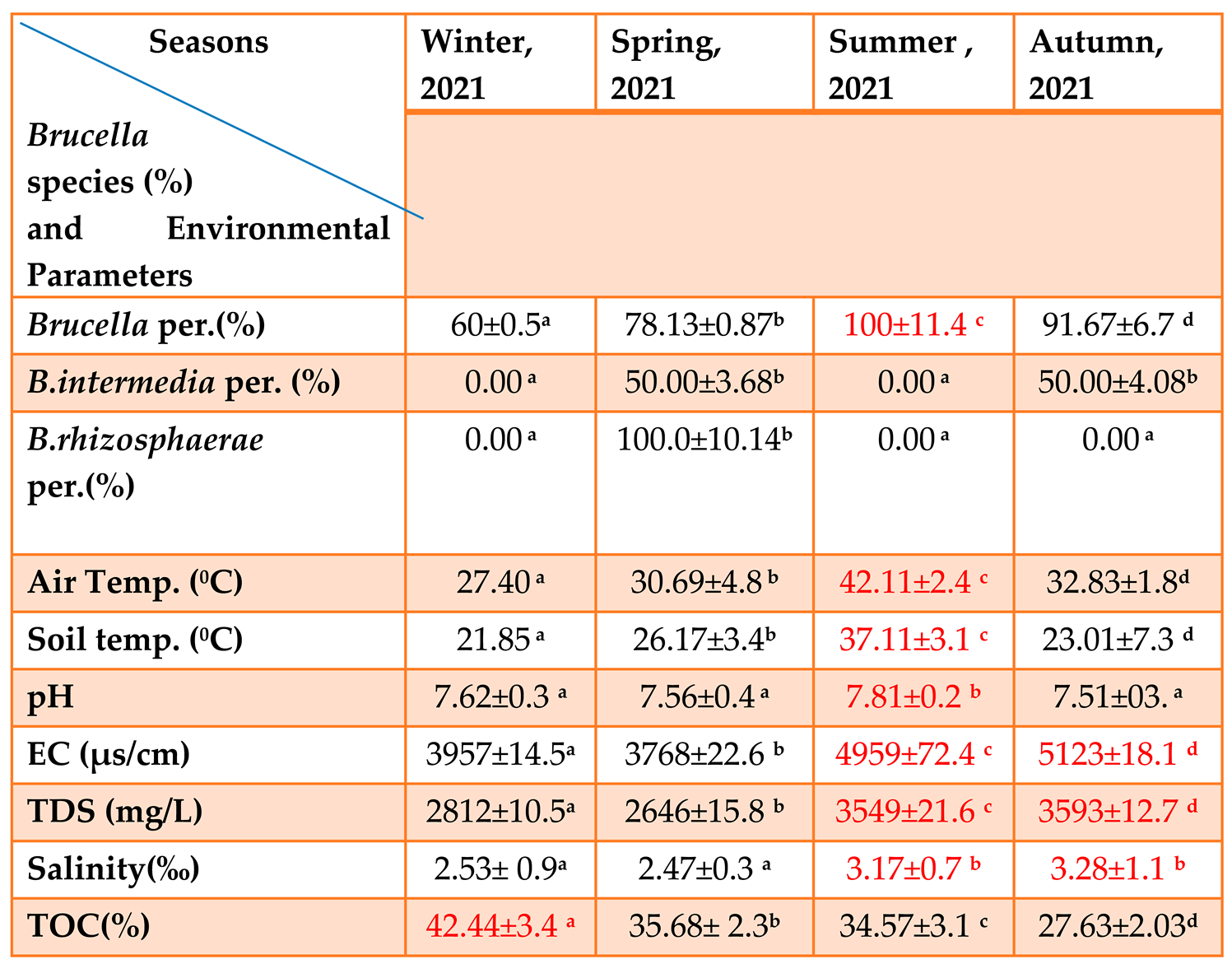
Table 1-6.
The Relationship between the Four Seasons and the Physical, Chemical Parameters and the Percentage of Brucella species (Mean ± S.D). p≤0.05.
Table 1-6.
The Relationship between the Four Seasons and the Physical, Chemical Parameters and the Percentage of Brucella species (Mean ± S.D). p≤0.05.
This table(1-6) noted that proportion of Brucella presence in summer is higher that it is in relation with environmental parameters which include (Air and Soil temperature, pH, EC, TDS and Salinity) that are high, except TOC is low while proportion of Brucella presence in winter is lower, also relate with environmental parameters include (Air and Soil temperature, pH, EC, TDS and Salinity), these the parameters (the physical and chemical) is low except TOC which is high. This indicated that variation of seasons affect to Brucella ratio in soil.
Discussion
A microbiological study is necessary for detecting cases of Brucella spp. because the symptoms are varied and non-specific, as shown by Bonaventura et al.,(202122. Typical zoonotic illnesses Brucella spp. was the source of the disease brucellosis, which continues to pose a serious health risk in many underdeveloped nations worldwide23. The 16S rRNA gene was the target of the PCR technique which was used to identify the bacteria in the samples24. Srinivasan et al., (2015)25 who utilized 10 ng of genomic DNA extracted from each strain to amplify this gene. Sanger sequences have been created. Isolated 16S rRNA sequences with a genus and species name (Isolated named- strains 16S aligned. fasta) were obtained from the Greengenes database26. PCR was used to diagnose the risk of brucellosis in people who were exposed to animals at work. It is a more accurate for finding Brucella spp. To tackle this potentially hazardous zoonotic disease in Pakistan, especially where brucellosis is common in animals, there is an urgent need for more accurate and focused diagnostic tools like PCR27, as mentioned in figures (1-1),(1-2). The 16S rRNA gene was the target of the PCR technique which was used to identify the bacteria in the samples28. A final diagnosis of the condition is made using either serological or cultural methods, or both29. As a result, PCR-based approaches have improved in accuracy, sensitivity, speed, and ability to work with DNA rather than highly contagious live cultures as a result of their increased using for detecting and identifying Brucella species30. In particular, for slow-growing bacteria like Brucella, Rahman et al., (2014)31 found that the PCR assays were efficient for speedy and precise diagnoses. It was shown by Manivannan et al., (2021)32 to be a sensitive and specific approach for identifying Brucella spp.
In table (1-4) showed that all isolates of B.intermedia has significant differences (p<0.05) between genotype and environmental factors, This indicates presence of genetic variation in them. This implies the presence of genetic variation in them and suggests that these bacteria changed as a result of polymorphism caused by environmental indicators and heavy metals. This caused differences between them, allowing them to adapt to and live in harsh environments. In addition, to survive in this soil, its resistance to the poisonous heavy metals may be changed into new strains. The psychological differences amongst microorganisms are caused by genetic changes. while table (1-5) mentioned that B.rhizosphaerae, does not exhibit significant differences (p>0.05) between genotype and environmental parameters, This may be because B. rhizosphaerae is related to roots, B. rhizosphaerae is a gram-negative, oxidase-positive bacterium discovered in the rhizosphere of a potato in Austria33.
There was many ways to become infected by the dangerous bacteria known as Brucella. Long-lasting resistance was possible in both inside and outside of mammalian hosts, even in harsh circumstances. It can linger in food for up to 15 months under adverse conditions such as acidity and temperatures ranging from 11 to 14 °C or for a few days if kept below 37 °C for up to two months in the winter and for only a few hours if exposed to direct sunshine, Brucella can also survive in contaminated manure and aborted infected feti34. B. intermedia is a bacterium in the genus Brucella35. Velasco et al., (1998)36 described it initially. Only one case of cholangitis following liver transplantation has been reported in humans37.
Exposure to Heavy metal persists and is rising in many regions of the world the fact that heavy metals were known to have several negative health impacts and that these effects could endure for a very long time. For ecological, evolutionary, nutritional and environmental reasons, heavy metals were significant environmental contaminants and their toxicity is a problem that is becoming more and more important38,39. The oxidative degradation of biological macromolecules were mostly caused by heavy metal binding to DNA where it displaced original metals from their native binding sites and caused malfunction to cells and finally became toxic40.
The phylogenetic tree of Ochrobactrum spp. already shows the presence of the A44T-related strains in a separate branch41. Ramette et al., (2011)42 based on the concatenated sequences of 16S rRNA and genes to acquire higher phylogenetic resolution within this group, which reveals that O. rhizosphaerae PR17T is the closest relative of A44T. The strain Ochrobactrum sp. was identified in the rhizosphere of a field-grown potato in Gelderland, the Netherlands. According to phylogenetic analysis based solely on the 16S rRNA gene as in figures (1-3),(1-4).
Conclusions
In this research, it was noted that proportion of Brucella spp. presence in summer was high and also environmental parameters.
Brucella spp. survived in local soils due to environmental conditions that being variations in Brucella species, which led to a change in their properties and make it resistance. Recording new nucleotides sequencing in soil samples and presence of new isolates of Brucella in local soils samples. Registered new strains in soil were two new species of Brucella (B. rhizosphaerae, and B. intermedia).
Phylogenetic analysis of the 16S rRNA sequences noted that B.intermedia was nearest to Pakistan, and the other B.intermedia was nearest to Russia, while B.rhizosphaerae was nearest to Ukraine and Pakistan in animals soils samples, these stated consider one of the sources for the infection.
References
- Leclercq, S.O.; Cloeckaert, A.; Zygmunt, M.S. Taxonomic organization of the family Brucellaceae based on a phylogenomic approach. Frontiers in microbiology 2020, 10, 3083. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Treanor, J.J.; Wallen, R.L.; White, P.J. Timing of parturition events in Yellowstone bison Bison bison: implications for bison conservation and brucellosis transmission risk to cattle. Wildlife Biology 2010, 16, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: the strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biology and Biochemistry 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Seleem, M.N.; Boyle, S.M.; Sriranganathan, N. Brucellosis: a re-emerging zoonosis. Veterinary microbiology 2010, 140, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Fouskis, I.; Sandalakis, V.; Christidou, A.; Tsatsaris, A.; Tzanakis, N.; Tselentis, Y.; Psaroulaki, A. The epidemiology of Brucellosis in Greece, 2007–2012: a ‘One Health’ approach. Transactions of The Royal Society of Tropical Medicine and Hygiene 2018, 112, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E. Retrospective and prospective perspectives on zoonotic brucellosis. Frontiers in microbiology 2014, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahab, A.E.W.; Hegazy, Y.; Wael, F.; Mikeal, A.; Kapaby, A.F.; Abdelfatah, M.; Eltholth, M.M. Knowledge, attitudes and practices (KAPs) and risk factors of brucellosis at the human-animal interface in the Nile Delta, Egypt. bioRxiv 2019, 607655. [Google Scholar]
- Motsi, T.R.; Tichiwangana, S.C.; Matope, G.; Mukarati, N.L. A serological survey of brucellosis in wild ungulate species from five game parks in Zimbabwe: research communication. Onderstepoort Journal of Veterinary Research 2013, 80, 1–4. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Frontiers in microbiology 2012, 3, 399. [Google Scholar] [CrossRef]
- Li, Y.; Li, X. , Liang, S.; Fang, L.; Cao, W. Epidemiological features and risk factors associated with the spatial and temporal distribution of human brucellosis in China. BMC Infectious Diseases 2013, 13, 547. [Google Scholar] [CrossRef]
- Morales, J.R.A. Climate change, climate variability and brucellosis. Recent patents on anti-infective drug discovery 2013, 8, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Muhammad, K.; Rabbani, M.; Khan, M.S.; Ali, M.A.; Naureen, S.; Kanwal, F.; Raza, S.; Aqib, A.I.; Sadia, H.; et al. Phylogenetic Analysis of Soil Borne Brucella Species by Targeting Insertion Sequence 711 Element in Punjab, Pakistan. International Journal of Agriculture and Biology 2017, 19, 1457–1462. [Google Scholar]
- Gaudette, H.; Flight, W.; Toner, L.; Folger, D. An inexpensive titration method for the determination of organic carbon in recent sediments. Journal of Sedimentary Petrology 1974, 44, 249–253. [Google Scholar]
- Sharidah, M.M.A. Heavy metals in mangrove sediments of the United Arab Emirates shoreline (Arabian Gulf). Water Air Soil Pollut 1999, 116, 523–534. [Google Scholar] [CrossRef]
- American Public health Association (APHA). (1985). Standard Methods For Examination of Water and Wastewater, 20th Edition, American Public Health Association. Washington D. C.
- Forbes, B.A.; Sahm, D.F.; Weissfeld, A.S. (2007). Bailey and Scotts' Diagnostic microbiology: A textbook for isolation and identification of pathogenic microorganisms. 12th. ed. St Louis, Mosby. 378.
- Alton, G.G.; Jones, L.M.; Angus, R.D.; Verger, J.M. (1988). Bacteriological methods. In: Techniques for the brucellosis laboratory. Institute National de la Recherche Agronomique, INRA, Paris France. 34-60.
- Dadar, M.; Alamian, S.; Behrozikhah, A.M., Yazdani, F.; Kalantari, A.; Etemadi, A.; Whatmore, A.M. (2019). Molecular identification of Brucella species and biovars associated with animal and human infection in Iran. Faculty of Veterinary Medicine, Urmia University, Urmia, Iran. In Veterinary research forum. 10:4-315.
- Dadar, M.; Alamian, S. Identification of main Brucella species implicated in ovine and caprine abortion cases by molecular and classical methods. Archives of Razi Institute 2021, 76, 51. [Google Scholar] [PubMed]
- Şahin, M.; Unver, A.; Otlu, S. Isolation and biotyping of B. melitensis from aborted sheep foetuses in Turkey. Bull Vet Inst Pulawy 2008, 52, 59–62. [Google Scholar]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PloS one 2015, 10, e0117617. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, D.; Angeletti, G.S.; Ianni, A.; Petitti, T.; Gherardi, G. Microbiological laboratory diagnosis of human brucellosis: An overview. Pathogens 2021, 10, 1623. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Zhang, Y.; Bai, L.; Zhao, Y.; Liu, C.; Ma, A.; Yu, H. Polymerase chain reaction–based assays for the diagnosis of human brucellosis. Annals of clinical microbiology and antimicrobials 2014, 13, 1–8. [Google Scholar] [CrossRef]
- Hamdy, M.E.; Zaki, H.M. Detection of virulence-associated genes in Brucella melitensis biovar 3, the prevalent field strain in different animal species in Egypt. Open Veterinary Journal 2018, 8, 112–117. [Google Scholar] [CrossRef]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PloS one 2015, 10, e0117617. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.; Waheed, U.; Farooq, M.; Ali, T.; Khan, Q.M. Frequency of Brucellosis in High Risk Human Groups in Pakistan Detected through Polymerase Chain Reaction and its Comparison with Conventional Slide Agglutination Test. International Journal of Agriculture & Biology 2014, 16. [Google Scholar]
- Hamdy, M.E.; Zaki, H.M. Detection of virulence-associated genes in Brucella melitensis biovar 3, the prevalent field strain in different animal species in Egypt. Open Veterinary Journal 2018, 8, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Etman, R.H.; Barsoum, S.A.; Ibrahim, I.G.A.; El-Ashmawy, W.R.; Abou-Gazia, K.A. Evaluation of efficacy of some serological tests used for diagnosis of brucellosis in cattle in Egypt using latent class analysis. Sokoto Journal of Veterinary Sciences 2014, 12, 1–7. [Google Scholar] [CrossRef]
- Moussa, I.M.; Omnia, M.E.; Amin, A.S.; Selim, S.A. Evaluation of the currently used polymerase chain reaction assays for molecular detection of Brucella species. African Journal of Microbiology Research 2011, 5, 1511–1520. [Google Scholar] [CrossRef]
- Rahman, M.S.; Sarker, M.A.S.; Rahman, A.K.M.A.; Sarker, R.R.; Melzer, F.; Sprague, L.D.; Neubauer, H. The prevalence of Brucella abortus DNA in seropositive bovine sera in Bangladesh. Afr. J. Microbiol. Res. 2014, 8, 3856–3860. [Google Scholar]
- Manivannan, K.; Mahmoud, S.M.; Ramasamy, M.; Shehata, A.A.; Ahmed, H.; Solaimuthu, C.; Dhandapani, K. Molecular detection of brucellosis in dromedary camels of Qatar by real-time PCR technique. Comparative Immunology, Microbiology and Infectious Diseases 2021, 78, 101690. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Sessitsch, A.; Schloter, M.; Huber, B.; Busse, H.J.; Scholz, H.C. Ochrobactrum rhizosphaerae sp. nov. and Ochrobactrum thiophenivorans sp. nov., isolated from the environment. International journal of systematic and evolutionary microbiology 2008, 58, 1426–1431. [Google Scholar] [CrossRef]
- Lucero, N.; Tenenbaum, M.; Jacob, N.; Escobar, G.; Groussaud, P.; Whatmore, A. Application of variable number of tandem repeats typing to describe familial outbreaks of brucellosis in Argentina. J Med Microb. 2010, 59, 648–652. [Google Scholar] [CrossRef]
- Liu, D. (Ed.). (2011). Molecular detection of human bacterial pathogens. CRC press.
- Velasco, J.; Romero, C.; López-Goni, I.; Leiva, J.; Díaz, R.; Moriyón, I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol 1998, 48, 759–768. [Google Scholar] [CrossRef]
- Möller, L.V.; Arends, J.P.; Harmsen, H.J.; Talens, A. , Terpstra, P.; Slooff, M.J. Ochrobactrum intermedium infection after liver transplantation. Journal of clinical microbiology 1999, 37, 241–244. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdisciplinary toxicology 2014, 7, 60. [Google Scholar] [CrossRef]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: a review. Environmental chemistry letters 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Flora, S.J.S.; Mittal, M.; Mehta, A. Heavy metal induced oxidative stress & its possible reversal by chelation therapy. Indian Journal of Medical Research 2008, 128, 501. [Google Scholar] [PubMed]
- Kämpfer, P.; Scholz, H.C.; Huber, B.; Falsen, E.; Busse, H.J. Ochrobactrum haematophilum sp. nov. and Ochrobactrum pseudogrignonense sp. nov., isolated from human clinical specimens. International Journal of Systematic and Evolutionary Microbiology 2007, 57, 2513–2518. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A.; Frapolli, M.; Le Saux, F.M.; Gruffaz, C.; Meyer, J.M.; De´fago, G; et al. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 2011, 34, 180–188. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).